Introduction
Glioma is the most common and aggressive type of
tumor of the human central nervous system (CNS). The annual
incidence of glioma has been reported as 30~80/1 million worldwide
(1). Regarding pathological grade,
glioma includes glioblastoma (GBM) and low-grade glioma (LGG).
Currently, standard treatments for glioma include chemoradiotherapy
and targeted therapy, which are administered following maximum
surgical resection (2–5). The prognosis of glioma was reported to
have a 5-year survival rate of ~10–20% (6). Therapeutic techniques for increasing the
survival rate and improving quality of life of patients with glioma
in the neurosurgical field (7,8).
Polypeptide-N-acetyl-galactosaminlytransferase 7
(GALNT7) is a member of the N-acetyl-D-galactosamine-transferase
family. The enzyme encoded by this gene controls the initiation
step of mucin-type O-linked protein glycosylation and transfers
N-acetyl galactosamine to serine and threonine amino acid residues
(9). Previous studies have indicated
that GALNT7 can influence the prognoses of multiple types of
malignant tumor, including cervical (10), esophageal (11) and liver cancer (12), as well as renal cell carcinoma
(13), by affecting cell
proliferation, metastasis, apoptosis, migration, differentiation
and invasion (14–17).
The Cancer Genome Atlas (TCGA) is a publicly funded
project that aims to catalogue and identify major cancer-causing
genomic alterations, to create a comprehensive ‘atlas’ of genomic
profiles of various types of cancer. To date, the program has
investigated >30 types of human tumor, including GBM and LGG,
through large-scale genome sequencing and integrated
multi-dimensional analyses (18). To
the best of our knowledge, the present study is the first to
investigate the role of GALNT7 in glioma, using GBM and LGG RNA-seq
data downloaded from the TCGA database.
Materials and methods
Data collection
The RNA-seq expression and clinical data of 174
patients with GBM and 529 patients with LGG were obtained from TCGA
(using the search terms: GBM and LGG, level 3 and RNA-seq;
http://cancergenome.nih.gov/). Complete
gene expression data, clinical data and definite pathological
diagnosis was available for all included patients. RNA-seq
expression data for 114 normal brain tissues were download from the
GTEx project (https://www.gtexportal.org/home/; Brain-Cortex-GTEx,
RNA-seq) and used as the control group (19).
Statistical analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was used for
data processing. Quantitative data are expressed as the mean ±
standard deviation (x¯±s). Two
samples were compared by t-test and multiple comparisons were
performed by one-way analysis of variance followed by
Least-Significant-Difference test. Qualitative data were expressed
as a percentage (%) and analyzed by χ2. Survival
analysis was performed using Cox regression and Kaplan-Meier
analysis (P<0.05). Gene-set enrichment analysis was performed
using GSEA (version, 2.2.1; http://software.broadinstitute.org/gsea/downloads.jsp)
from the Molecular Signatures Database (v6.3 MsigDB; Broad
Institute, Inc., Massachusetts Institute of Technology, and Regents
of the University of California). According to default-weighted
enrichment, the cut-offs for the number of random combinations was
1,000, for nominal P-values (NOM P-val) <0.05, and false
discovery rates (FDR), <0.25. Pearson correlation coefficient
was used to assess the association between the expression of 2
genes. P<0.05 was considered to indicate a statistically
significant difference.
Results
GALNT7 expression is positively
associated with glioma malignancy
GALNT7 expression levels were significantly higher
in glioma, particularly in GBM, compared with the negative control
samples (P<0.05). This indicates a positive association between
GALNT7 expression and glioma malignancy (Table I; Fig.
1A).
 | Table I.Differential analysis of GLANT7
RNA-seq expression. |
Table I.
Differential analysis of GLANT7
RNA-seq expression.
| Group | No. | mean ± standard
deviation | F | P-value |
|---|
| GBM | 174 | 3.46±1.38 | 15.37 | <0.05 |
| LGG | 529 | 2.92±1.58 |
|
|
| Normal | 114 | 2.46±1.58 |
|
|
Association between GALNT7 expression
and clinical characteristics
GALNT7 was expressed at different levels in normal
brain tissues and glioma tissues. In order to investigate the
effect of its expression in glioma, GALNT7 expression in normal
brain tissue from GTEx was employed as the cut-off value, and TCGA
data regarding expression of GALNT7 in glioma tissue were divided
into high (HIGH) and low (LOW) groups. The association between
GALNT7 expression levels and clinical characteristics of patients
with glioma was analyzed using t-test. The results demonstrated
that expression level was not associated with sex, but it was
associated with age, tumor grade and survival rate (Table II).
 | Table II.Clinical characteristics of patients
with glioma. |
Table II.
Clinical characteristics of patients
with glioma.
|
Characteristics | No. | Low GALNT7
expression (%) | High GALNT7
expression (%) | χ2 | P-value |
|---|
| Age (years) |
|
|
| 10.46 | <0.05 |
|
<40 | 274 | 132 (48.2) | 142 (51.8) |
|
|
|
≥42 | 414 | 149 (36.0) | 265 (64.0) |
|
|
| Sex |
|
|
| 0.05 | 0.82 |
|
Male | 393 | 162 (41.2) | 231 (58.8) |
|
|
|
Female | 295 | 119 (40.3) | 176 (59.7) |
|
|
| Grade |
|
|
| 38.37 | <0.05 |
| G2 | 257 | 138 (53.7) | 119 (46.3) |
|
|
| G3 | 263 | 104 (39.5) | 159 (60.5) |
|
|
| G4 | 168 | 39 (23.2) | 129 (76.8) |
|
|
| Survival
status |
|
|
| 38.456 | <0.05 |
| Not
alive | 416 | 209 (50.2) | 207 (49.8) |
|
|
|
Alive | 272 | 72 (26.5) | 200 (73.5) |
|
|
Negative association between GALNT7
expression and survival time
Using the median GALNT7 expression value as the
standard, GALNT7 expression data were divided into HIGH GALNT7
transcripts per million (TPM) and LOW GALNT7 TPM groups. Cox
survival analysis demonstrated a regression coefficient (B) of
0.102 (P<0.05; Table III).
Together with Kaplan-Meier survival analysis, this suggests that
patients with glioma exhibiting high expression of GALNT7 have
relatively shorter disease-free (Fig.
1B) and overall (Fig. 1C)
survival times than those exhibiting low expression.
 | Table III.Cox regression analysis of GALNT7
expression. |
Table III.
Cox regression analysis of GALNT7
expression.
| GALNT7 | B | SE | Wald | df | P-value | HR | 95% Confidence
intervals |
|---|
|
| 0.10 | 0.02 | 30.94 | 1 | <0.05 | 1.11 | (1.07,1.15) |
GSEA and prediction of co-expressed
genes
Based on the KEGG database (20), the results of GSEA (NOM P-val
<0.05; FDR<0.25) indicated 6 pathways associated with glioma,
including ‘regulation of the actin cytoskeleton’ (Fig. 2A), ‘natural killer cell-mediated
cytotoxicity’ (Fig. 2B), ‘JAK-STAT
signaling’ (Fig. 2C), ‘cell adhesion
molecules’ (‘CAMs’) (Fig. 2D) and
‘ECM-receptor interaction’ (Fig. 2E).
In order to predict genes that are co-expressed, Pearson's
correlation analysis was used to determine the correlation between
GALNT7 expression and all genes associated with these signaling
pathways. As a result, Crk (Fig. 2F),
RAC1 (Fig. 2G), STAT3 (Fig. 2H), PVR (Fig.
2I) and Tnc (Fig. 2J) were
identified as potential target genes. Crk participates in
regulating the ‘actin cytoskeleton’ pathway, RAC1 is involved in
the ‘natural killer cell mediated cytotoxicity’ pathway and STAT3
is associated with the ‘JAK-STAT’ signaling pathway. Also, PVR
participates in the ‘CAMs’ pathway and Tnc is involved in the
‘ECM-receptor interaction’ pathway.
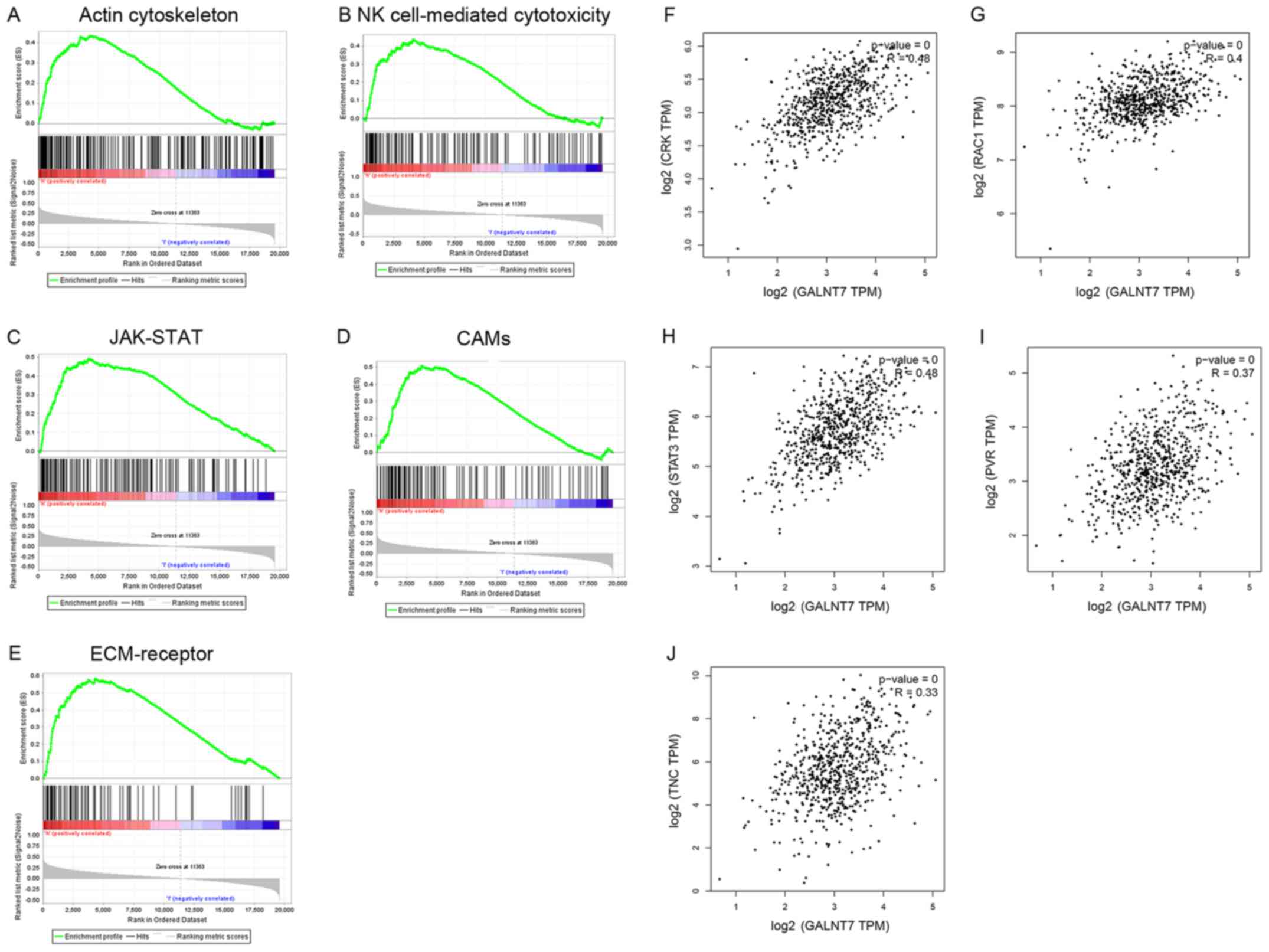 | Figure 2.Gene-set enrichment analysis and
co-expression prediction. The results of GSEA (Nominal P-value
<0.05, false discovery rate <0.25) indicated six pathways
up-regulated by GALNT7 and associated with glioma, including (A)
‘regulation of actin cytoskeleton’, N (B) ‘natural killer
cell-mediated cytotoxicity’, (C) JAK-STAT signaling pathway, (D)
‘Cell adhesion molecules’, (E) ‘ECM-receptor interaction’. The
potential target genes identified by Pearson's correlation
coefficient included (F) ‘Crk’, (G) RAC1, (H) STAT3, (I) PVR, and
(J) Tnc (P<0.05). Crk functions in the ‘regulation of actin
cytoskeleton’ pathway, RAC1 functions in the ‘natural killer cell
mediated cytotoxicity’ pathway, STAT3 functions in the ‘JAK-STAT
signaling’ pathway, PVR functions in the ‘Cell adhesion molecules’
pathway, and Tnc functions in the ‘ECM-receptor interaction’
pathway. GSEA, gene set enrichment analysis; GALNT7,
polypeptide-N-acetyl-galactosaminlytransferase 7; JAK, janus
kinase; STAT, signal transducer and activator of transcription;
ECM, extracellular matrix; PVR, poliovirus receptor; Tnc, Tenascsin
C. |
Differential and survival analysis of
co-expressed genes in glioma
Differential analyses demonstrated that Crk
(Fig. 3A) expression is positively
associated with the malignancy of glioma; positive association was
also observed with RAC1 (Fig. 3B),
STAT3 (Fig. 3C), and Tnc (Fig. 3E). PVR (Fig.
3D) highly expressed in GBM, but expressed lower than normal
brain tissue in LGG (P<0.05). Survival analyses demonstrate that
disease-free time was negatively correlated with the expression of
RAC1 (Fig. 4B), STAT3 (Fig. 4C), PVR (Fig.
4D) and Tnc (Fig. 4E), but not
Crk (Fig. 4A). Meanwhile, the
expression levels of Crk (Fig. 4F),
RAC1 (Fig. 4G), STAT3 (Fig. 4H), PVR (Fig.
4I) and Tnc (Fig. 4J) were
negatively correlated with overall survival time (P<0.05).
 | Figure 3.Differential analysis of co-expressed
genes in glioma. Tumor tissue, red; normal tissue, blue.
Differential analyses revealed that (A) Crk, (B) RAC1, (C) STAT3
and (E) Tnc expression was positively associated with the
malignancy of glioma. (D) PVR was highly expressed in GBM, but
expressed at a lower level than in normal brain tissue in LGG
(P<0.05). RAC1, Rac family amall GTPase 1; STAT3, signal
transducer and activator of transcription 3; Tnc, Tenascsin 3; PVR,
poliovirus receptor; GBM, glioblastoma; LGG, low-grade glioma. |
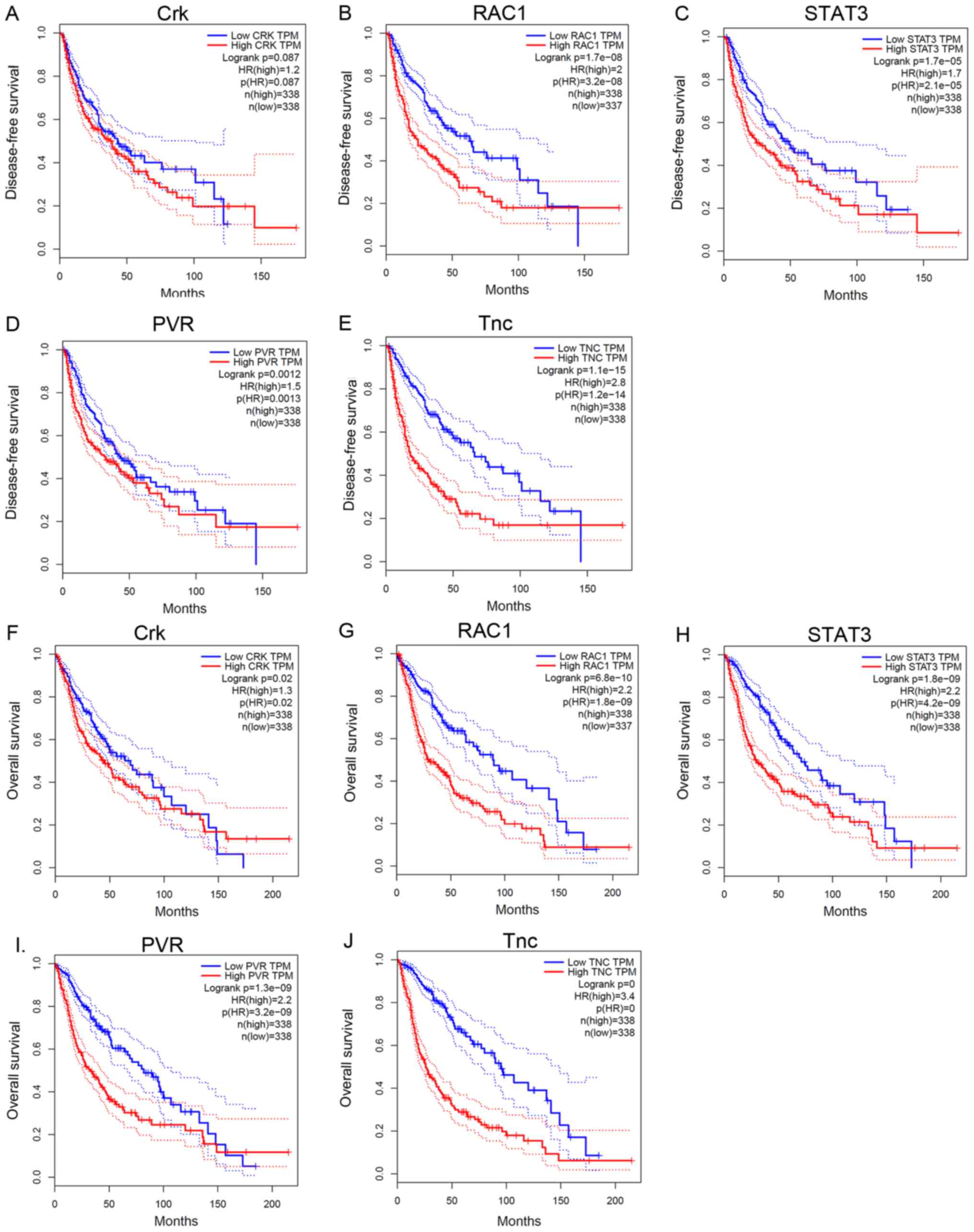 | Figure 4.Survival analysis of co-expressed
genes in glioma. (A) Kaplan-Meier survival analysis revealed that
the expression of Crk was not associated with disease-free survival
time (P>0.05), whereas that of (B) RAC1, (C) STAT3, (D) PVR, and
(E) Tnc were negatively associated with disease-free survival time
(P<0.05). Overall survival time was negatively associated with
the expression of (F) Crk, (G) RAC1, (H) STAT3, (I) PVR and (J) Tnc
(P<0.05). RAC1, Rac family amall GTPase 1; STAT3, signal
transducer and activator of transcription 3; PVR, poliovirus
receptor; Tnc, Tenascsin 3. |
Discussion
Glioma is divided into 4 grades according to the
World Health Organization standard (1): Grade I, pilocytic astrocytoma, which
manifests as a benign tumor and patients may have a full recovery
following total tumor resection; Grade II, includes
oligoastrocytoma and diffuse astrocytoma, it has a poorer prognosis
compared with Grade I, but is still considered to be LGG (2); Grade III, includes anaplastic
astrocytoma, and Grade IV, GBM. Grade III and IV tumors are
associated with high degrees of malignancy, strong invasive
abilities, poor prognosis and multiple differentiation potentials
(3,21). The present study aimed to reveal the
association between GALNT7 expression and glioma by identifying
co-expressed genes and relevant signaling pathways.
Firstly, a positive correlation between GALNT7
expression and the malignancy of glioma was assessed by analyzing
TCGA glioma datasets. Cox and Kaplan-Meier survival analyses
demonstrated that patients with glioma exhibiting high expression
levels of GALNT7 had relatively short disease-free and overall
survival times. Therefore, high expression levels of GALNT7 may
affect the prognosis of glioma.
Correlation analyses suggested that Crk, RAC1,
STAT3, PVR and Tnc are genes that are co-expressed with GALNT7.
Survival and differential analyses demonstrated that the expression
of Crk, RAC1, STAT3, PVR and Tnc are positively associated with the
malignancy of glioma, and are negatively associated with survival
time. These results were consistent with those of previous studies,
which indicated that high expression levels of Crk, RAC1, STAT3,
PVR and Tnc were associated with poor prognosis by promoting the
invasion, proliferation and other biological functions of glioma
(22–27). Therefore, we speculate that GALNT7 may
affect the progression of glioma by upregulating the expression of
target genes through relevant signaling pathways. Based on this
hypothesis, GSEA (28,29) was used to identify such relevant
signaling pathways. As a result, 5 signaling pathways associated
with GALNT7 and glioma were identified, including ‘regulation of
the actin cytoskeleton’, ‘natural killer cell-mediated
cytotoxicity’, ‘JAK-STAT signaling’, ‘CAMs’ and ‘ECM-receptor
interaction’ pathways. Specific target genes of GALNT7 participate
in each signaling pathway, and were subsequently evaluated.
There are a variety of intracellular actin binding
proteins that regulate multiple cellular functions, including
changes in the structure of the actin cytoskeleton, which occur
through the binding or dissociating of the proteins with actin
(30). Regulation of the actin
cytoskeleton pathway governs the cell motility of glioma by
regulating the locomotion of the cytoskeleton, and slows the
recovery of patients with glioma (31,32). Crk
participates in regulation of the ‘actin cytoskeleton pathway’, and
its expression is associated with overall survival time of patients
with glioma. Tsuda et al (22)
demonstrate that overexpression of Crk can increase the invasive
potential of cancer cells by notably inducing tyrosine
phosphorylation of scaffolding molecules, including p130 (Cas) and
paxillin through Src family tyrosine kinases, and stimulating the
activation loop of intracellular signaling. Therefore, as a gene
that is co-expressed with Crk, GALNT7 promotes the invasion of
glioma by upregulating Crk expression through regulation of the
actin cytoskeleton pathway.
NK cells form the first line of immune defense
against tumors (33,34). Such cells destroy tumors by
controlling cell proliferation, cytotoxicity and cytokine
production in the early stages of tumor formation (35,36). RAC1
has been reported to be involved in the NK cell-mediated
cytotoxicity pathway (37). Former
research suggests that high expression levels of RAC1 inhibit the
cytotoxic effects of NK cells by two mechanisms: Decreased
interaction between NK and target cells; NK cells that do interact
have a reduced ability to polarize their effector molecules towards
target cells (38). Based on these
results, coexpression with RAC1 and high expression of GALNT7
inhibits the cytotoxic effects of NK cells by regulating the
natural killer cell-mediated cytotoxicity pathway, which leads to
loss of control of glioma cell proliferation and poor
prognosis.
The JAK-STAT signaling pathway is mainly formed of
the tyrosine kinase associated receptor, JAK and STAT (39). Previous studies reported that the
activation of the JAK-STAT signaling pathway is associated with
poor prognosis of glioma (40,41).
Additionally, a previous study verified that high expression levels
of STAT3 enhance the proliferation of glioma cells (24). In the present study, GALNT7 was
coexpressed with STAT3, and high expression levels of STAT3 were
associated with poor prognosis of glioma. This suggests that GALNT7
may trigger and increase in glioma cell proliferation by activating
the JAK-STAT signaling pathway through upregulation of STAT3
expression.
CAMs are membrane and transmembrane glycoproteins
that regulate intercellular, cell and extracellular matrix
interactions, which are closely associated with cell adhesion,
migration, differentiation and signal transduction (42,43). CAMs
are widely expressed in glioma and are regulated by the CAMs
signaling pathway (44). PVR is
involved in the CAMs signaling pathway, and can enhance the
metastasis of glioblastoma through its over-expression (45). As GALNT7 is co-expressed with PVR,
GALNT7 may regulate the CAMs signaling pathway by up-regulating
PVR, thereby influencing tumor cell migration and affecting
prognosis.
ECM is mainly formed of insoluble components that
contribute to the behavior and structure of stromal cells and
epithelial vessels, respectively. The ECM constitutes collagen,
elastin, proteoglycan and glycoprotein. ECM can influence cell
differentiation, proliferation, adhesion, morphogenesis and
phenotypic expression (46). Brösicke
et al (47,48) proposed Tnc as a key gene in the
ECM-receptor interaction pathway and that high coexpression with
GALNT7 occurs within glioma tissue, which is consistent with our
research. Accumulating evidence also suggests that Tnc serves a
crucial role in cell migration and invasion, the most malignant
characteristics of glioma. Therefore, GALNT7 can affect the
ECM-receptor interaction pathway by upregulating the expression of
Tnc; this mechanism may affect the invasion of glioma cells and
lead to a poor prognosis of glioma.
In conclusion, high expression of GALNT7 was
demonstrated to be associated with poor prognosis of glioma, likely
via promoting invasion and proliferation through multiple signaling
pathways. Therefore, GALNT7 may be employed as a novel molecular
target for the early detection, diagnosis and treatment of
glioma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated or analyzed during the
current study are available in the TCGA/KEGG/GTEx repository,
[https://cancergenome.nih.gov/]
[https://www.kegg.jp/] [https://www.gtexportal.org/home/].
Authors' contributions
SH contributed to writing the manuscript, and was
responsible for project design and data interpretation; YC and CD
were responsible for data collection and preliminary analysis; HL
was responsible for all data analyses; YL and JZ conceived and
designed the present study given final approval of the version to
be published.
Ethical approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GALNT7
|
polypeptide-N-acetyl-galactosaminlytransferase 7
|
|
TCGA
|
The Cancer Genome Atlas
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes database
|
|
GTEx
|
Genotype-Tissue Expression project
|
|
GBM
|
glioblastoma
|
|
LGG
|
low grade glioma
|
|
NOM P-val
|
nominal P-values
|
|
FDR
|
false discovery rates
|
|
MsigDB
|
Molecular Signatures Database
|
|
NK
|
natural killer
|
|
CAMS
|
cell adhesion molecules
|
|
CNS
|
central nervous system
|
|
MIF
|
macrophage migration inhibitory
factor
|
|
JAK
|
janus kinase
|
|
STAT
|
signal transducer and activator of
transcription
|
|
GSEA
|
gene-set enrichment analysis
|
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forst DA, Nahed BV, Loeffler JS and
Batchelor TT: Low-grade gliomas. Oncologist. 19:403–413. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Groot JF: High-grade gliomas. Continuum
(Minneap Minn. ). 21:332–344. 2015.PubMed/NCBI
|
|
4
|
Kim IA, Yang YJ, Yoon SC, Choi IB, Kay CS,
Kwon HC, Kim CM, Joe YA, Kang JK and Hong YK: Potential of
adenoviral p53 gene therapy and irradiation for the treatment of
malignant gliomas. Int J Oncol. 19:1041–1047. 2001.PubMed/NCBI
|
|
5
|
Maris D, Nica D, Mohan D, Moisa H and
Ciurea AV: Multidisciplinary management of adult low grade gliomas.
Chirurgia (Bucur). 109:590–599. 2014.PubMed/NCBI
|
|
6
|
Pekmezci M and Perry A: Genetic markers in
adult high-grade gliomas. Semin Radiat Oncol. 24:235–239. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duffy A, Le J, Sausville E and Emadi A:
Autophagy modulation: A target for cancer treatment development.
Cancer Chemother Pharmacol. 75:439–447. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin Q, Mao KL, Tian FR, Yang JJ, Chen PP,
Xu J, Fan ZL, Zhao YP, Li WF, Zheng L, et al: Brain tumor-targeted
delivery and therapy by focused ultrasound introduced
doxorubicin-loaded cationic liposomes. Cancer Chemother Pharmacol.
77:269–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bennett EP, Hassan H, Hollingsworth MA and
Clausen H: A novel human UDP-N-acetyl-D-galactosamine: Polypeptide
N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for
partial GalNAc-glycosylated acceptor substrates. FEBS Lett.
460:226–230. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu H, Chen J, Li D, Liu X, Li L and Wang
K: MicroRNA-30e functions as a tumor suppressor in cervical
carcinoma cells through targeting GALNT7. Transl Oncol. 10:876–885.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu Q, Xu L, Li C, Yuan Y, Huang S and Chen
H: miR-214 inhibits invasion and migration via downregulating
GALNT7 in esophageal squamous cell cancer. Tumour Biol.
37:14605–14614. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shan SW, Fang L, Shatseva T, Rutnam ZJ,
Yang X, Du W, Lu WY, Xuan JW, Deng Z and Yang BB: Mature miR-17-5p
and passenger miR-17-3p induce hepatocellular carcinoma by
targeting PTEN, GalNT7 and vimentin in different signal pathways. J
Cell Sci. 126:1517–1530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Li Y, Chen D, Jin L, Su Z, Liu J,
Duan H, Li X, Qi Z, Shi M, et al: miR30a5p in the tumorigenesis of
renal cell carcinoma: A tumor suppressive microRNA. Mol Med Rep.
13:4085–4094. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Duan HF, Li XQ, Hu HY, Li YC, Cai Z, Mei
XS, Yu P, Nie LP, Zhang W, Yu ZD and Nie GH: Functional elucidation
of miR-494 in the tumorigenesis of nasopharyngeal carcinoma. Tumour
Biol. 36:6679–6689. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li W, Ma H and Sun J: MicroRNA34a/c
function as tumor suppressors in Hep2 laryngeal carcinoma cells and
may reduce GALNT7 expression. Mol Med Rep. 9:1293–1298. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gaziel-Sovran A, Segura MF, Di Micco R,
Collins MK, Hanniford D, de Miera Vega-Saenz E, Rakus JF, Dankert
JF, Shang S, Kerbel RS, et al: miR-30b/30d regulation of GalNAc
transferases enhances invasion and immunosuppression during
metastasis. Cancer Cell. 20:104–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mayer A, Schneider F, Vaupel P, Sommer C
and Schmidberger H: Differential expression of HIF-1 in
glioblastoma multiforme and anaplastic astrocytoma. Int J Oncol.
41:1260–1270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gonda DD, Cheung VJ, Muller KA, Goyal A,
Carter BS and Chen CC: The cancer genome atlas expression profiles
of low-grade gliomas. Neurosurg Focus. 36:E232014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
The Genotype-Tissue Expression (GTEx)
project. Nat Genet. 45:580–585. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Du J, Yuan Z, Ma Z, Song J, Xie X and Chen
Y: KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway
analysis using a path analysis model. Mol Biosyst. 10:2441–2447.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pouleau HB, Sadeghi N, Baleriaux D, Melot
C, De Witte O and Lefranc F: High levels of cellular proliferation
predict pseudoprogression in glioblastoma patients. Int J Oncol.
40:923–928. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsuda M and Tanaka S: Roles for crk in
cancer metastasis and invasion. Genes Cancer. 3:334–340. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yukinaga H, Shionyu C, Hirata E, Ui-Tei K,
Nagashima T, Kondo S, Okada-Hatakeyama M, Naoki H and Matsuda M:
Fluctuation of Rac1 activity is associated with the phenotypic and
transcriptional heterogeneity of glioma cells. J Cell Sci.
127:1805–1815. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Priester M, Copanaki E, Vafaizadeh V,
Hensel S, Bernreuther C, Glatzel M, Seifert V, Groner B, Kögel D
and Weissenberger J: STAT3 silencing inhibits glioma single cell
infiltration and tumor growth. Neuro Oncol. 15:840–852. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chandramohan V, Bryant JD, Piao H, Keir
ST, Lipp ES, Lefaivre M, Perkinson K, Bigner DD, Gromeier M and
McLendon RE: Validation of an Immunohistochemistry Assay for
Detection of CD155, the poliovirus receptor, in malignant gliomas.
Arch Pathol Lab Med. 141:1697–1704. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia S, Lal B, Tung B, Wang S, Goodwin CR
and Laterra J: Tumor microenvironment tenascin-C promotes
glioblastoma invasion and negatively regulates tumor proliferation.
Neuro Oncol. 18:507–517. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brosicke N, van Landeghem FK, Scheffler B
and Faissner A: Tenascin-C is expressed by human glioma in vivo and
shows a strong association with tumor blood vessels. Cell Tissue
Res. 354:409–430. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Machado CM, Freitas AT and Couto FM:
Enrichment analysis applied to disease prognosis. J Biomed
Semantics. 4:212013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mueller C, deCarvalho AC, Mikkelsen T,
Lehman NL, Calvert V, Espina V, Liotta LA and Petricoin EF III:
Glioblastoma cell enrichment is critical for analysis of
phosphorylated drug targets and proteomic-genomic correlations.
Cancer Res. 74:818–828. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hou M, Liu X, Cao J and Chen B: SEPT7
overexpression inhibits glioma cell migration by targeting the
actin cytoskeleton pathway. Oncol Rep. 35:2003–2010. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Klopocka W, Korczynski J and Pomorski P:
Cytoskeleton and nucleotide signaling in glioma C6 cells. Adv Exp
Med Biol. 986:103–119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Uzdensky A, Kristiansen B, Moan J and
Juzeniene A: Dynamics of signaling, cytoskeleton and cell cycle
regulation proteins in glioblastoma cells after sub-lethal
photodynamic treatment: Antibody microarray study. Biochim Biophys
Acta. 1820:795–803. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang BY, Zhan YP, Zong WJ, Yu CJ, Li JF,
Qu YM and Han S: The PD-1/B7-H1 pathway modulates the natural
killer cells versus mouse glioma stem cells. PLoS One.
10:e01347152015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miller JS: Biology of natural killer cells
in cancer and infection. Cancer Invest. 20:405–419. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
He W, Kuang Y, Xing X, Simpson RJ, Huang
H, Yang T, Chen J, Yang L, Liu E, He W and Gu J: Proteomic
comparison of 3D and 2D glioma models reveals increased HLA-E
expression in 3D models is associated with resistance to NK
cell-mediated cytotoxicity. J Proteome Res. 13:2272–2281. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Orozco-Morales M, Sanchez-Garcia FJ,
Golan-Cancela I, Hernández-Pedro N, Costoya JA, de la Cruz VP,
Moreno-Jiménez S, Sotelo J and Pineda B: RB mutation and RAS
overexpression induce resistance to NK cell-mediated cytotoxicity
in glioma cells. Cancer Cell Int. 15:572015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mainiero F, Soriani A, Strippoli R,
Jacobelli J, Gismondi A, Piccoli M, Frati L and Santoni A: RAC1/P38
MAPK signaling pathway controls beta1 integrin-induced
interleukin-8 production in human natural killer cells. Immunity.
12:7–16. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Billadeau DD, Brumbaugh KM, Dick CJ,
Schoon RA, Bustelo XR and Leibson PJ: The Vav-Rac1 pathway in
cytotoxic lymphocytes regulates the generation of cell-mediated
killing. J Exp Med. 188:549–559. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nicolas CS, Amici M, Bortolotto ZA,
Doherty A, Csaba Z, Fafouri A, Dournaud P, Gressens P, Collingridge
GL and Peineau S: The role of JAK-STAT signaling within the CNS.
JAKSTAT. 2:e229252013.PubMed/NCBI
|
|
40
|
Tu Y, Zhong Y, Fu J, Cao Y, Fu G, Tian X
and Wang B: Activation of JAK/STAT signal pathway predicts poor
prognosis of patients with gliomas. Med Oncol. 28:15–23. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng Q, Han L, Dong Y, Tian J, Huang W,
Liu Z, Jia X, Jiang T, Zhang J, Li X, et al: JAK2/STAT3 targeted
therapy suppresses tumor invasion via disruption of the
EGFRvIII/JAK2/STAT3 axis and associated focal adhesion in
EGFRvIII-expressing glioblastoma. Neuro Oncol. 16:1229–1243. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao WJ and Schachner M: Neuregulin 1
enhances cell adhesion molecule l1 expression in human glioma cells
and promotes their migration as a function of malignancy. J
Neuropathol Exp Neurol. 72:244–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J, Liu X, Duan Y, Liu Y, Wang H, Lian
S, Zhuang G and Fan Y: Combined blockade of T cell immunoglobulin
and mucin domain 3 and carcinoembryonic antigen-related cell
adhesion molecule 1 results in durable therapeutic efficacy in mice
with intracranial gliomas. Med Sci Monit. 23:3593–3602. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen X, Ma WY, Xu SC, Liang Y, Fu YB, Pang
B, Xin T, Fan HT, Zhang R, Luo JG, et al: The overexpression of
epithelial cell adhesion molecule (EpCAM) in glioma. J Neurooncol.
119:39–47. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sloan KE, Stewart JK, Treloar AF, Matthews
RT and Jay DG: CD155/PVR enhances glioma cell dispersal by
regulating adhesion signaling and focal adhesion dynamics. Cancer
Res. 65:10930–10937. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Brösicke N and Faissner A: Role of
tenascins in the ECM of gliomas. Cell Adh Migr. 9:131–140. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Brosicke N, Sallouh M, Prior LM, Job A,
Weberskirch R and Faissner A: Extracellular matrix
glycoprotein-derived synthetic peptides differentially modulate
glioma and sarcoma cell migration. Cell Mol Neurobiol. 35:741–753.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shimizu T, Kurozumi K, Ishida J, Ichikawa
T and Date I: Adhesion molecules and the extracellular matrix as
drug targets for glioma. Brain Tumor Pathol. 33:97–106. 2016.
View Article : Google Scholar : PubMed/NCBI
|


















