Introduction
Ranking fifth most common malignancies,
hepatocellular carcinoma (HCC) is diagnosed in >500,000
individuals worldwide (2011) (1).
With an increasing incidence, HCC has become the third most common
cause of cancer-associated mortality worldwide (2010) (2). The majority of patients with HCC exhibit
aggressive symptoms upon diagnosis and the median 1-year survival
rate is <30% (3,4). Traditionally, HCC is detected using
biomarkers, including α-fetoprotein (5,6), with
39–65% sensitivity and 76–94% specificity (7). Several biomarkers, such as miR-101-1,
miR-221, miR-638 can be used as diagnostic and prognostic
indicators of HCC (8–10). However, there are still a number of
biomarkers that may be biologically relevant for the development of
HCC. Therefore, a more efficient biomarker with improved accuracy
is urgently required.
MicroRNAs (miRNAs) are small non-coding RNA
molecules with an approximate length of 23 nucleotides. miRNAs
serve as post-transcriptional regulators of gene expression by
binding to the 3′-untranslated region of mRNA (11,12).
Previous studies have demonstrated that the dysregulation of
specific miRNAs contributes to the progression of
hepatocarcinogenesis (13,14). Therefore, these molecules may
represent promising biomarkers for the diagnosis of HCC. A previous
study demonstrated that miRNA (miR)-122-5p was downregulated in
patients with HCC (15); however, its
molecular mechanism and diagnostic significance remain to be
elucidated.
The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) is a program, which
contains RNA sequencing (RNA-Seq) data on 32 types of tumor without
limitations or restrictions by the National Cancer Institute (NCI)
and the National Human Genome Research Institute (NHGRI). The Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) is an online public
resource provided by the National Center for Biotechnology
Information (NCBI) in 2000. The GEO covers ample gene chip
expression data, as well as data from non-chip technologies, such
as serial analysis of gene and mass spectrometry. GEO provides
large quantities of high-throughput data and is one of the most
comprehensive public gene expression data resources available
currently.
Therefore, the present study examined the accuracy
of miR-122-5p in distinguishing HCC patients from healthy controls
based on data collected from TCGA and GEO. The association between
distinct clinicopathological features and the expression of
miR-122-5p was also investigated. To further clarify the regulatory
mechanism of miR-122-5p in HCC, subsequent bioinformatic analyses
were conducted. The present study aims to perform a quantitative
evaluation of the association between miR-122-5p in HCC and
pathophysiological processes, specifically investigating the type
of genes and signaling pathways that are associated with
downregulated miR-122-5p levels in HCC specimens.
Materials and methods
Collection of TCGA and GEO data
TCGA provided miRNA raw sequencing data from 370 HCC
cases and 50 adjacent liver tissue samples. The publicly available
RNA-Seq data were downloaded directly from the TCGA launch data
portal via bulk download of the liver hepatocellular carcinoma
(LIHC), RNASeqV2 (data type) and level 3 (data level) cancer
tissues collected on January 1, 2017 (16). Additionally, the GEO database was
searched and GEO datasets (GSE6857, GSE12717, GSE10694, GSE21279,
GSE22058, GSE21362, GSE20971, GSE39678, GSE31383, GSE40744,
GSE50013, GSE36915, GSE54751, GSE57555, GSE67882, GSE74618,
GSE65708 and GSE64989) (17)
containing an expression profile or fold-change value of miR-122-5p
in HCC and control samples were included. There was no restriction
on the specific type of non-cancerous control. Cell line assays or
assays without the expression value were excluded. The following
information obtained from Genomic Spatial Event (GSE) (https://www.ncbi.nlm.nih.gov/geo/) (18) chips was recorded: Main contributor
(the name that ranked first), publishing year, country, sample
type, experiment type, the platform of the GSE chips, and the
number of patients with HCC and controls. All expression values for
miR-122-5p in GEO were log2 scaled. The number, mean and standard
deviation for the control and experimental groups were calculated
based on each single gene chip.
Literature search
Pertinent studies were retrieved in PubMed
(https://www.ncbi.nlm.nih.gov/pubmed),
Wiley online library (http://onlinelibrary.wiley.com/), Sciencedirect
(http://www.sciencedirect.com/), Web of
science (http://login.webofknowledge.com/), Cochrane Central
Register of Controlled Trials (http://www.cochranelibrary.com/), EMSCO (http://emsco.com/), China National Knowledge
Infrastructure (CNKI) (http://www.cnki.net/), China Biology Medicine disc
(http://www.sinomed.ac.cn/), Chongqing
VIP (http://www.cqvip.com/) and Wan Fang Data
(http://www.wanfangdata.com.cn/) using
the following key terms: (malignant* OR cancer OR tumor OR tumor OR
neoplasm* OR carcinoma) AND (hepatocellular OR liver OR hepatic OR
HCC) AND (miR-122 OR miRNA-122 OR microRNA-122 OR miR122 OR
miRNA122 OR microRNA122 OR ‘miR 122’ OR ‘miRNA 122’ OR ‘microRNA
122’ OR miR-122-5p OR miRNA-122-5p OR microRNA-122-5p). Studies
that did not provide true positive (TP), false positive (FP), true
negative (TN) and false negative (FN) results were excluded. The
author, publishing year, sample type, number of HCC patients and
controls, and the number of TP, FP, TN and FN results were recorded
as basic information.
Selection of prospective target genes
of miR-122-5p
The potential target mRNAs of miR-122-5p were
predicted based on 12 online prediction databases, including
miRWalk, TargetScan and miRMap (19).
Genes recorded in >5 of the 12 prediction databases were
selected (20). The selected
predicted target genes were further intersected with TCGA and GEO
differentially expressed genes. The overlapping genes were
considered as potential target genes of miR-122-5p.
Bioinformatic analyses using Gene
Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and
annotation and the protein-protein interaction (PPI) network
Using the Database for Annotation, Visualization and
Integrated Discovery (DAVID; david.abcc.ncifcrf.gov), GO annotation was
subsequently conducted based on the prospective target mRNAs. A
total of three GO terms, including biological process, cellular
component and molecular function, were used to identify the
enrichment of target genes. KEGG pathway analysis (www.genome.jp/kegg) was applied to identify noteworthy
pathways associated with the selected target genes. The Bingo
plug-in component from Cytoscape 3.4.0 (www.cytoscape.org) and GraphPad Prism 7 software
(GraphPad Software, Inc., La Jolla, CA, USA) were employed to
visualize the network of GO terms and KEGG pathways, respectively
(21). GO, KEGG and Protein Analysis
Through Evolutionary Relationships (PANTHER) pathway analyses were
implemented in DAVID to summarize the essential functions of
potential target genes of miR-122-5p (22). To explicitly determine the
interactions and connections between hub genes, the PPI network was
applied using Search Tool for the Retrieval of Interacting
Genes/Proteins (www.string-db.org) (23). The nodes and edges presented the genes
and reciprocal actions between genes, whose number would reveal the
dominant hub genes of miR-122-5p in HCC. P<0.05 was considered
to indicate a statistically significant difference.
Statistical analysis
Student's t-test for independent samples and one way
analysis of variance (with Bonferroni's post-hoc test) was
performed using SPSS 19 software (IBM Corp., Armonk, NY, USA) to
determine the association between the miR-122-5p expression profile
and various clinicopathological parameters. P<0.05 was
considered to indicate a statistically significant difference.
SPSS 19 software was applied to generate a receiver
operator characteristic (ROC) curve to assess diagnostic efficacy.
The relative expression level of miR-122-5p from the microarray
included in the GEO was processed separately. All samples were
pooled with HCC and controls to conduct another diagnostic trial.
Meta-DiSc 1.4 (http://www.hrc.es/investigacion/metadisc_en.htm) was
used to generate combined effect and forest plots using a random
effects model. The sensitivity (SEN), specificity (SPE), positive
likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic
score (DS) and diagnostic odds ratio (DOR) were recorded. The area
under the curve (AUC) and point where sensitivity and specificity
are equal (index Q*) were calculated. Threshold and meta-regression
analyses were conducted to determine heterogeneity in the present
study, and P<0.1 or I2>50% were considered to
represent marked heterogeneity.
The analysis was repeated as aforementioned, and the
threshold and meta-regression analyses were performed in Stata14
(StataCorp LLC, College Station, TX, USA). Deek's funnel plot and
Fagan plot analyses were performed to examine publication bias and
to detect the diagnostic value of miR-122-5p in HCC.
Results
miR-122-5p expression levels and
associated clinicopathological features in TCGA and GEO
datasets
The average expression level of miR-122-5p in the
TCGA cohort was decreased in the patients with HCC compared with
the controls (P<0.001; Fig. 1A).
There were 375 patients with HCC and 50 controls in the TCGA
cohort, and 955 patients with HCC and 685 controls in the GEO
cohort. From the expression data in 20 GSE chips, all the
clinicopathological features mentioned in chips were gathered and
associations with miR-122-5p expression levels were investigated.
The downregulation of miR-122-5p was associated with HCC tissues
(P<0.001; Fig. 1B), tumor vascular
invasion (P<0.001), metastasis (P=0.001), tissue (compared with
serum) (P<0.001), virus infection state (P=0.001) and sex
(P=0.006) in the GEO cohort (Table
I).
 | Table I.Association between the expression of
miR-122-5p and clinicopathological parameters in HCC based on the
Gene Expression Omnibus database. |
Table I.
Association between the expression of
miR-122-5p and clinicopathological parameters in HCC based on the
Gene Expression Omnibus database.
|
|
| miR-122-5p relevant
expression |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | n | Mean ± SD | P-value |
|---|
| Tissue |
|
|
|
|
Non-cancerous | 685 | 15.35±1.40 | <0.001 |
|
HCC | 955 | 14.18±1.58 |
|
| Age, years |
|
|
|
|
<50 | 145 | 15.40±1.60 | 0.894 |
|
≥50 | 117 | 15.38±1.09 |
|
| Sex |
|
|
|
|
Male | 453 | 15.24±1.54 | 0.006 |
|
Female | 105 | 14.78±1.39 |
|
| Clinical TNM
stage |
|
|
|
| Early
stage | 82 | 15.25±0.53 | 0.979 |
|
Advanced stage | 66 | 15.25±0.44 |
|
| Metastasis |
|
|
|
| − | 466 | 15.36±0.80 | 0.001 |
| + | 171 | 15.09±0.99 |
|
| Tumor vascular
invasion |
|
|
|
| − | 91 | 14.13±0.38 | <0.001 |
| + | 81 | 12.81±1.09 |
|
| Alcohol abuse |
|
|
|
| − | 120 | 15.26±0.48 | 0.860 |
| + | 36 | 15.25±0.49 |
|
| Virus infection
state |
|
|
|
| − | 51 | 14.55±1.64 | 0.001 |
|
HBV | 175 | 15.47±1.87 |
|
|
HCV | 55 | 14.82±1.13 |
|
| Cirrhosis |
|
|
|
| − | 80 | 15.11±0.82 | 0.178 |
| + | 97 | 14.98±1.07 |
|
| Chemotherapy |
|
|
|
| − | 4 | 12.35±0.41 | 0.599 |
| + | 42 | 12.01±1.25 |
|
| Sample |
|
|
|
|
Tissue | 1,626 | 14.56±1.51 | <0.001 |
|
Serum | 14 | 20.10±2.55 |
|
Diagnostic value based on data from
TCGA and GEO
The relative expression level of miR-122-5p data
collected from TCGA demonstrated 81.07% sensitivity and 84.00%
specificity. The area under the ROC curve (AUC) was 0.865 [95%
confidence interval (CI), 0.829–0.896; Fig. 1C]. In the GEO cohort, the overall SEN
and the SPE were 66.77 and 79.70%, respectively, with an AUC of
0.749 (95% CI, 0.726–0.771; Fig. 1D).
The diagnostic value of every individual chip was also calculated
(Table II) (15,24–40).
 | Table II.Summary of every individual GEO chip
of miR-122-5p in HCC. |
Table II.
Summary of every individual GEO chip
of miR-122-5p in HCC.
| Accession no. | Country | Sample type | Experiment
type | Platform | HCC/Control | TP | FP | FN | TN | (Refs.) |
|---|
| GSE6857 | USA | Tissue | Non-coding RNA
profiling by array | GPL4700 | 241/241 | 115 | 41 | 126 | 200 | (24) |
| GSE12717 | China | Tissue | Non-coding RNA
profiling by array | GPL7274 | 10/6 | 3 | 0 | 7 | 6 | (25) |
| GSE10694 | China | Tissue | Non-coding RNA
profiling by array | GPL6542 | 78/88 | 57 | 39 | 21 | 49 | (26) |
| GSE21279 | China | Tissue | Non-coding RNA
profiling by high throughput sequencing | GPL9052 | 4/11 | 1 | 7 | 3 | 4 | (27) |
| GSE22058 | USA | Tissue | Expression
profiling by array | GPL6793 | 96/96 | 61 | 13 | 35 | 83 | (15) |
| GSE21362 | Japan | Tissue | Non-coding RNA
profiling by array | GPL10312 | 73/73 | 12 | 7 | 61 | 66 | (28) |
| GSE20971 | France | Tissue | Non-coding RNA
profiling by array | GPL10231 | 49/9 | 33 | 1 | 16 | 8 | (29) |
| GSE39678 | South Korea | Tissue | Non-coding RNA
profiling by array | GPL15852 | 16/8 | 2 | 6 | 14 | 2 | (30) |
| GSE31383 | USA | Tissue | Non-coding RNA
profiling by array | GPL10122 | 9/10 | 2 | 0 | 7 | 10 | (31) |
| GSE40744 | USA | Tissue | Non-coding RNA
profiling by array | GPL14613 | 39/37 | 12 | 0 | 27 | 37 | (32) |
| GSE50013 | USA | Plasma | Expression
profiling by RT-PCR | GPL15497 | 10/4 | 4 | 4 | 6 | 0 | (33) |
| GSE36915 | Taiwan | Tissue | Non-coding RNA
profiling by array | GPL8179 | 68/21 | 57 | 13 | 11 | 8 | (34) |
| GSE54751 | USA | Tissue | Expression
profiling by RT-PCR | GPL18262 | 10/10 | 8 | 2 | 2 | 8 | (35) |
| GSE57555 | Japan | Tissue | Non-coding RNA
profiling by array | GPL18044 | 5/16 | 5 | 8 | 0 | 8 | (36) |
| GSE67882 | India | Tissue | Non-coding RNA
profiling by array | GPL10850 | 4/8 | 4 | 1 | 0 | 7 | (37) |
| GSE74618 | Spain | Tissue | Non-coding RNA
profiling by array | GPL14613 | 218/20 | 80 | 1 | 138 | 19 | (38) |
| GSE65708 | China | Tissue | Other | GPL16850 | 6/8 | 5 | 0 | 1 | 8 | (39) |
| GSE64989 | Germany | Tissue | Non-coding RNA
profiling by array | GPL16384 | 8/10 | 6 | 5 | 2 | 5 | (40) |
Meta-analysis based on the previous
literature
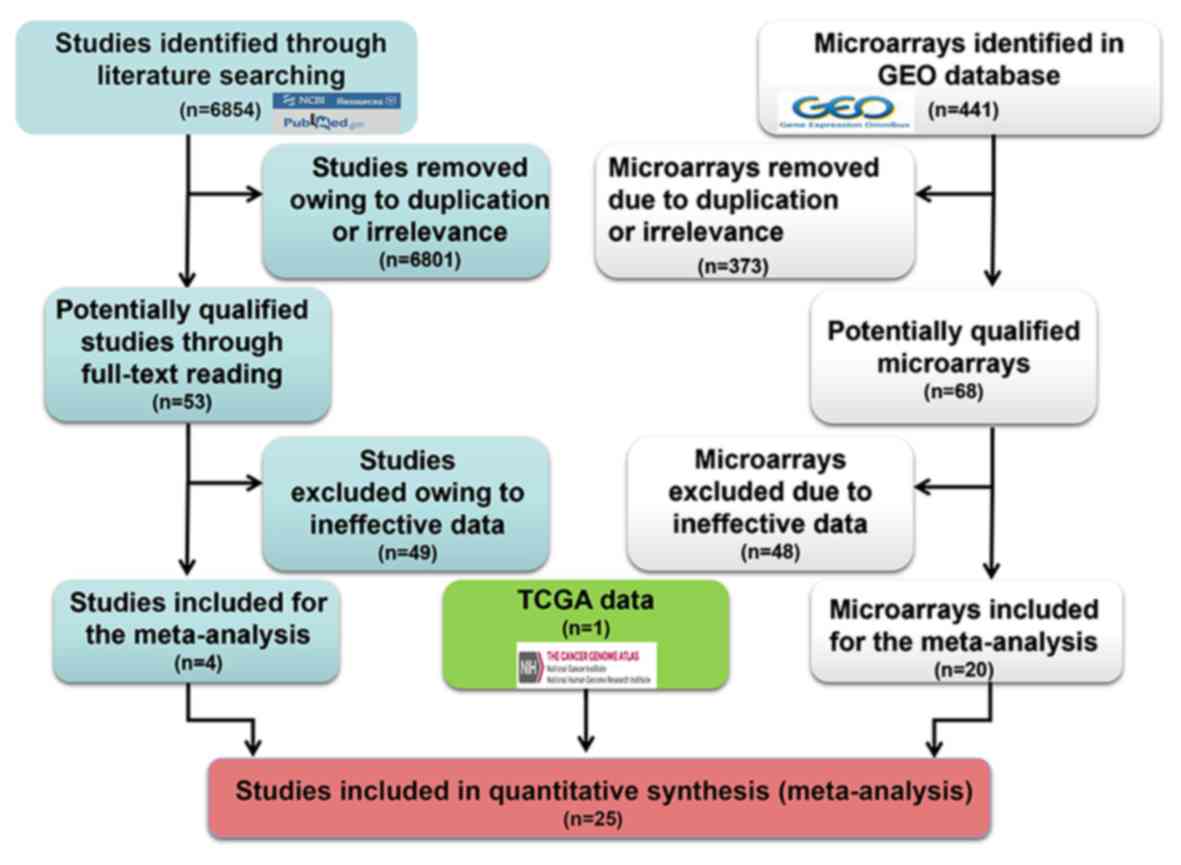
In total, 6,854 studies were collected from the
literature. Following exclusion of repeated and irrelevant
articles, 53 full texts were read. Finally, 4 studies were included
(Table III; Fig. 2) (41–44). In
the meta-analysis, the pooled SEN, SPE, PLR, NLR, DOR and AUC
values were 0.73 (95% CI, 0.56–0.86), 0.79 (95% CI, 0.69–0.86),
3.43 (95% CI, 2.38–4.95), 0.34 (95% CI, 0.20–0.59), 10.11 (95% CI,
4.70–21.77) and 0.83 (95% CI, 0.79–0.86), respectively, when
Stata14 was applied (Fig. 3). No
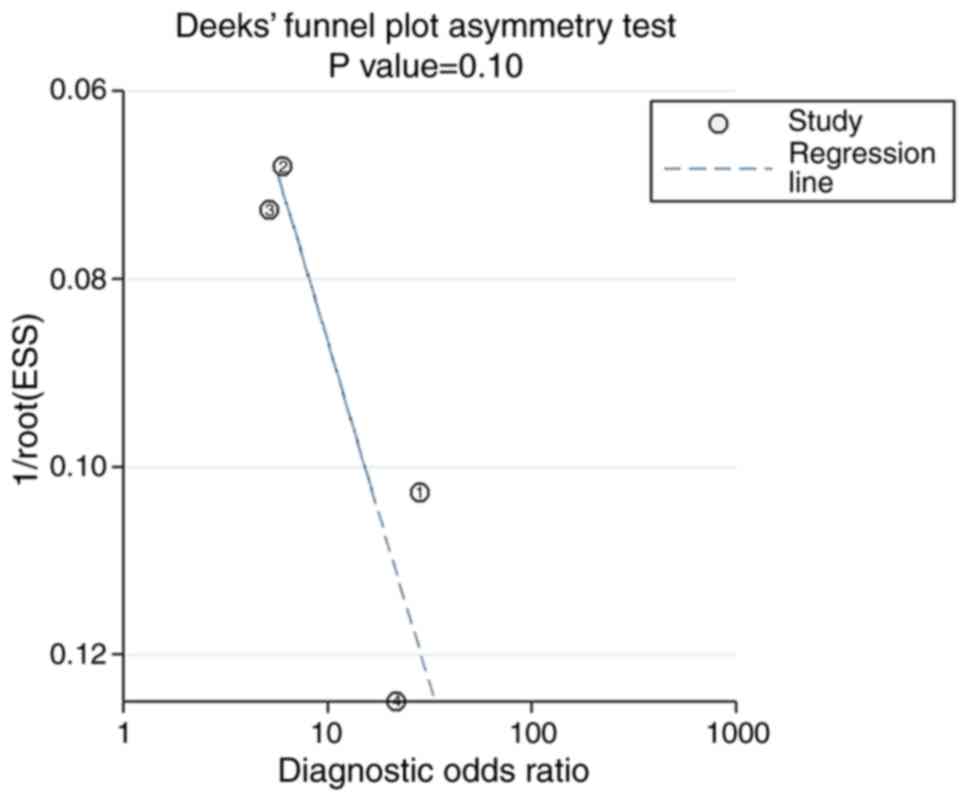
significant publication bias was observed in the meta-analysis
using Deek's funnel plot (P=0.10; Fig.
4). The pre-test probability value of miR-122-5p in patients
with HCC was 20%, and the post-test probability values, considering
PLR and NLR results, were 46 and 8%, respectively, based on Fagan's
nomogram (Fig. 5). The results were
also calculated using Meta-DiSc 1.4 (data not shown).
 | Figure 5.Fagan's nomogram detecting the
clinical use of miR-122-5p in the qualified studies in HCC based on
Stata14. The pre-test probability value of miR-122-5p in HCC was
20%, and the post-test probability values, considering the
LR_positive and LR_negative results, were 46 and 8%, respectively.
miR, microRNA; HCC, hepatocellular carcinoma; LR_positive, positive
likelihood ratio; LR_negative, negative likelihood ratio;
post_prob_pos, post-test positive probability; post_prob_neg,
post-test negative probability. |
 | Table III.Basic information of included studies
in this meta-analysis. |
Table III.
Basic information of included studies
in this meta-analysis.
| Country | Age, mean ± SD
(HCC/Control) | Sex,
female/male | Sample type | Experiment
type | Cut-off value | nHCC/Control | TP | FP | FN | TN | QUADAS | (Refs.) |
|---|
| China | Not shown | 9/36 | Serum | RT-PCR | 2.6963 | 45/50 | 40 | 11 | 5 | 39 | 5 | (41) |
| China |
53.57±11.59/40.20±6.61 | 49/176 | Serum | RT-qPCR | NR | 135/90 | 65 | 12 | 70 | 78 | 8 | (42) |
| China | Not shown | 44/146 | Serum | RT-qPCR | 0.7 | 101/89 | 71 | 28 | 30 | 61 | 9 | (43) |
| China | Not shown | Not shown | Serum | RT-qPCR | 0.475 | 48/24 | 39 | 4 | 9 | 20 | 8 | (44) |
Overall assessment of diagnostic
value
The data from GEO, TCGA and eligible studies were
combined in order to perform a pooled diagnostics test. In Stata14,
the pooled SEN, SPE, PLR, NLR and DOR values were 0.60 (95% CI,
0.48–0.71), 0.81 (95% CI, 0.70–0.89), 3.20 (95% CI, 1.93–5.30),
0.49 (95% CI, 0.37–0.65) and 6.49 (95% CI, 3.23–13.08). The AUC
value was 0.76 (95% CI, 0.72–0.80; Fig.
6). No significant publication bias was observed in the
meta-analysis using Deek's funnel plot (P=0.96; Fig. 7). The pre-test probability value for
miR-122-5p expression in patients with HCC was 63%, and the
post-test probability values, considering PLR and NLR results, were
84 and 45%, respectively, based on Fagan's nomogram (Fig. 8). In Meta-DiSc1.4, the pooled SEN,
SPE, PLR, NLR and DOR values were also calculated by Meta-DiSc 1.4
(data not shown).
 | Figure 8.The Fagan plot was displayed to test
the diagnostic value of miR-122-5p based on The Cancer Genome
Atlas, Gene Expression Omnibus and eligible studies in HCC by
Stata14. The pre-test probability value of miR-122-5p in HCC was
63%, and the post-test probability values, considering the
LR_positive and LR_negative results, were 84 and 45%, respectively.
miR, microRNA; HCC, hepatocellular carcinoma; LR_positive, positive
likelihood ratio; LR_negative, negative likelihood ratio;
post_prob_pos, post-test positive probability; post_prob_neg,
post-test negative probability. |
Identification of potential target
mRNAs of miR-122-5p
A total of 61,399 target genes were predicted by 12
online software programs, and 5,657 genes that were detected ≥5
times were regarded as potential target genes of miR-122-5p.
Additionally, differentially expressed genes assembled from the
TCGA and GEO databases were integrated to generate the intersection
of the two databases. The 5,657 potential target genes intersected
198 genes, with 3,278 differentially expressed genes from the TCGA
database and 5,122 differentially expressed genes from the GEO
database. These overlapping genes were used in subsequent
bioinformatic analyses.
Bioinformatic analyses of the
potential target genes of miR-122-5p
Further investigations were conducted to determine
the functional mechanism of miR-122-5p in the progression of HCC.
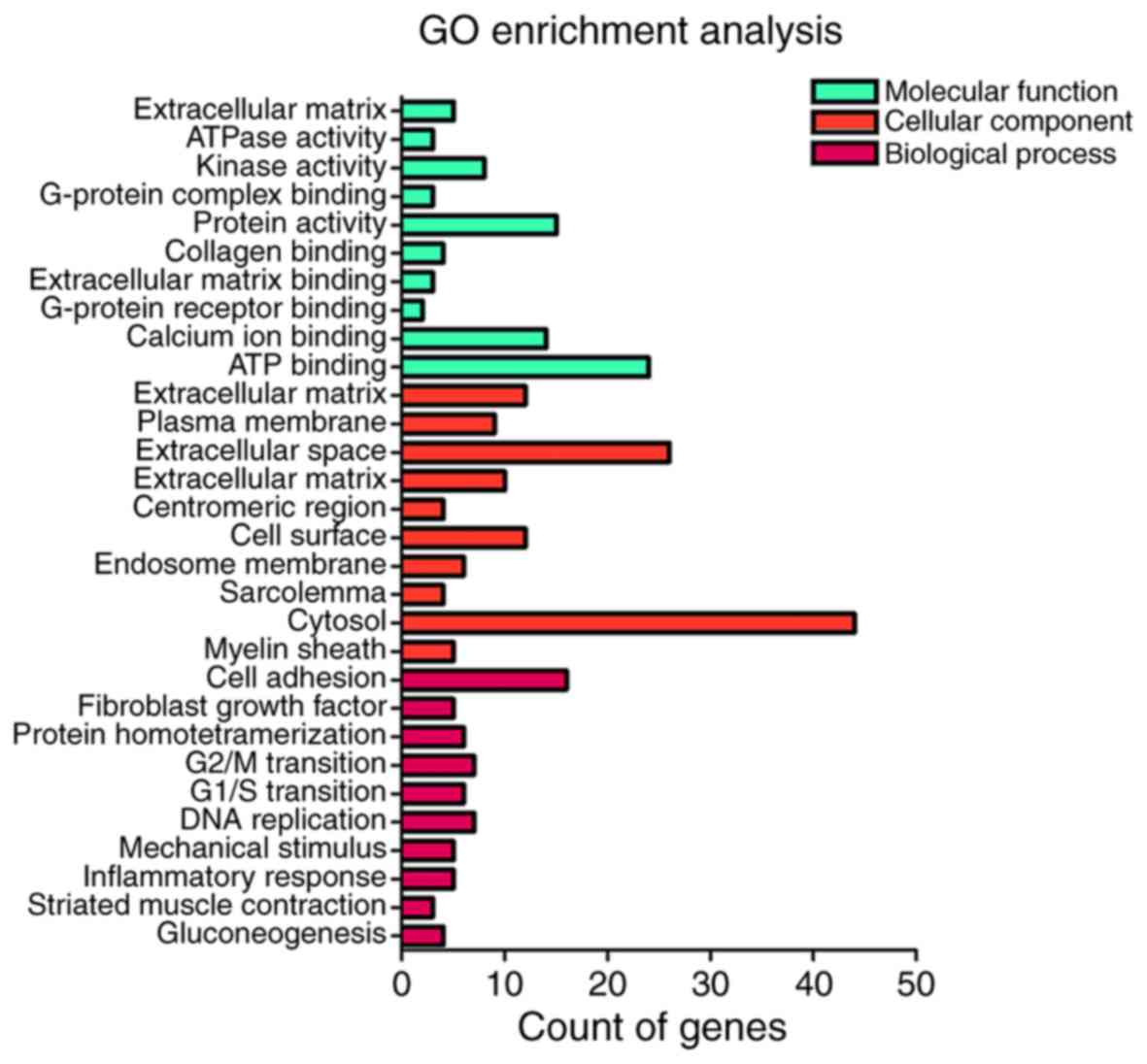
For the results of GO pathway analysis in DAVID, the potential
targets of miR-122-5p were notably associated with the regulation
of cell proliferation for cell adhesion (P=1.22×10−4),
proteinaceous extracellular matrix (P=1.20×10−4) and
extracellular matrix structural constituent
(P=5.57×10−3; Table IV;
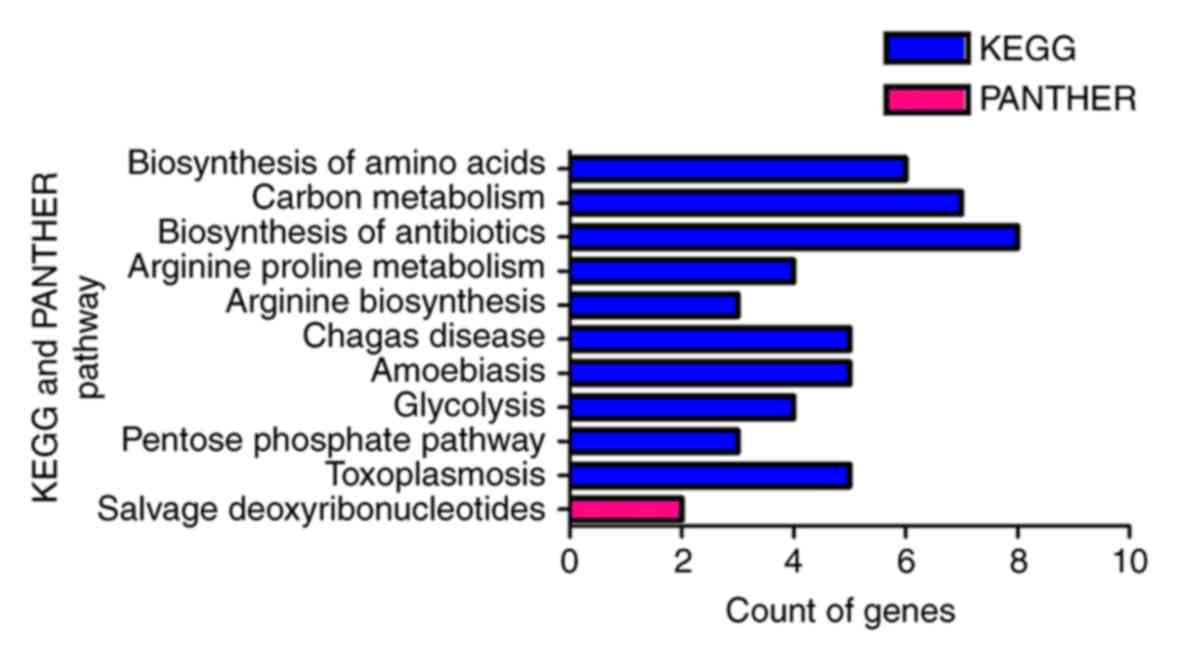
Fig. 9). Furthermore, KEGG pathway
analysis identified biosynthesis of amino acids, carbon metabolism,
biosynthesis of antibiotics, arginine and proline metabolism,
arginine biosynthesis, Chagas disease (American trypanosomiasis),
amoebiasis, glycolysis/gluconeogenesis, the pentose phosphate
pathway and toxoplasmosis as significant (P<0.05). In addition,
the most enriched term in the PANTHER analysis was salvage pathways
of pyrimidine deoxyribonucleotides (Table
V; Fig. 10). In the PPI
analysis, the network demonstrated 198 nodes and 57 edges (Fig. 11A). Genes with a combined score
>0.900, based on available experimental data and importing known
protein complexes from curated databases (23), including minichromosome maintenance
complex component (MCM)4, MCM8, MCM10, chromatin licensing and DNA
replication factor 1, cell division cycle (CDC) 6, CDC25A and
checkpoint kinase 1, were defined as hub genes. CDC6, MCM4 and
MCM8, with 24, 24 and 22 interactions with other hub genes in the
network, respectively, were recognized as hub genes of the highest
significance (Fig. 11B).
 | Table IV.The top 10 most enriched terms in
every GO section, based on the target genes of miR-122-5p in
hepatocellular carcinoma. |
Table IV.
The top 10 most enriched terms in
every GO section, based on the target genes of miR-122-5p in
hepatocellular carcinoma.
| Category | Term | Count | P-value |
|---|
| GOTERM_BP | GO:0007155~cell
adhesion | 16 |
1.22×10−4 |
| GOTERM_BP | GO:0044344~cellular
response to fibroblast growth factor stimulus | 5 |
2.76×10−4 |
| GOTERM_BP | GO:0051289~protein
homotetramerization | 6 |
4.42×10−4 |
| GOTERM_BP | GO:0000086~G2/M
transition of mitotic cell cycle | 7 |
3.51×10−3 |
| GOTERM_BP | GO:0000082~G1/S
transition of mitotic cell cycle | 6 |
4.71×10−3 |
| GOTERM_BP | GO:0006260~DNA
replication | 7 |
6.38×10−3 |
| GOTERM_BP | GO:0071260~cellular
response to mechanical stimulus | 5 |
6.96×10−3 |
| GOTERM_BP | GO:0050729~positive
regulation of inflammatory response | 5 |
7.67×10−3 |
| GOTERM_BP | GO:0006941~striated
muscle contraction | 3 |
8.16×10−3 |
| GOTERM_BP |
GO:0006094~gluconeogenesis | 4 |
1.14×10−2 |
| GOTERM_CC |
GO:0005578~proteinaceous extracellular
matrix | 12 |
1.20×10−4 |
| GOTERM_CC | GO:0009897~external
side of plasma membrane | 9 |
1.80×10−3 |
| GOTERM_CC |
GO:0005615~extracellular space | 26 |
3.52×10−3 |
| GOTERM_CC |
GO:0031012~extracellular matrix | 10 |
3.98×10−3 |
| GOTERM_CC |
GO:0000775~chromosome, centromeric
region | 4 |
2.16×10−2 |
| GOTERM_CC | GO:0009986~cell
surface | 12 |
2.70×10−2 |
| GOTERM_CC | GO:0010008~endosome
membrane | 6 |
4.47×10−2 |
| GOTERM_CC |
GO:0042383~sarcolemma | 4 |
5.92×10−2 |
| GOTERM_CC |
GO:0005829~cytosol | 44 |
6.95×10−2 |
| GOTERM_CC | GO:0043209~myelin
sheath | 5 |
7.45×10−2 |
| GOTERM_MF |
GO:0005201~extracellular matrix structural
constituent | 5 |
5.57×10−3 |
| GOTERM_MF | GO:0043225~anion
transmembrane-transporting ATPase activity | 3 |
5.77×10−3 |
| GOTERM_MF | GO:0016301~kinase
activity | 8 |
1.45×10−2 |
| GOTERM_MF |
GO:0031683~G-protein beta/gamma-subunit
complex binding | 3 |
1.87×10−2 |
| GOTERM_MF | GO:0042803~protein
homodimerization activity | 15 |
2.36×10−2 |
| GOTERM_MF | GO:0005518~collagen
binding | 4 |
2.58×10−2 |
| GOTERM_MF |
GO:0050840~extracellular matrix
binding | 3 |
3.08×10−2 |
| GOTERM_MF |
GO:0031821~G-protein coupled serotonin
receptor binding | 2 |
4.17×10−2 |
| GOTERM_MF | GO:0005509~calcium
ion binding | 14 |
4.20×10−2 |
| GOTERM_MF | GO:0005524~ATP
binding | 24 |
4.48×10−2 |
 | Table V.KEGG and PANTHER Pathway analysis of
potential target genes of miR-122-5p. |
Table V.
KEGG and PANTHER Pathway analysis of
potential target genes of miR-122-5p.
| Category | Term | Count | P-value |
|---|
| KEGG_PATHWAY |
hsa01230:Biosynthesis of amino acids | 6 |
1.67×10−3 |
| KEGG_PATHWAY | hsa01200:Carbon
metabolism | 7 |
2.00×10−3 |
| KEGG_PATHWAY |
hsa01130:Biosynthesis of antibiotics | 8 |
1.18×10−2 |
| KEGG_PATHWAY | hsa00330:Arginine
and proline metabolism | 4 |
2.05×10−2 |
| KEGG_PATHWAY | hsa00220:Arginine
biosynthesis | 3 |
2.25×10−2 |
| KEGG_PATHWAY | hsa05142:Chagas
disease (American trypanosomiasis) | 5 |
3.33×10−2 |
| KEGG_PATHWAY |
hsa05146:Amoebiasis | 5 |
3.54×10−2 |
| KEGG_PATHWAY |
hsa00010:Glycolysis/Gluconeogenesis | 4 |
4.35×10−2 |
| KEGG_PATHWAY | hsa00030:Pentose
phosphate pathway | 3 |
4.49×10−2 |
| KEGG_PATHWAY |
hsa05145:Toxoplasmosis | 5 |
4.93×10−2 |
|
PANTHER_PATHWAY | P02774:Salvage
pyrimidine deoxyribonucleotides | 2 |
5.89×10−2 |
Discussion
A number of studies have confirmed that the
downregulation of miR-122-5p in HCC was associated with metastasis
and a poor prognosis (45,46). Therefore, the present study further
investigated these conclusions. The expression of miR-122-5p was
decreased in patients with HCC compared with that in controls in
the TCGA and GEO databases. Additionally, decreased miR-122-5p
expression was significantly associated with metastasis and
vascular invasion in the GEO dataset, suggesting that the
downregulation of miR-122-5p may lead to a poor prognosis, and that
miR-122-5p may act as a tumor suppressor in HCC. Furthermore,
miR-122-5p expression varied in different virus infection states,
indicating that miR-122-5p may be associated with virus-associated
HCC. In tissues, miR-122-5p expression was decreased compared with
that in serum. However, reflecting the limited number of serum
samples, further studies are required to confirm distinctions in
the diagnostic accuracy between tissues and serum.
With regards to diagnostic value, the pooled AUC was
0.76 (95% CI, 0.72–0.80), suggesting that miR-122-5p possessed a
moderate degree of accuracy in diagnostic tests and that it may be
a promising biomarker for the diagnosis of patients with HCC. A
discrepancy in sample type and patients may reflect the observed
differences in the AUC among TCGA, GEO and meta-analysis studies.
Li et al (47), He et
al (48) and Huang et al
(49) conducted integrated research
to examine the diagnostic value of miR-122-5p, and the AUC values
were 0.81, 0.78 and 0.77, respectively, consistent with the results
of the present study. In search strategy, Wu et al (46) applied 2 public databases: PubMed and
Embase, Qi et al (44) applied
Medline and CancerLit Embase prior to July 31, 2015. Huang et
al (49) analyzed the diagnostic
value of miRNAs in patients with HCC utilizing the Medline, Embase
and CNKI databases. The present study aimed to explore the
potential clinical value of miR-122-5p in HCC. In addition to
pertinent literature regarding miR-122-5p, data from TCGA and GEO
public databases were also extracted. The SEN, SPE, PLR, NLR, DS
and DOR values were determined in Meta-DiSc 1.4. The aforementioned
study was repeated, excluding threshold analysis and
meta-regression in Stata14. Deek's funnel plot and Fagan plot were
displayed to test the publication bias and to detect the diagnostic
value of miR-122-5p in HCC. The heterogeneity in the meta-analysis
of these studies did not reflect a threshold effect or clinical
features. Subgroup and meta-regression analyses were not performed,
reflecting the limited number of studies collected. The source of
heterogeneity in the pooled meta-analysis was not determined by
meta-regression.
In the PPI network, CDC6, MCM4 and MCM8 were
highlighted as the most significant hub genes with multiple
interactions in the network. Further examination of these notable
genes would reveal the function for miR-122-5p in HCC. CDC6 is a
necessary regulator of DNA replication and is inextricably
associated with tumorigenesis (50).
High CDC6 expression has been associated with various types of
cancer. In ovarian cancer, Deng et al (51) discovered that the elevated expression
of CDC6 may accelerate cell proliferation and worsen prognosis. In
addition, CDC6 may participate in chemotherapy resistance in
bladder cancer (52). Previous
studies have also demonstrated oncogenetic functions for CDC6 in
HCC (53,54). Xiong et al (55) demonstrated that CDC6 overexpression
may increase susceptibility to HCC, and the Cdc6-515A>G
polymorphism may attenuate CDC6 expression to decrease the risk of
carcinogenesis. Therefore, studies focused on approaches to
downregulate CDC6 have been reported. HKH40A was utilized to
disrupt the cell cycle and to promote apoptosis, during which CDC6
was downregulated (56). MCM4 is also
an essential replication modulator. In cervical cancer (57), non-small cell lung cancer (58) and laryngeal squamous cell carcinoma
(59), the upregulation of MCM4 was
also observed. Studach et al (60) observed upregulation of MCM4 in HCC in
X/c-myc bitransgenics, indicating the identical expression profile
of MCM4 in HCC. However, the MCM4 polymorphism may generate
contradictory results. For example, Nan et al (61) reported that MCM4 may decrease the risk
of HCC. Another notable hub gene of miR-122-5p, MCM8, which is also
a gene replication regulator, may modulate DNA replication through
interactions with other MCM proteins, including the aforementioned
MCM4 (62). MCM8 was overexpressed in
chronic myelogenous leukemia (63)
and the overexpression of MCM8 was associated with the poor
prognosis in pancreatic cancer (64).
However, to the best of our knowledge, there are no available
studies regarding the function of MCM8 in HCC. As the hub genes of
miR-122-5p with the most interactions with other hub genes, CDC6,
MCM4 and MCM8 all exerted the function of genome replication.
Consistently, regulation of cell proliferation for cell adhesion
was also highlighted in the GO analysis. Therefore, we hypothesized
that miR-122-5p may be involved in the biological process of gene
replication. Considering that miR-122-5p exhibited significantly
decreased expression in HCC, whereas CDC6 and MCM4 were
upregulated, miR-122-5p may represent a therapeutic target and
biomarker for HCC.
In conclusion, the downregulation of miR-122-5p in
HCC demonstrated diagnostic value worthy of further attention.
Furthermore, this molecule may function as a tumor suppressor by
modulating genome replication. Additional experiments and studies
are required to verify this discovery.
Acknowledgements
The present study was partly supported by the Fund
of National Natural Science Foundation of China (grant nos.
NSFC81260222 and NSFC81060202), the Fund of Youth Science
Foundation of Guangxi Medical University (grant no. GXMUYSF201624)
and the Guangxi Medical University Students Innovative Project
(grant no. WLXSZX17001).
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GEO
|
Gene Expression Omnibus
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PANTHER
|
Protein Analysis Through Evolutionary
Relationships
|
|
PPI
|
and protein-protein interaction
network
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the curve
|
|
TP
|
true positive
|
|
FP
|
false positive
|
|
TN
|
true negative
|
|
FN
|
false negative
|
References
|
1
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferenci P, Fried M, Labrecque D, Bruix J,
Sherman M, Omata M, Heathcote J, Piratsivuth T, Kew M, Otegbayo JA,
et al: Hepatocellular carcinoma (HCC): A global perspective. J Clin
Gastroenterol. 44:239–245. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oliveri RS, Wetterslev J and Gluud C:
Transarterial (chemo)embolisation for unresectable hepatocellular
carcinoma. Cochrane Database Syst Rev. 3:CD0047872011.
|
|
4
|
Feng M and Ben-Josef E: Radiation therapy
for hepatocellular carcinoma. Semin Radiat Oncol. 21:271–277. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marrero JA, Feng Z, Wang Y, Nguyen MH,
Befeler AS, Roberts LR, Reddy KR, Harnois D, Llovet JM, Normolle D,
et al: Alpha-fetoprotein, des-gamma carboxyprothrombin, and
lectin-bound alpha-fetoprotein in early hepatocellular carcinoma.
Gastroenterology. 137:110–118. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baig JA, Alam JM, Mahmood SR, Baig M,
Shaheen R, Sultana I and Waheed A: Hepatocellular carcinoma (HCC)
and diagnostic significance of A-fetoprotein (AFP). J Ayub Med Coll
Abbottabad. 21:72–75. 2009.PubMed/NCBI
|
|
7
|
Collier J and Sherman M: Screening for
hepatocellular carcinoma. Hepatology. 27:273–278. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shaker O, Alhelf M, Morcos G and
Elsharkawy A: miRNA-101-1 and miRNA-221 expressions and their
polymorphisms as biomarkers for early diagnosis of hepatocellular
carcinoma. Infect Genet Evol. 51:173–181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye W, Li J, Fang G, Cai X, Zhang Y, Zhou
C, Chen L and Yang W: Expression of microRNA 638 and
sex-determining region Y-box 2 in hepatocellular carcinoma:
Association between clinicopathological features and prognosis.
Oncol Lett. 15:7255–7264. 2018.PubMed/NCBI
|
|
10
|
Li F, Wang F, Zhu C, Wei Q, Zhang T and
Zhou YL: miR-221 suppression through nanoparticle-based miRNA
delivery system for hepatocellular carcinoma therapy and its
diagnosis as a potential biomarker. Int J Nanomed. 13:2295–2307.
2018. View Article : Google Scholar
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hua HW, Jiang F, Huang Q, Liao Z and Ding
G: MicroRNA-153 promotes Wnt/β-catenin activation in hepatocellular
carcinoma through suppression of WWOX. Oncotarget. 6:3840–3847.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu LM, Ji JS, Yang Z, Xing CY, Pan TT, Xie
HY, Zhang F, Zhuang L, Zhou L and Zheng SS: Oncogenic role of
microRNA-423-5p in hepatocellular carcinoma. Hepatobiliary Pancreat
Dis Int. 14:613–618. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Burchard J, Zhang C, Liu AM, Poon RT, Lee
NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al:
microRNA-122 as a regulator of mitochondrial metabolic gene network
in hepatocellular carcinoma. Mol Syst Biol. 6:4022010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hutter C and Zenklusen JC: The cancer
genome atlas: Creating lasting value beyond its data. Cell.
173:283–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Danford T, Rolfe A and Gifford D: GSE: A
comprehensive database system for the representation, retrieval,
and analysis of microarray data. Pac Symp Biocomput. 1–550.
2008.
|
|
19
|
Lee M and Lee H: DMirNet: Inferring direct
microRNA-mRNA association networks. BMC Syst Biol. 10:1252016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang S, Xie Y, Yang P, Chen P and Zhang
L: HCV core protein-induced down-regulation of microRNA-152
promoted aberrant proliferation by regulating Wnt1 in HepG2 cells.
PLoS One. 9:e817302014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiao X, Sherman BT, da Huang W, Stephens
R, Baseler MW, Lane HC and Lempicki RA: DAVID-WS: A stateful web
service to facilitate gene/protein list analysis. Bioinformatics.
28:1805–1806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li W, Xie L, He X, Li J, Tu K, Wei L, Wu
J, Guo Y, Ma X, Zhang P, et al: Diagnostic and prognostic
implications of microRNAs in human hepatocellular carcinoma. Int J
Cancer. 123:1616–1622. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong
Q, Qin L, Wu X, Zheng Y, Yang Y, et al: Identification of miRNomes
in human liver and hepatocellular carcinoma reveals miR-199a/b-3p
as therapeutic target for hepatocellular carcinoma. Cancer Cell.
19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sato F, Hatano E, Kitamura K, Myomoto A,
Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S and
Shimizu K: MicroRNA profile predicts recurrence after resection in
patients with hepatocellular carcinoma within the Milan Criteria.
PLoS One. 6:e164352011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cairo S, Wang Y, de Reyniès A, Duroure K,
Dahan J, Redon MJ, Fabre M, McClelland M, Wang XW, Croce CM and
Buendia MA: Stem cell-like micro-RNA signature driven by Myc in
aggressive liver cancer. Proc Natl Acad Sci USA. 107:20471–20476.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK,
Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al: MiR-145 functions
as a tumor suppressor by directly targeting histone deacetylase 2
in liver cancer. Cancer Lett. 335:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han K, Li J, Zhao H, Liang P, Huang X,
Zheng L, Li Y, Yang T and Wang L: Identification of the typical
miRNAs and target genes in hepatocellular carcinoma. Mol Med Rep.
10:229–235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Diaz G, Melis M, Tice A, Kleiner DE,
Mishra L, Zamboni F and Farci P: Identification of microRNAs
specifically expressed in hepatitis C virus-associated
hepatocellular carcinoma. Int J Cancer. 133:816–824. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen J, Wang A, Wang Q, Gurvich I, Siegel
AB, Remotti H and Santella RM: Exploration of genome-wide
circulating microRNA in hepatocellular carcinoma: MiR-483-5p as a
potential biomarker. Cancer Epidemiol Biomarkers Prev.
22:2364–2373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang DH, Wang GY, Zhang JW, Li Y, Zeng XC
and Jiang N: miR-501-5p regulates CYLD expression and promotes cell
proliferation in human hepatocellular carcinoma. Jpn J Clin Oncol.
45:738–744. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao L and Zhang Y: miR-342-3p affects
hepatocellular carcinoma cell proliferation via regulating NF-κB
pathway. Biochem Biophys Res Commun. 457:370–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murakami Y, Kubo S, Tamori A, Itami S,
Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T and Taguchi YH:
Comprehensive analysis of transcriptome and metabolome analysis in
intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci
Rep. 5:162942015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ding W, Yang H, Gong S, Shi W, Xiao J, Gu
J, Wang Y and He B: Candidate miRNAs and pathogenesis investigation
for hepatocellular carcinoma based on bioinformatics analysis.
Oncol Lett. 13:3409–3414. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Martinez-Quetglas I, Pinyol R, Dauch D,
Torrecilla S, Tovar V, Moeini A, Alsinet C, Portela A,
Rodriguez-Carunchio L, Solé M, et al: IGF2 Is up-regulated by
epigenetic mechanisms in hepatocellular carcinomas and is an
actionable oncogene product in experimental models.
Gastroenterology. 151:1192–1205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ,
Zhang Q, Li SP, Xiong Y, Yuan Y, Min J, et al: A serum microRNA
classifier for early detection of hepatocellular carcinoma: A
multicentre, retrospective, longitudinal biomarker identification
study with a nested case-control study. Lancet Oncol. 16:804–815.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liese J, Peveling-Oberhag J, Doering C,
Schnitzbauer AA, Herrmann E, Zangos S, Hansmann ML, Moench C,
Welker MW, Zeuzem S, et al: A possible role of microRNAs as
predictive markers for the recurrence of hepatocellular carcinoma
after liver transplantation. Transpl Int. 29:369–380. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu LN, Huang DF, Li F, et al: Diagnostic
value of serum miRNA-122 and miRNA-221 expression in patients with
primary liver cancer. Jiangsu Med J. 41:1285–1288. 2015.(In
Chinese).
|
|
42
|
Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X,
Zhou X and Gan J: A serum microRNA panel as potential biomarkers
for hepatocellular carcinoma related with hepatitis B virus. PLoS
One. 9:e1079862014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu J, Wu C, Che X, Wang L, Yu D, Zhang T,
Huang L, Li H, Tan W, Wang C and Lin D: Circulating microRNAs,
miR-21, miR-122, and miR-223, in patients with hepatocellular
carcinoma or chronic hepatitis. Mol Carcinog. 50:136–142. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qi P, Cheng SQ, Wang H, Li N, Chen YF and
Gao CF: Serum microRNAs as biomarkers for hepatocellular carcinoma
in Chinese patients with chronic hepatitis B virus infection. PLoS
One. 6:e284862011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hsu SH, Wang B, Kota J, Yu J, Costinean S,
Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al: Essential
metabolic, anti-inflammatory, and anti-tumorigenic functions of
miR-122 in liver. J Clin Invest. 122:2871–2883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang
WJ, Yang L, Fu Q, Xu JJ and Gu JX: Decreased expression of
hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in
hepatitis B virus-associated hepatocellular carcinoma enhances
potential oncogenic GALNT10 protein activity. J Biol Chem.
290:1170–1185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li G, Shen Q, Li C, Li D, Chen J and He M:
Identification of circulating MicroRNAs as novel potential
biomarkers for hepatocellular carcinoma detection: A systematic
review and meta-analysis. Clin Transl Oncol. 17:684–693. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
He S, Hu XW, Wang D, Han LF, Zhang DC and
Wei C: Accuracy of microRNAs for the diagnosis of hepatocellular
carcinoma: A systematic review and meta-analysis. Clin Res Hepatol
Gastroenterol. 40:405–417. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang JT, Liu SM, Ma H, Yang Y, Zhang X,
Sun H, Zhang X, Xu J and Wang J: Systematic review and
meta-analysis: Circulating miRNAs for diagnosis of hepatocellular
carcinoma. J Cell Physiol. 231:328–335. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Borlado LR and Méndez J: CDC6: From DNA
replication to cell cycle checkpoints and oncogenesis.
Carcinogenesis. 29:237–243. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Deng Y, Jiang L, Wang Y, Xi Q, Zhong J,
Liu J, Yang S, Liu R, Wang J, Huang M, et al: High expression of
CDC6 is associated with accelerated cell proliferation and poor
prognosis of epithelial ovarian cancer. Pathol Res Pract.
212:239–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen S, Chen X, Xie G, He Y, Yan D, Zheng
D, Li S, Fu X, Li Y, Pang X, et al: Cdc6 contributes to
cisplatin-resistance by activation of ATR-Chk1 pathway in bladder
cancer cells. Oncotarget. 7:40362–40376. 2016.PubMed/NCBI
|
|
53
|
Lee CF, Ling ZQ, Zhao T, Fang SH, Chang
WC, Lee SC and Lee KR: Genomic-wide analysis of lymphatic
metastasis-associated genes in human hepatocellular carcinoma.
World J Gastroenterol. 15:356–365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Notas G, Alexaki VI, Kampa M, Pelekanou V,
Charalampopoulos I, Sabour-Alaoui S, Pediaditakis I, Dessirier V,
Gravanis A, Stathopoulos EN, et al: APRIL binding to BCMA activates
a JNK2-FOXO3-GADD45 pathway and induces a G2/M cell growth arrest
in liver cells. J Immunol. 189:4748–4758. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xiong XD, Fang JH, Qiu FE, Zhao J, Cheng
J, Yuan Y, Li SP and Zhuang SM: A novel functional polymorphism in
the Cdc6 promoter is associated with the risk for hepatocellular
carcinoma. Mutat Res. 643:70–74. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kosakowska-Cholody T, Cholody WM,
Hariprakasha HK, Monks AP, Kar SP, Wang M, Michejda CJ and Carr B
1: Growth inhibition of hepatocellular carcinoma cells in vitro and
in vivo by the 8-methoxy analog of WMC79. Cancer Chemother
Pharmacol. 63:769–778. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gan N, Du Y, Zhang W and Zhou J: Increase
of Mcm3 and Mcm4 expression in cervical squamous cell carcinomas.
Eur J Gynaecol Oncol. 31:291–294. 2010.PubMed/NCBI
|
|
58
|
Kikuchi J, Kinoshita I, Shimizu Y, Kikuchi
E, Takeda K, Aburatani H, Oizumi S, Konishi J, Kaga K, Matsuno Y,
et al: Minichromosome maintenance (MCM) protein 4 as a marker for
proliferation and its clinical and clinicopathological significance
in non-small cell lung cancer. Lung Cancer. 72:229–237. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lian M, Fang J, Han D, Ma H, Feng L, Wang
R and Yang F: Microarray gene expression analysis of tumorigenesis
and regional lymph node metastasis in laryngeal squamous cell
carcinoma. PLoS One. 8:e848542013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Studach LL, Menne S, Cairo S, Buendia MA,
Hullinger RL, Lefrançois L, Merle P and Andrisani OM: Subset of
Suz12/PRC2 target genes is activated during hepatitis B virus
replication and liver carcinogenesis associated with HBV X protein.
Hepatology. 56:1240–1251. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Nan YL, Hu YL, Liu ZK, Duan FF, Xu Y, Li
S, Li T, Chen DF and Zeng XY: Relationships between cell cycle
pathway gene polymorphisms and risk of hepatocellular carcinoma.
World J Gastroenterol. 22:5558–5567. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Stolk L, Zhai G, van Meurs JB, Verbiest
MM, Visser JA, Estrada K, Rivadeneira F, Williams FM, Cherkas L,
Deloukas P, et al: Loci at chromosomes 13, 19 and 20 influence age
at natural menopause. Nat Genet. 41:645–647. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cai L, Zhao K and Yuan X: Expression of
minichromosome maintenance 8 in chronic myelogenous leukemia. Int J
Clin Exp Pathol. 8:14180–14188. 2015.PubMed/NCBI
|
|
64
|
Peng YP, Zhu Y, Yin LD, Zhang JJ, Guo S,
Fu Y, Miao Y and Wei JS: The expression and prognostic roles of
MCMs in pancreatic cancer. PLoS One. 11:e01641502016. View Article : Google Scholar : PubMed/NCBI
|