Introduction
Lung cancer is one of the most common types of
cancer worldwide and it remains the leading cause of
cancer-associated mortality among all human malignancies (1). In developing and developed countries,
the morbidity and mortality rates of lung cancer have markedly
increased, particularly in China (2).
Non-small cell lung cancer (NSCLC) accounts for 80–85% of lung
cancer cases according to the World Health Organization (WHO)
classification (3). Over the past
decade, the treatment strategy for NSCLC has changed significantly
and the therapeutic efficacy has improved with the application of
novel individualized therapy. This is based on specific molecular
alterations that define biological characteristics of lung cancer
and may be used to predict treatment response (4). The epidermal growth factor receptor
(EGFR) gene is considered the most valuable driver oncogene in the
clinical management of NSCLC. EGFR gene mutations are identifiable
in 10–50% of later stage NSCLC cases and are frequently detected in
female non-smoker Asian patients with adenocarcinoma (5–7). EGFR
tyrosine kinase inhibitors (TKIs) have exhibited promising clinical
efficacy for the treatment of advanced NSCLC harboring sensitive
EGFR mutations (8). Phase III
randomized trials have provided robust evidence that EGFR TKIs
could be applied as first-line therapy for advanced NSCLC with
particular EGFR mutations (9–12).
EGFR mutation detection is being established as a
prerequisite for the tailored treatment of late stage NSCLC. The
diagnosis of early-stage lung adenocarcinoma has increased and
stage I lung adenocarcinoma is the most common type of lung cancer
in China, including MIA (13).
However, few studies have investigated the characteristics of EGFR
mutation or its significance in lung MIA (14,15). The
present study described the EGFR mutation spectrum in surgically
resected MIA specimens from Chinese patients and further evaluated
the association between clinicopathological features and EGFR
mutation status. Furthermore, the present study aimed to
investigate the differences in clinicopathological parameters
between the two most common EGFR mutation subtypes (deletion in
exon 19 and point mutation in exon 21 L858R), and to analyze the
clinical relevance of this.
Materials and methods
Ethics approval
The study protocol was approved by the Institutional
Review Board of Anhui Provincial Hospital, The First Affiliated
Hospital of University of Science and Technology of China (Anhui,
China; reference no. 20160183). Written informed consent was
obtained from all patients and all patients agreed to the use of
tissue samples for EGFR mutation analysis. The present study was
performed according to the approved guidelines and principles of
good clinical practice.
Patients and samples
Between January 2015 and June 2017, 79 patients (32
male and 47 female; (median, 58; range 39–82 years of age) who
underwent pulmonary resection for a small pulmonary nodule
following low-dose computed tomography (LD-CT) examination, were
enrolled at Anhui Provincial Hospital. Patients included in the
present study met the following inclusion criteria: i)
Pathologically-confirmed pulmonary microinvasive adenocarcinoma
diagnosis, according to the WHO Classification of Lung Tumors (4th
edition) (16), and ii) surgically
resected specimens available for EGFR mutation detection. Patients
were excluded from the present study if they had received
chemotherapy, radiotherapy or other therapy prior to surgery.
All resected tissue samples were fixed in 10%
neutral-buffered formalin for 24 h at room temperature and embedded
in paraffin. Clinical and pathological data were collected from the
medical records of each patient. Histological subtype and the
maximum size of the microinvasion component were evaluated by 3
pathologists, (ML, MZ and CL) from the Department of Pathology,
Anhui Provincial Hospital, The First Affiliated Hospital University
of Science and Technology (Hefei, China), according to the new
International Association for the Study of Lung Cancer/American
Thoracic Society/European Respiratory Society adenocarcinoma
classification and WHO criteria (16,17). The
patient demographic characteristics are summarized in Table I.
 | Table I.Demographic characteristics of 79
Chinese patients with lung microinvasion adenocarcinoma. |
Table I.
Demographic characteristics of 79
Chinese patients with lung microinvasion adenocarcinoma.
| Factor | n | Proportion, % |
|---|
| Total cases | 79 |
|
| Age |
|
|
|
≤50 | 16 | 20.25 |
|
51–60 | 31 | 39.24 |
|
61–70 | 21 | 26.58 |
|
>70 | 11 | 13.92 |
| Sex |
|
|
|
Male | 32 | 40.51 |
|
Female | 47 | 59.49 |
| Smoking status |
|
|
| Never
smoked | 71 | 89.87 |
|
Smoker | 8 | 10.13 |
| Tumor number |
|
|
|
Single | 65 | 82.28 |
|
Multiple | 14 | 17.72 |
| Tumor site |
|
|
|
Left | 28 | 35.44 |
|
Right | 49 | 62.03 |
|
Bilateral | 2 |
2.53 |
Immunohistochemical (IHC) analysis and
TTF-1 scoring
IHC staining was performed according to a previously
described method (18). In brief,
4-µm thick formalin-fixed, paraffin-embedded tissue sections were
deparaffinized in xylene and rehydrated in gradient ethanol, which
included 100, 95, and 85% ethanol. Tissue sections were treated
with 3% hydrogen peroxide at room temperature for 10 min and washed
with PBS (pH 7.4), and antigens were retrieved at a
high-temperature of ~102°C and high-pressure for 2 min in citrate
buffer (pH 6.0; cat. no. 14746; Santa Cruz; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The slides were subsequently
washed with PBS (pH 7.4) and blocked in 3% bovine serum albumin
(cat. no. 9998; Santa Cruz; Santa Cruz Biotechnology, Inc) at room
temperature for 30 min. A TTF-1 monoclonal antibody (dilution,
1:200; clone 8G7G3/1; cat. no. IS05630; Dako; Agilent Technologies,
Inc., Santa Clara, CA, USA) was used as the primary antibody for
TTF-1 detection. The slides were incubated with the primary
antibody for 2 h at room temperature. The TTF-1 IHC staining was
performed according to the manufacturer's protocol of the Envision
Two-step Detection kit (cat. no. GK500705; Dako; Agilent
Technologies, Inc.). The IHC slides were analyzed under a light
microscope Olympus BX 51 (magnification, ×200; Olympus Corporation,
Tokyo, Japan). TTF-1 expression was indicated by nuclear
staining.
Previous studies (19,20) have
performed TTF-1 expression scoring based on the staining intensity.
The intensity was scored as follows: 0, no expression; 1, weak
expression; 2, intermediate expression; or 3, strong expression. A
sample was defined as negative for tumor cells with a score of
<2 and positive for tumor cells with a score of ≥2.
EGFR mutation analysis
Genomic DNA was isolated and purified from
formalin-fixed paraffin-embedded tissues using a QIAamp DNA FFPE
Tissue kit (cat. no. 56404; Qiagen GmbH, Hilden, Germany),
according to the manufacturer's protocol. Macrodissection of
materials from tissue sections was performed if necessary for
specimens containing insufficient tumor components (the proportion
of tumor cells was <50%). The extracted DNA purity and
concentration were evaluated by a NanoDrop 2000 ultraviolet
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). EGFR mutation status was detected using
Human EGFR Gene Mutation Detection kit (cat. no. 20143402001;
ADx-ARMS; Amoy Diagnostics Co., Ltd., Amoy, Fujian, China),
according to the manufacturer's protocol for the amplification
refractory mutation system quantitative polymerase chain reaction
(PCR). A total of 29 mutations were detected in the present study,
including G719A, G719S, G719C, T790M, L858R, L861Q and S768I, three
types of insertions in exon 20, and 19 types of deletions in exon
19. Positive and negative controls were set in each experiment. The
positive controls were the plasmids containing mutated EGFR gene
sequences and negative control were the plasmids containing the
wild-type EGFR gene sequences. An external control was used for
each sample to guarantee the loading of sufficient DNA template and
an internal control was used in each reaction tube to exclude the
presence of PCR inhibitors.
Statistical analysis
Statistical analysis was performed using SPSS
software for Windows (version 13.0; SPSS, Inc., Chicago, IL, USA).
The association between various clinicopathological parameters and
EGFR mutation status was evaluated using the χ2 and
Fisher's exact tests, as appropriate. Subgroup analysis aimed to
investigate differences in EGFR gene status. All tests were
two-sided and P<0.05 was considered to indicate a statistically
significant difference.
Results
Descriptive characteristics of
patients
A total of 79 patients with lung MIA were included
in the present study and the demographic characteristics of these
patients are summarized in Table I.
All pathological diagnoses were determined by three professional
pulmonary pathologists according to the criteria of the WHO/IASLC
Histological Classification of Lung and Pleural Tumors. The median
age at diagnosis was 58 years (range, 39–82 years). There were 32
(40.51%) male and 47 female (59.49%) patients. A total of 8
(10.12%) of the enrolled patients were smokers and 71 (89.87%) had
never smoked. With respect to the number of pulmonary tumors, 65
(82.28%) patients had a single tumor and 14 (17.72%) patients had
multiple tumors. Tumors of 28 (35.44%) cases were located in the
left lung, 49 (62.03%) cases in right lung and 2 (2.53%) cases had
bilateral tumors. Of all 79 patients included in the present study,
MIA occurred more frequently in females, patients who had never
smoked, patients aged 51–60 years and patients with a single tumor
located in the right lung (Table
I).
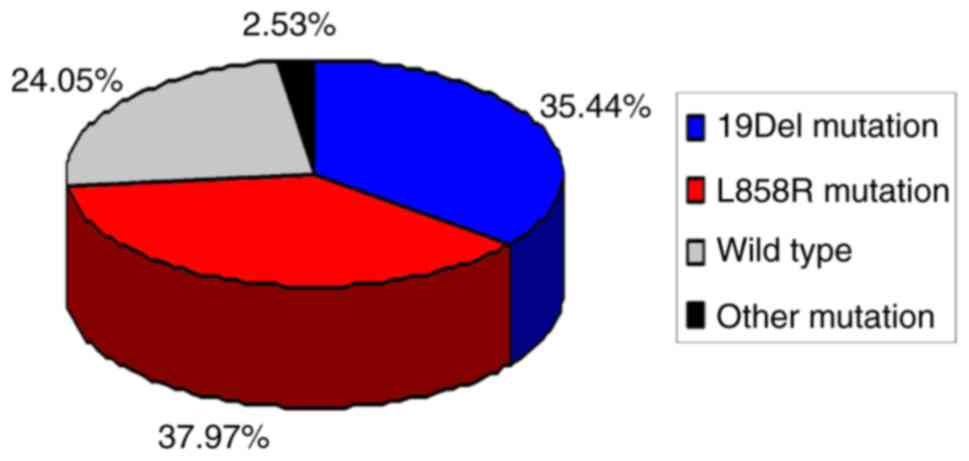
An EGFR mutation was identified in 60/79 patients
with MIA, with a 75.95% mutation rate. A total of 28 cases (35.44%)
had a deletion in exon 19 (19Del), 30 cases (37.97%) had a point
mutation in exon 21 (L858R) and 19 wild-type EGFR gene cases
(24.05%) were identified. The remaining 2 cases (2.53%) harbored
rare mutations, one of exon 18 G719X, and the other was a
concomitant double mutation of exon 18 G719X and exon 20 S768I. The
mutation spectrum of EGFR in the 79 patients with MIA is described
in Fig. 1.
Association between EGFR mutation
status and clinicopathological characteristics
As presented in Table
II, the distribution of EGFR mutations did not differ
significantly according to sex, age at diagnosis, smoking status or
tumor site/size/number in the 79 patients with MIA in the present
study. The maximum diameter of microinvasion, percentage of
lymphocytes in the peripheral blood and radiological appearance
with or without ground glass opacity (GGO) were not associated with
EGFR mutation. With respect to the histological subtype of
microinvasion component according to the IASLC/ATS/ERS
classification, the EGFR mutation occurred more frequently in
lepidic and acinar predominant subtypes (P<0.01). However, the
distribution of patients with EGFR was not significantly different
between these two histological subtypes. In addition, EGFR
mutations occurred more frequently in patients with intratumoral
fibrosis and intratumoral inflammatory cell infiltration
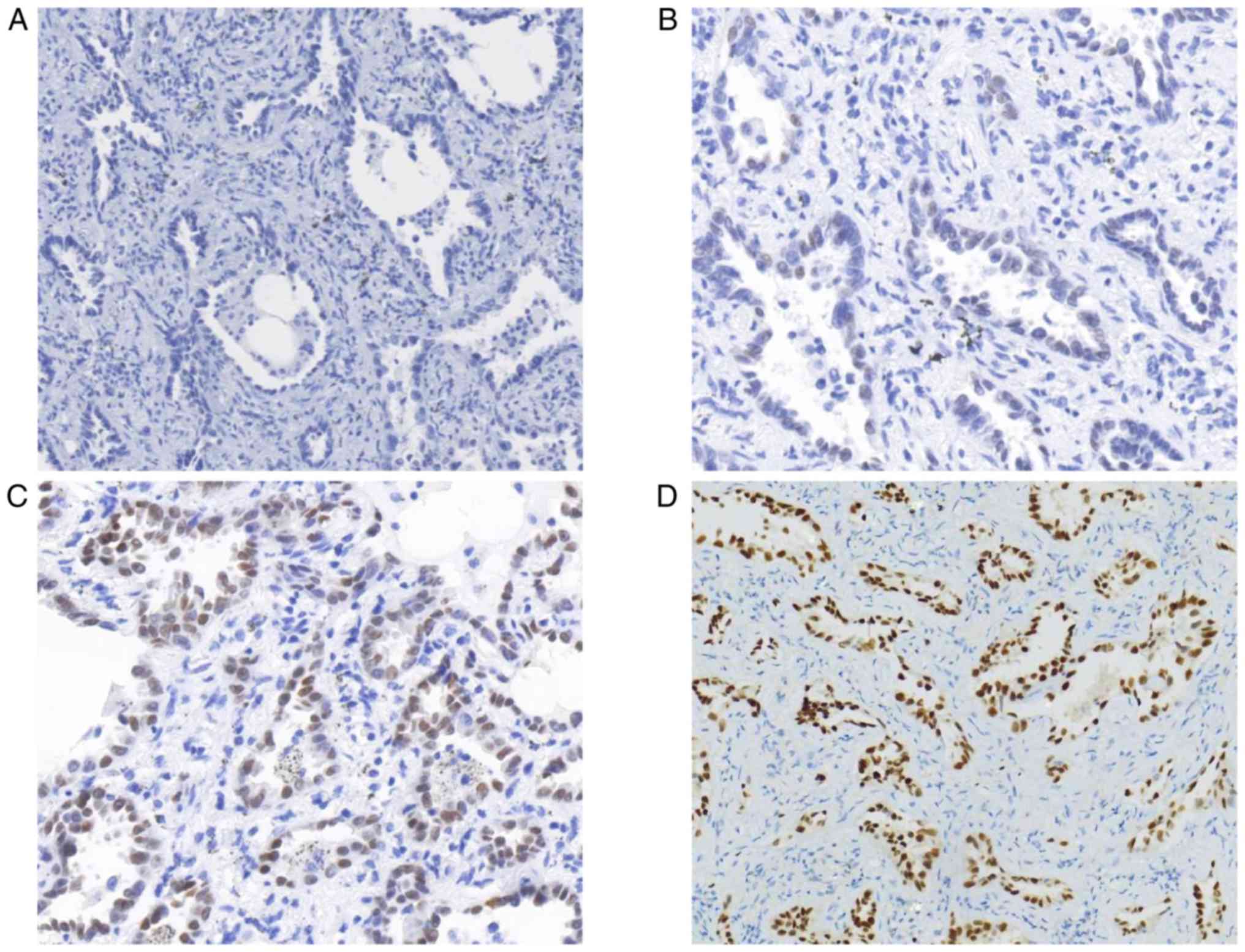
(P<0.01). On the other hand, the present study identified 63
cases (79.75%, defined as staining score ≥2) with MIA that were
TTF-1-positive and 16 cases (20.25%, defined as staining score
<2) that were negative (Fig. 2).
EGFR mutations occurred more frequently in MIA patients with TTF-1
expression (Table II;
P<0.01).
 | Table II.Association between the epidermal
growth factor receptor mutation status and clinicopathological
features. |
Table II.
Association between the epidermal
growth factor receptor mutation status and clinicopathological
features.
| Factor | n | Mutation (%) | P-value |
|---|
| Total cases | 79 | 60 (75.95) |
|
| Age |
|
| 0.303 |
|
≤50 | 16 | 13 (81.25) |
|
|
51–60 | 31 | 26 (83.87) |
|
|
61–70 | 21 | 13 (61.90) |
|
|
>70 | 11 | 8 (72.73) |
|
| Sex |
|
| 0.871 |
|
Male | 32 | 24 (75.00) |
|
|
Female | 47 | 36 (76.59) |
|
| Smoking status |
|
| 0.348 |
| Never
smoked | 71 | 55 (77.46) |
|
|
Smoker | 8 | 5 (62.50) |
|
| Tumor number |
|
| 0.260 |
|
Single | 65 | 51 (78.46) |
|
|
Multiple | 14 | 9 (64.28) |
|
| Tumor site |
|
| 0.495 |
|
Left | 28 | 20 (71.43) |
|
|
Right | 49 | 39 (79.59) |
|
|
Bilateral | 2 | 1 (50.00) |
|
| Tumor size, cm |
|
| 0.301 |
| ≤1 | 46 | 33 (71.74) |
|
| >1,
≤2 | 33 | 27 (81.82) |
|
| Presence of
GGO |
|
| 0.660 |
|
Yes | 73 | 55 (75.34) |
|
| No | 6 | 5 (83.33) |
|
| Maximum diameter of
tumor microinvasion, mm |
|
| 0.133 |
| ≤2 | 34 | 23 (67.65) |
|
| >2,
≤5 | 45 | 37 (82.22) |
|
| Histological
subtype of microinvasion component |
|
| 0.001a |
| Lepidic
predominant | 43 | 33 (76.74) |
|
| Acinar
predominant | 32 | 27 (84.38) |
|
| Other
sub-type | 4 | 0 (0.00) |
|
| Intratumoral
fibrosis |
|
| 0.007a |
|
Presence | 46 | 40 (86.96) |
|
|
Absence | 33 | 20 (60.61) |
|
| Intratumoral
inflammatory cell infiltration |
|
| 0.007a |
|
Presence | 38 | 34 (89.47) |
|
|
Absence | 41 | 26 (63.41) |
|
| Percentage of
lymphocytes in peripheral blood |
|
| 0.134 |
|
≤30 | 19 | 12 (63.16) |
|
|
>30 | 60 | 48 (80.00) |
|
| Expression of TTF-1
protein |
|
| 0.007a |
|
Positive | 63 | 52 (82.54) |
|
|
Negative | 16 | 8 (50.00) |
|
Comparison of clinicopathological
characteristics among the EGFR mutation subtypes
As shown in Table
III, subgroup analysis was conducted to analyze the association
between the EGFR mutation subtype and clinicopathological
parameters. The distribution of patients with 19Del and L858R in
the present study cohort was similar with respect to sex, age at
diagnosis, smoking status, tumor site and number, histological
subtype of the microinvasive component, percentage of lymphocytes
in the peripheral blood and radiological appearance with or without
GGO. Nevertheless, there was a significant association between the
19Del mutation status and tumor size (P<0.01), diameter of tumor
microinvasion (P<0.05), presence of intratumoral fibrosis
(P<0.05) and inflammatory cell infiltration (P<0.01). There
was a significant association between the L858R mutation and the
aforementioned features. Furthermore, the expression status of
TTF-1 protein was significantly associated with the L858R mutation
status (P<0.05), but not with the 19Del mutation (Table III).
 | Table III.Association between the two subtypes
of tyrosine kinase inhibitor-sensitive EGFR mutation and
clinicopathological features. |
Table III.
Association between the two subtypes
of tyrosine kinase inhibitor-sensitive EGFR mutation and
clinicopathological features.
|
|
| EGFR mutation
status |
|---|
|
|
|
|
|---|
| Factor | n | 19Del | P-value | L858R | P-value |
|---|
| Sex |
|
| 0.427 |
| 0.310 |
|
Male | 32 | 13 (40.63) |
| 10 (31.25) |
|
|
Female | 47 | 15 (31.91) |
| 20 (42.55) |
|
| Age |
|
| 0.062 |
| 0.902 |
|
≤50 | 16 | 8 (50.00) |
| 5 (31.25) |
|
|
51–60 | 31 | 14 (45.16) |
| 12 (38.71) |
|
|
61–70 | 21 | 5 (23.81) |
| 8 (38.10) |
|
|
>70 | 11 | 1 (9.09) |
| 5 (45.45) |
|
| Smoking status |
|
| 0.515 |
| 0.977 |
|
Smoked | 8 | 2 (25.00) |
| 3 (37.50) |
|
| Never
smoked | 71 | 26 (36.62) |
| 27 (38.03) |
|
| Presence of
GGO |
|
| 0.096 |
| 0.263 |
|
Presence | 73 | 24 (32.88) |
| 29 (39.73) |
|
|
Absence | 6 | 4 (66.66) |
| 1 (16.67) |
|
| Tumor number |
|
| 0.227 |
| 0.678 |
|
Single | 65 | 25 (38.46) |
| 24 (36.92) |
|
|
Multiple | 14 | 3 (21.43) |
| 6 (42.86) |
|
| Tumor site |
|
| 0.343 |
| 0.533 |
|
Left | 28 | 7 (25.00) |
| 11 (39.29) |
|
|
Right | 49 | 20 (40.82) |
| 19 (38.78) |
|
|
Bilateral | 2 | 1 (50.00) |
| 0 (0.00) |
|
| Tumor size, cm |
|
| 0.007a |
|
<0.0001a |
| ≤1 | 46 | 22 (47.83) |
| 9 (19.57) |
|
| >1,
≤2 | 33 | 6 (18.18) |
| 21 (63.64) |
|
| Maximum diameter of
tumor microinvasion, mm |
|
| 0.019b |
| 0.001a |
| ≤2 | 34 | 17 (50.00) |
| 6 (17.65) |
|
| >2,
≤5 | 45 | 11 (24.44) |
| 24 (53.33) |
|
| Histological
sub-type of microinvasion component |
|
| 0.112 |
| 0.084 |
| Lepidic
predominant | 43 | 19 (44.19) |
| 14 (32.56) |
|
| Acinar
predominant | 32 | 9 (28.13) |
| 16 (50.00) |
|
| Other
subtype | 4 | 0 (0.00) |
| 0 (0.00) |
|
| Intratumoral
fibrosis |
|
| 0.011b |
|
<0.0001a |
|
Presence | 46 | 11 (23.91) |
| 27 (58.69) |
|
|
Absence | 33 | 17 (51.52) |
| 3 (9.09) |
|
| Intratumoral
inflammatory cells infiltration |
|
| 0.002a |
|
<0.0001a |
|
Presence | 38 | 7 (18.42) |
| 25 (65.79) |
|
|
Absence | 41 | 21 (51.22) |
| 5 (12.20) |
|
| Percentage of
lymphocytes in peripheral blood |
|
| 0.686 |
| 0.510 |
|
≤30 | 19 | 6 (31.58) |
| 6 (31.58) |
|
|
>30 | 60 | 22 (36.67) |
| 24 (40.00) |
|
| TTF-1 |
|
| 0.847 |
| 0.030b |
|
Positive | 63 | 22 (34.92) |
| 29 (46.03) |
|
|
Negative | 16 | 6 (37.50) |
| 1 (6.25) |
|
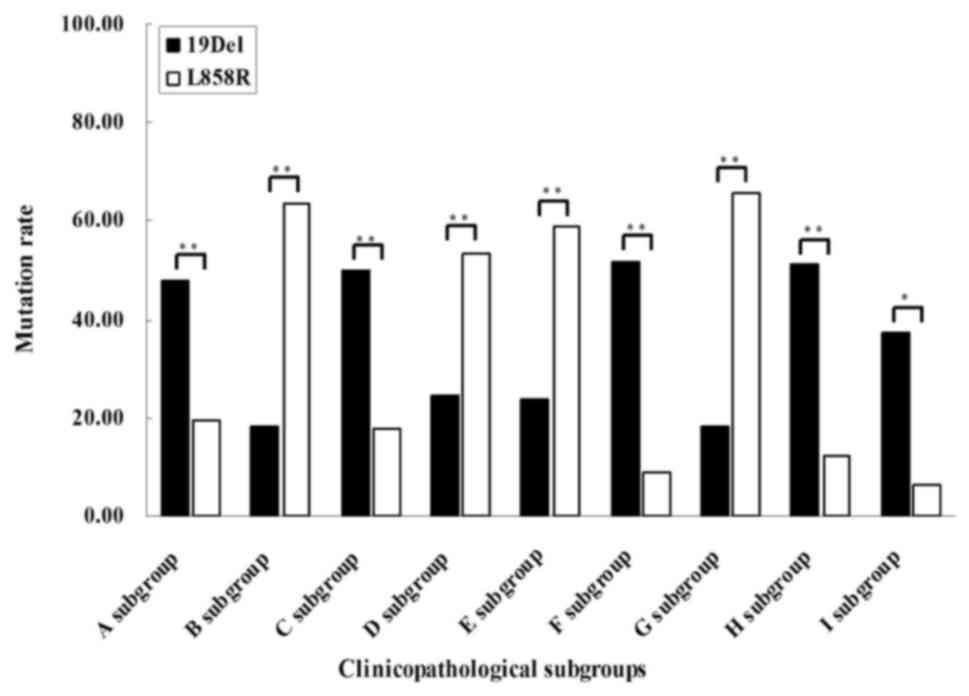
As demonstrated in Fig.
3 and Table IV, the present
study also demonstrated that 19Del occurred more frequently in
patients with MIA exhibiting small tumor size (P=0.004) and smaller
diameter of microinvasive area (P=0.005). 19Del was also observed
more frequently in patients without intratumoral fibrosis
(P=0.000), inflammatory cell infiltration (P<0.001) and TTF-1
expression (P=0.033). By contrast, the subgroup analysis revealed
that the L858R mutation was more frequently detected in patients
with MIA with larger tumors (P<0.001), larger diameters of
microinvasive area (P=0.005), intratumoral fibrosis (P=0.001) and
inflammatory cell infiltration (P<0.001). Furthermore,
significant differences were not observed between the 19Del and
L858R subtypes with regard to other clinicopathological feature
subgroups.
 | Table IV.Comparison of clinicopathological
characteristics between the 19Del and L858R EGFR status. |
Table IV.
Comparison of clinicopathological
characteristics between the 19Del and L858R EGFR status.
|
| A sub-group
(n=46) |
| B sub-group
(n=33) |
| C sub-group
(n=34) |
|
|---|
|
|
|
|
|
|
|
|
|---|
| EGFR mutation
subtype | − | + | P-value | − | + | P-value | − | + | P-value |
|---|
| 19Del | 24 | 22 | 0.004a | 27 | 6 |
<0.0001a | 17 | 17 | 0.005a |
| L858R | 37 | 9 |
| 12 | 21 |
| 28 | 6 |
|
|
|
| D sub-group
(n=45) |
| E sub-group
(n=46) |
| F sub-group
(n=33) |
|
|
|
|
|
|
|
|
|
|
| − | + | P-value | − | + | P-value | − | + | P-value |
|
| 19Del | 34 | 11 | 0.005a | 35 | 11 | 0.001a | 16 | 17 |
<0.0001a |
| L858R | 21 | 24 |
| 19 | 27 |
| 30 | 3 |
|
|
|
| G sub-group
(n=38) |
| H sub-group
(n=41) |
| I sub-group
(n=16) |
|
|
|
|
|
|
|
|
|
|
| − | + | P-value | − | + | P-value | − | + | P-value |
|
| 19Del | 31 | 7 |
<0.0001a | 20 | 21 |
<0.0001a | 10 | 6 | 0.033b |
| L858R | 13 | 25 |
| 36 | 5 |
| 15 | 1 |
|
Discussion
The present study aimed to improve the understanding
of the EGFR mutation features, and their clinicopathological
relevance, in MIA. MIA is a newly defined subtype of lung
adenocarcinoma with distinctive clinicopathological features
(21). MIA is a very common type of
stage I lung cancer and its prognosis remains controversial. The
present study retrospectively detected EGFR mutations in patients
with MIA from a Chinese population and evaluated the association
between the mutation status and clinicopathological
characteristics. The results indicated that lung MIA occurred more
frequently in females, patients who had never smoked, patients aged
51–60 years and patients with a single tumor in the right lung. The
results of the present study revealed the clinical characteristics
of MIA in a relatively large cohort. Previous studies have
suggested that EGFR mutations are more frequently observed (40–60%)
in lung adenocarcinomas of Asian patients (22–26). It
has been revealed that the EGFR mutations are significantly
associated with MIA and more frequently detected in Japanese
patients with MIA (14,15). To the best of our knowledge, the
present study was the first to evaluate EGFR mutation status in a
relatively large cohort of surgically resected lung MIAs from the
Chinese population. The results indicated that the EGFR mutation
rate was 75.95% in MIA, which was increased compared with the
previously reported prevalence. The results of the present study
also revealed that 19Del and L858R are the two dominant mutation
subtypes, which was consistent with a previous report regarding
patients with IA (27,28). Additionally, a rare single mutation
(exon 18 G719X) and a rare concomitant mutation (exon 18 G719X and
exon 20 S768I) were observed in the present cohort. According to
the EGFR mutation features in the present study, it was concluded
that MIA may harbor specific molecular characteristics, and further
large-scale studies are required to confirm this.
EGFR is a member of the ErbB/HER family of
transmembrane receptor tyrosine kinases expressed in several human
malignancies, including lung cancer (29–31).
EGFR-associated signaling pathways can be deregulated through
different mechanisms, the most important of which is EGFR mutation
(32). The most common EGFR mutations
are 19Del and L858R, which are known as activating EGFR mutations
in NSCLC (33). Activating EGFR
mutations may result in constitutive activation of the receptor and
diverse downstream signaling pathways, independent of ligand
binding (34,35). The constitutive activation of
EGFR-associated signaling pathways elicited by EGFR mutations are
thought to be a significant contributor to the tumorigenesis of
NSCLC. Numerous TKIs have been developed to inhibit
EGFR-sensitizing mutations for the molecular-targeted therapy of
NSCLC (36,37). EGFR mutations may be predictive
biomarkers of EGFR TKI responsiveness (38–43).
Previous studies have focused on investigating the applications of
EGFR TKIs in early resectable NSCLC, and several research groups
have concluded its feasibility and safety (44–47). A
number of ongoing studies have aimed to provide further evidence
for guiding the extended application of targeted therapy for
advanced and early stage lung cancer. Therefore, a complete
understanding of EGFR mutation features and the potential clinical
significance in MIA may provide additional information regarding
the clinical management of early-stage lung cancer. However, due to
the small tumor size in patients with MIA, the quantity and quality
of tumor samples is not always sufficient for the purpose of
mutation analysis in clinical practice. It would be beneficial to
identify efficient alternative indicators of EGFR mutation status
prior to testing. Therefore, the present study evaluated the
association between the EGFR mutation and various
clinicopathological features in MIA (Table II). Analysis of the unselected group
of patients suggested no association between the EGFR mutation and
sex, age at diagnosis, smoking history, or tumor site/size/number
in the 79 patients with MIA. With respect to IA, previous reports
have demonstrated that EGFR mutations are commonly observed in
females and patients who have never smoked (48,49). The
results of the present study for MIA were not consistent with those
of previous studies of IA. However, a study by Lai et al
(28) also revealed that there was no
significant association between EGFR mutation subtype and sex,
smoking history or tumor histology in IA. The discrepancy may be
caused by the intrinsic molecular characteristics of MIA or could
be explained by variations between selected and unselected tumor
stages or sampling error. Further studies are required to shed
light on these discrepancies and their underlying causes. On the
other hand, the results of the present study suggested that EGFR
mutations were more frequently observed in lepidic and acinar
predominant microinvasive component subtypes of MIA, which was
consistent with the previously obtained results for IA (44,50).
In addition, the results of the present study
indicated that EGFR mutations were significantly associated with
TTF-1 expression in MIA. Previous studies have suggested a
significant association between EGFR mutation and TTF-1 protein
expression in advanced lung adenocarcinoma (51–53),
particularly for exon 21 mutations (54). It was concluded that TTF-1 may be
regarded not only as a significant marker for the diagnosis of lung
adenocarcinoma, but also as useful guidance regarding EGFR mutation
status prior to molecular testing. Furthermore, previous data
revealed the potential interaction signal between TTF-1 and EGFR in
lung adenocarcinoma (55). It can be
hypothesized that the interactivity between TTF-1 expression and
EGFR mutation may serve key roles in the initiation of lung
adenocarcinoma. Therefore, further studies are required to
investigate this interaction in lung adenocarcinoma, particularly
in early stage tumors. With respect to the expression of TTF-1 in
MIA, the present study identified 16 patients with MIA who were
TTF-1-negative (Fig. 2). Previous
studies had reported several TTF-1-negative patients with MIA in
their cohorts (18,56). The exact expression profile of TTF-1
and the associated significance requires further investigation in
patients with MIA. The results of the present study suggested that
the EGFR mutation occurred more frequently in patients with MIA
with intratumoral fibrosis and inflammatory cell infiltration. To
the best of our knowledge, the association between these two
pathological features and the EGFR mutation status has not been
previously revealed.
The present study concluded that intratumoral
fibrosis and inflammatory cell infiltration could be regarded as
alternative indicators for the identification of EGFR mutations in
patients with MIA, or even IA. Previous studies have also indicated
that tumor cell proliferation and invasiveness could be affected by
alterations in the tumor microenvironment, including intratumoral
fibrosis and inflammatory cell infiltration (57,58). Based
on the results of the present study, we hypothesize an association
between the clinical outcome of MIA and EGFR mutation status.
Further studies are required to validate this hypothesis.
The present study conducted subgroup analysis
(Table III), which suggested that
19Del and L858R mutations were associated with pathological
features, including tumor size, diameter of tumor microinvasion,
intratumoral fibrosis and inflammatory cell infiltration. The
differential results between the group and subgroup analyses
suggested that lung MIA harboring different EGFR mutation subtypes
may exhibit distinctive clinicopathological characteristics. In
addition, the results of the present study suggested that TTF-1w
expressionwwaswwsignificantlyw associated with the L858R mutation,
but not with the 19Del mutation. Taken together, the present study
indicated that it is meaningful to consider MIA as a group of
different subsets based on the EGFR mutation subtype.
The present study subsequently conducted a
stratification analysis regarding the association of 19Del and
L858R, respectively with certain clinicopathological features
(Fig. 3; Table IV). The present data indicated that
the 19Del mutation was more frequently detected in MIA with the
following features: Smaller tumor size, smaller area of
microinvasion, no intratumoral fibrosis, inflammatory cell
infiltration and TTF-1 expression. By contrast, the L858R mutation
is more frequently observed in MIA with entirely different
characteristics compared with 19Del, including larger tumor size,
larger area of microinvasion, presence of intratumoral fibrosis and
inflammatory cell infiltration. Previous studies have consistently
demonstrated that 19Del and L858R mutations show different
prognostic and predictive roles in IA (5,59). Data
from these studies suggested that lung IA harboring 19Del and L858R
should be regarded as different diseases. However, there have been
no studies evaluating the differences between the 19Del and L858R
subgroups in surgically resected MIA. The present results revealed
that there were significant differences in tumor
microenvironment-associated histological characteristics, including
intratumoral fibrosis and inflammatory cell infiltration, between
MIA cases with 19Del and L858R mutations. Previous studies proposed
that these two histological characteristics could be considered as
prognostic markers for a number of tumor types (57,58).
Considering these data, it can be hypothesized that clinical
outcomes of MIA may be affected by EGFR mutation subtype. The
prognosis of MIA remains unclear (60). It has been reported that EGFR mutation
status has no effect on the prognosis of patients with early stage
lung adenocarcinoma (61). However,
the present data provide preliminary evidence to support
consideration of EGFR mutation status as indication of prognostic
stratification in future clinical studies involving MIA. Follow-up
data are being collected and outcome analysis will be investigated
in our future work. Furthermore, an increased number of MIA cases
will also be included the future studies.
In conclusion, to the best of our knowledge, the
present study was the first to reveal that surgically resected MIA
tissues with different EGFR gene statuses exhibit distinct
clinicopathological features. The results further suggest that
there were significant differences in tumor
microenvironment-associated histological characteristics between
MIA with 19Del and L858R. Therefore, the present study concluded
that the clinical outcomes of MIA may be affected by EGFR mutation
subtypes and that EGFR mutation analysis should be considered for
prognostic evaluation and clinical management of MIA.
Acknowledgements
The authors would like to thank Dr Bo Meng and Dr
Jingjing Chen (Department of Pathology, Anhui Provincial Hospital)
for providing technical assistance.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no.
81602605).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ML conceived and designed the experiments. ML and
MLZ performed the experiments. LK, CYL and MC analyzed the data.
ML, LK and MC contributed reagents/materials/analysis tools. ML,
CYL and MLZ performed clinicopathological assessment. ML and CYL
wrote the manuscript. The final version of the manuscript was read
and approved by all authors.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Anhui Provincial Hospital, The First Affiliated
Hospital of University of Science and Technology of China
(reference no. 20160183), and written informed consent was obtained
from all participants.
Patient consent for publication
Written informed consent for the publication of the
clinical details was obtained from the patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
She J, Yang P, Hong Q and Bai C: Lung
cancer in China: Challenges and interventions. Chest.
143:1117–1126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 world health organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim
Biophys Acta. 1856:189–210. 2015.PubMed/NCBI
|
|
5
|
Rosell R, Moran T, Queralt C, Porta R,
Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M,
et al: Screening for epidermal growth factor receptor mutations in
lung cancer. N Engl J Med. 361:958–967. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang SF, Liu HP, Li LH, Ku YC, Fu YN,
Tsai HY, Chen YT, Lin YF, Chang WC, Kuo HP, et al: High frequency
of epidermal growth factor receptor mutations with complex patterns
in non-small cell lung cancers related to gefitinib responsiveness
in Taiwan. Clin Cancer Res. 10:8195–8203. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pao W and Miller VA: Epidermal growth
factor receptor mutations, small-molecule kinase inhibitors, and
non-small-cell lung cancer: Current knowledge and future
directions. J Clin Oncol. 23:2556–2568. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Giaccone G and Rodriguez JA: EGFR
inhibitors: What have we learned from the treatment of lung cancer?
Nat Clin Pract Oncol. 2:554–561. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang
Y, Li W, Hou M, Shi JH, Lee KY, et al: Afatinib versus cisplatin
plus gemcitabine for first-line treatment of Asian patients with
advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-
Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol.
15:213–222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sequist LV, Yang JC, Yamamoto N, O'Byrne
K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, et al:
Phase III study of afatinib or cisplatin plus pemetrexed in
patients with metastatic lung adenocarcinoma with EGFR mutations. J
Clin Oncol. 31:3327–3334. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Inoue A, Kobayashi K, Maemondo M, Suqawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Updated overall survival results from a randomized phase III
trial comparing gefitinib with carboplatin-paclitaxel for
chemo-naïve non-small cell lung cancer with sensitive EGFR gene
mutations (NEJ002). Ann Oncol. 24:54–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H and Hirashima T:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small cell lung cancer harboring mutations of the epidermal
growth factor receptor (WJTOG3405): An open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao W, Wang H, Xie J and Tian B: A
clinicopathological study of small lung adenocarcinoma 1 cm or less
in size: Emphasis on histological subtypes associated with lymph
node metastasis and recurrence. Int J Surg Pathol. 26:4–11. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoshizawa A, Sumiyoshi S, Sonobe M,
Kobayashi M, Fujimoto M, Kawakami F, Tsuruyama T, Travis WD, Date H
and Haga H: Validation of the IASLC/ATS/ERS lung adenocarcinoma
classification for prognosis and association with EGFR and KRAS
gene mutations: Analysis of 440 Japanese patients. J Thorac Oncol.
8:52–61. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yanagawa N, Shiono S, Abiko M, Ogata SY,
Sato T and Tamura G: The correlation of the international
association for the study of lung cancer (IASLC)/American thoracic
society (ATS)/European respiratory society (ERS) classification
with prognosis and EGFR mutation in lung adenocarcinoma. Ann Thorac
Surg. 98:453–458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Travis WD, Brambilla E, Burke AP, Marx A
and Nicholson AG: WHO classification of tumors of lung, pleural,
thymus and heart. J Thorac Oncol. 10:1240–1242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G,
Garg K, et al: International association for the study of lung
cancer/American thoracic society/European respiratory society:
International multidisciplinary classification of lung
adenocarcinoma: Executive summary. Proc Am Thorac Soc. 8:381–385.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou C, Zhao J, Shao J and Li W:
Prognostic relevance of TTF-1 expression in stage I adenocarcinoma.
Oncotarget. 8:107462–107468. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kadota K, Nitadori J, Sarkaria IS, Sima
CS, Jia X, Yoshizawa A, Rusch VW, Travis WD and Adusumilli PS:
Thyroid transcription factor-1 expression is an independent
predictor of recurrence and correlates with the IASLC/ATS/ERS
histologic classification in patients with stage I lung
adenocarcinoma. Cancer. 119:931–938. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barletta JA, Perner S, Iafrate AJ, Yeap
BY, Weir BA, Johnson LA, Johnson BE, Meyerson M, Rubin MA, Travis
WD, et al: Clinical significance of TTF-1 protein expression and
TTF-1 gene amplification in lung adenocarcinoma. J Cell Mol Med.
13:1977–1986. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G,
Garg K, et al: International association for the study of lung
cancer/American thoracic society/European respiratory society
international multidisciplinary classificatiosn of lung
adenocarcinoma. J Thorac Oncol. 6:244–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shigematsu H, Lin L, Takahashi T, Nomura
M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, et
al: Clinical and biological features associated with epidermal
growth factor receptor gene mutations in lung cancers. J Natl
Cancer Inst. 97:339–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fridlender ZG, Sun J, Kim S, Kapoor V,
Cheng G, Ling L, Worthen GS and Albelda SM: Polarization of
tumor-associated neutrophil phenotype by TGF-beta: ‘N1’ versus ‘N2’
TAN. Cancer Cell. 16:183–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cedres S, Torrejon D, Martinez A, Martinez
P, Navarro A, Zamora E, Mulet-Margalef N and Felip E: Neutrophil to
lymphocyte ratio (NLR) as an indicator of poor prognosis in stage
IV non-small cell lung cancer. Clin Transl Oncol. 14:864–869. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
D'Angelo SP, Janjiqian YY, Ahye N, Riely
GJ, Chaft JE, Sima CS, Shen R, Zheng J, Dycoco J, Kris MG, et al:
Distinct clinical course of EGFR-mutant resected lung cancers:
Results of testing of 1118 surgical specimens and effects of
adjuvant gefitinib and erlotinib. J Thorac Oncol. 7:1815–1822.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshizawa A, Motoi N, Riely GJ, Sima CS,
Gerald WL, Kris MG, Park BJ, Rusch VW and Travis WD: Impact of
proposed IASLC/ATS/ERS classification of lung adenocarcinoma:
Prognostic subgroups and implications for further revision of
staging based on analysis of 514 stage I cases. Mod Pathol.
24:653–664. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ning H, Liu M, Wang L, Yang Y, Song N, Xu
X, Ju J and Jiang G: Clinicopathological features of Chinese lung
cancer patients with epidermal growth factor receptor mutation. J
Thorac Dis. 9:796–801. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lai Y, Zhang Z, Li J, Sun D, Zhou Y, Jiang
T, Han Y, Huang L, Zhu Y, Li X and Yan X: EGFR mutations in
surgically resected fresh specimens from 697 consecutive Chinese
patients with non-small cell lung cancer and their relationships
with clinical features. Int J Mol Sci. 14:24549–24559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mendelsohn J and Baselga J: Epidermal
growth factor receptor targeting in cancer. Semin Oncol.
33:369–385. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ciardiello F and Tortora G: EGFR
antagonists in cancer treatment. N Engl J Med. 358:1160–1174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thakur MK and Gadgeel SM: Predictive and
prognostic biomarkers in non-small cell lung cancer. Semin Respir
Crit Care Med. 37:760–770. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gazdar AF and Minna JD: Deregulated EGFR
signaling during lung cancer progression: Mutations, amplicons, and
autocrine loops. Cancer Prev Res (Phila). 1:156–160. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shroff GS, de Groot PM,
Papadimitrakopoulou VA, Truong MT and Carter BW: Targeted therapy
and immunotherapy in the treatment of non-small cell lung cancer.
Radiol Clin North Am. 56:485–495. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ciuffreda L, Incani UC, Steelman LS,
Abrams SL, Falcone I, Curatolo AD, Chappell WH, Franklin RA, Vari
S, Cognetti F, et al: Signaling intermediates (MAPK and PI3K) as
therapeutic targets in NSCLC. Curr Pharm Des. 20:3944–3957. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luo J, Solimini NL and Elledge SJ:
Principles of cancer therapy: Oncogene and non-oncogene addiction.
Cell. 136:823–837. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cooper WA, O'toole S, Boyer M, Horvath L
and Mahar A: What's new in non-small cell lung cancer for
pathologists: The importance of accurate subtyping, EGFR mutations
and ALK rearrangements. Pathology. 43:103–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gadgeel SM, Ramalingam SS and Kalemkerian
GP: Treatment of lung cancer. Radiol Clin North Am. 50:961–974.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu YL, Zhou C, Liam CK, Wu G, Liu X, Zhong
Z, Lu S, Cheng Y, Han B, Chen L, et al: First-line erlotinib versus
gemcitabine/cisplatin in patients with advanced EGFR
mutation-positive non-small-cell lung cancer: Analyses from the
phase III, randomized, open-label, ENSURE study. Ann Oncol.
26:1883–1889. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lungcancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han JY, Park K, Kim SW, Lee DH, Kim HY,
Kim HT, Ahn MJ, Yun T, Ahn JS, Suh C, et al: First-SIGNAL:
First-line single-agent iressa versus gemcitabine and cisplatin
trial in never-smokers with adenocarcinoma of the lung. J Clin
Oncol. 30:1122–1128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Boolell V, Alamgeer M, Watkins DN and
Ganju V: The evolution of therapies in non-small cell lung cancer.
Cancers (Basel). 7:1815–1846. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ryska A, Dziadziuszko R, Olszewski W,
Berzinec P, Oz B, Gottfried M, Cufer T, Samarzija M, Plank L,
Ostoros G and Timar J: Molecular diagnostics of lung cancer. Magy
Onkol. 59:259–266. 2015.PubMed/NCBI
|
|
44
|
Janjigian YY, Park BJ, Zakowski MF,
Ladanyi M, Pao W, D'Angelo SP, Kris MG, Shen R, Zheng J and Azzoli
CG: Impact on disease-free survival of adjuvant erlotinib or
gefitinib in patients with resected lung adenocarcinomas that
harbor EGFR mutations. J Thorac Oncol. 6:569–575. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Q, Wang H, Li P, Zhu H, He C, Wei B,
Ma J and Ma Z: Erlotinib-based perioperative adjuvant therapy for a
case of unresectable stage IIIA (N2) nonsmall cell lung cancer. Am
J Med Sci. 340:321–325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lara-Guerra H, Chung CT, Schwock J,
Pintilie M, Hwang DM, Leighl NB, Waddell TK and Tsao MS:
Histopathological and immunohistochemical features associated with
clinical response to neoadjuvant gefitinib therapy in early stage
non-small cell lung cancer. Lung Cancer. 76:235–241. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schaake EE, Kappers I, Codrington HE,
Valdes Olmos RA, Teertstra HJ, van Pel R, Burgers JA, van Tinteren
H and Klomp HM: Tumor response and toxicity of neoadjuvant
erlotinib in patients with early-stage non-small-cell lung cancer.
J Clin Oncol. 30:2731–2738. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tokumo M, Toyooka S, Kiura K, Shigematsu
H, Tomii K, Aoe M, Ichimura K, Tsuda T, Yano M, Tsukuda K, et al:
The relationship between epidermal growth factor receptor mutations
and clinicopathologic features in non-small cell lung cancers. Clin
Cancer Res. 11:1167–1173. 2005.PubMed/NCBI
|
|
49
|
Sonobe M, Manabe T, Wada H and Tanaka F:
Mutations in the epidermal growth factor receptor gene are linked
to smoking-independent, lung adenocarcinoma. Br J Cancer.
93:355–363. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Usuda K, Sagawa M, Motono N, Ueno M,
Tanaka M, Machida Y, Matoba M, Taniguchi M, Tonami H, Ueda Y and
Sakuma T: Relationships between EGFR mutation status of lung cancer
and preoperative factors-Are they predictive? Asian Pac J Cancer
Prev. 15:657–662. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chung KP, Huang YT, Chang YL, Yu CJ, Yang
CH, Chang YC, Shih JY and Yang PC: Clinical significance of thyroid
transcription factor-1 in advanced lung adenocarcinoma under
epidermal growth factor receptor tyrosine kinase inhibitor
treatment. Chest. 141:420–428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang Y, Wang R, Li Y, Pan Y, Hu H, Zhang
Y, Li H, Shen L, Yu Y, Sun Y and Chen H: Negative thyroid
transcription factor 1 expression defines an unfavorable subgroup
of lung adenocarcinomas. J Thorac Oncol. 10:1444–1450. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sun PL, Seol H, Lee HJ, Yoo SB, Kim H, Xu
X, Jheon S, Lee CT, Lee JS and Chung JH: High incidence of EGFR
mutations in Korean men smokers with no intratumoral heterogeneity
of lung adenocarcinomas: Correlation with histologic subtypes,
EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol.
7:323–330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shanzhi W, Yiping H, Ling H, Jianming Z
and Qiang L: The relationship between TTF-1 expression and EGFR
mutations in lung adenocarcinomas. PLoS One. 9:e954792014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yamaguchi T, Yanagisawa K, Sugiyama R,
Hosono Y, Shimada Y, Arima C, Kato S, Tomida S, Suzuki M, Osada H
and Takahashi T: NKX2-1/ TITF1/ TTF-1-induced ROR1 is required to
sustain EGFR survival signaling in lung adenocarcinoma. Cancer
Cell. 21:348–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang TW, Lin KF, Lee CH, Chang H, Lee SC
and Shieh YS: The role of thyroid transcription factor-1 and tumor
differentiation in resected lung adenocarcinoma. Sci Rep.
7:142222017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lee HO, Mullins SR, Franco-Barraza J,
Valianou M, Cukierman E and Cheng JD: FAP-overexpressing
fibroblasts produce an extracellular matrix that enhances invasive
velocity and directionality of pancreatic cancer cells. BMC Cancer.
11:2452011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Brambilla E, Le Teuff G, Marguet S,
Lantuejoul S, Dunant A, Graziano S, Pirker R, Douillard JY, Le
Chevalier T, Filipits M, et al: Prognostic effect of tumor
lymphocytic infiltration in resectable non-small-cell lung cancer.
J Clin Oncol. 34:1223–1230. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang JC, Wu YL, Schuler M, Sebastian M,
Popat S, Yamamoto N, Zhou C, Hu CP, O Byrne K, Feng J, et al:
Afatinib versus cisplatin-based chemotherapy for EGFR
mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6):
Analysis of overall survival data from two randomized, phase 3
trials. Lancet Oncol. 16:141–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kadota K, Villena-Vargas J, Yoshizawa A,
Motoi N, Sima CS, Riely GJ, Rusch VW, Adusumilli PS and Travis WD:
Prognostic significance of adenocarcinoma in situ, minimally
invasive adenocarcinoma, and nonmucinous lepidic predominant
invasive adenocarcinoma of the lung in patients with stage I
disease. Am J Surg Pathol. 38:448–460. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kaseda K, Asakura K, Kazama A and Ozawa Y:
Clinicopathological and prognostic features of surgically resected
pathological stage I lung adenocarcinoma harboring epidermal growth
factor receptor and K-ras mutation. Thorac Cancer. 8:229–237. 2017.
View Article : Google Scholar : PubMed/NCBI
|