Introduction
Inhibitor of growth family 5 (ING5) is a member of
the ING protein family and functions as a type-II tumor suppressor
gene. Human ING5 is mapped to chromosome 2q37.3 and encodes a
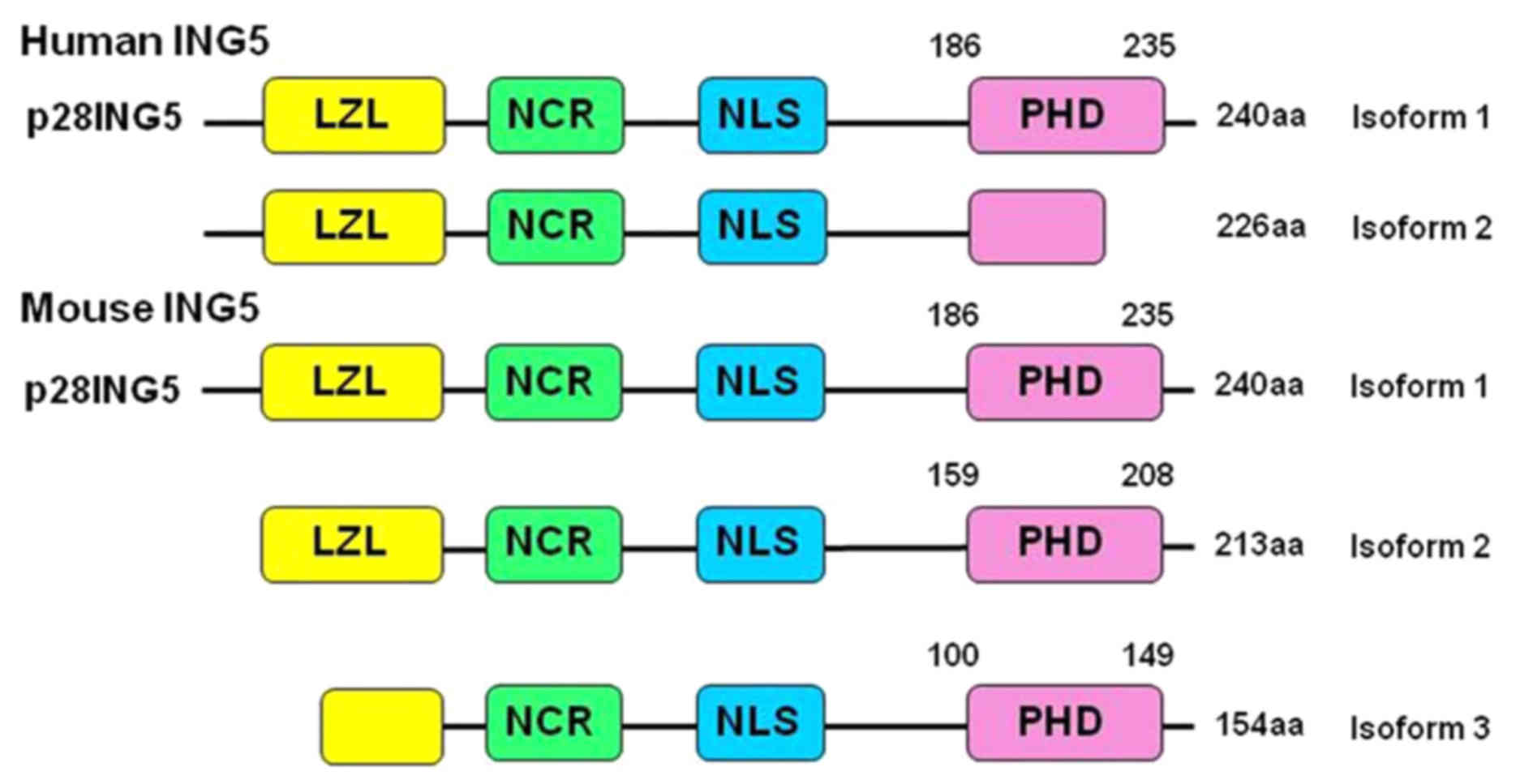
28-kDa protein with 240 amino acids (1). As indicated in Fig. 1, ING5 protein consists of a number of
domains, including leucine zipper like (LZL), novel conserved
region (NCR), nuclear localization signal (NLS) and plant
homeodomain (PHD). Among them, the LZL domain is important in DNA
repair, apoptotic induction and chromatin remodeling, whereas the
NCR domain can bind to histone acetyl transferase (HAT) complexes
during chromatin remodeling and the regulation of gene expression
(2,3).
ING5 interacts with histone H3K4me3 and is involved in the
formation of HAT complexes (ING5-histone acetyltransferase
KAT7-JADE and ING5-monocytic leukemia zinc finger
protein-MOZ-related factor-bromodomain-PHD finger protein (1,2).
ING5 protein is able to bind to mini-chromosome
maintenance proteins, which is important for DNA replication via
the formation of a pre-replicative complex (3). ING5 was reported to activate the
cyclin-dependent kinase inhibitor p21/cyclin dependent kinase
inhibitor 1A promoter to induce its expression, promote the
acetylation of p53 at Lys-382 residues and interact with the p300
protein of HAT complexes (2–6). ING5 is a cofactor of Tip60 for the
acetylation of p53 at K120 in response to DNA damage (7). Previously, reduced ING5 expression was
detected in pancreatic carcinoma cells transfected with a microRNA
(miR)-196a precursor, accompanied by decreased apoptosis, increased
invasion and proliferation compared with control cells (8). Additionally, low-level laser irradiation
treatment was able to induce the pro-proliferation effects of
miR-193 on bone mesenchymal stem cells, which is mediated via an
ING5 inhibitor (9).
In the RKO colorectal cancer cell line, ING5
overexpression results in decreased colony-forming efficiency,
decreased cell population in the S phase and p53-dependent
apoptosis induction (10,11). In a previous study, the loss of
heterozygosity on the long arm of chromosome 2, where the
ING5 gene is located, was detected in 85% (33/39) of oral
carcinoma cases. Reduced ING5 mRNA expression in 61% of oral
squamous cell carcinoma cases with missense mutations located
within the LZL finger and NCR domains of the ING5 protein has also
been reported (12). In head and neck
squamous cell carcinoma (HNSCC), nuclear ING5 may modulate the
transactivation of target genes, and promote apoptosis and cell
cycle arrest by interacting with p300 and p21 (13,14).
Additionally, two truncated fragments of ING5 (aa 1–184 and aa
107–226) are able to induce cellular senescence via the
downregulated expression of cyclin E and cyclin dependent kinase 2
(13). Reduced nuclear expression and
cytoplasmic translocation of ING5 was observed in HNSCC, gastric
and colorectal carcinoma tumorigenesis (14–16).
Identification of tissues or cell types which
express ING5 will contribute towards the elucidation of its
physiological function. Additionally, the clarification of the
expression pattern of ING5 and heterogeneity between tumor cases
will contribute to the development of target gene therapy and
conditional animal knockout models of ING5. In the present study,
an intermittent microwave irradiation for immunohistochemistry of
ING5 was employed, where the microwave irradiation causes minute
vibrations (>2.4 billion times/sec) and increases the
probability of specific antibody-antigen reactions (17). The expression profiling of ING5
protein has been investigated in normal mouse and human tissues, as
well as in human cancer tissues.
Materials and methods
Amino acid sequence alignment
Amino acid sequences of human and mouse ING5 were
obtained from GenBank (18),
including their isoforms. These sequences were aligned using
Genetyx 7 from Genetyx Corporation (Tokyo, Japan).
Tissue specimens and tissue
microarray
Written informed consent was obtained for the use of
tumor tissues (n=986) for clinical research, and ethical approval
was obtained from the Ethical and Animal Experimentation committees
at Jinzhou Medical University (Jinzhou, China).
C57BL/6 mice (3 males and 3 females; 8 weeks old)
were purchased from Beijing HFK Bioscience Co., Ltd. (Beijing,
China) and housed in pathogen-free conditions in a
temperature-controlled animal room with a 12-h light/dark
illumination cycle. All had ad libitum access to standard
rodent food and water. They were sacrificed under sodium
pentobarbital anesthesia, and the resected samples included brain,
heart, liver, spleen, lung, kidney, breast, stomach and intestine.
All tissues were fixed in 10% neutral formalin, embedded in
paraffin and cut into 4 mm sections. The tissue arrays of human
normal tissues (cerebrum, cerebellum, brain stem, aorta, tongue,
thyroid, esophagus, stomach, intestine, liver, pancreas, lung,
trachea, appendix, smooth muscle, skeletal muscle, heart, testis,
bladder and prostate) and cancer tissues [hepatocellular carcinoma
(n=62), renal clear cell carcinoma (n=62), pancreatic carcinoma
(n=62), esophageal squamous cell carcinoma (n=45) and cervical
squamous cell carcinoma (n=31)] were purchased from Shanghai Outdo
Biotech Co., Ltd (Shanghai, China). The human cervix, endometrium,
ovary and breast tissues were obtained from surgical samples in The
First Affiliated Hospital of Jinzhou Medical University (Jinzhou,
China). Breast (n=144), gastric (n=196), colorectal (n=96),
endometrial (n=96) and lung carcinoma (n=192) tissues were also
collected from The First Affiliated Hospital of Jinzhou Medical
University. Dissected and collected specimens from The First
Affiliated Hospital of Jinzhou Medical University were subjected to
routine preparation of formalin (10%)-fixed (room temperature for
48 h) and paraffine-embedded tissue block, and the performance of
tissue microarray using tissue microarrayer (KIN-1; Azumaya Co.,
Ltd., Warabi, Japan). None of the patients with cancer underwent
chemotherapy, radiotherapy or adjuvant treatment prior to surgical
resection between January 1, 2008 and Dececember 30, 2015.
Immunohistochemistry
Consecutive sections were dewaxed with xylene,
rehydrated with graded alcohol (100, 90, 80, 70 and 60%) to water
and subjected to antigen retrieval in a bioled target retrieval
solution (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA)
in a microwave oven (Panasonic Corporation, Osaka, Japan) for 15
min at 100°C. Sections were subsequently blocked in 5% bovine serum
albumin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 5 min
to prevent non-specific antibody binding. The sections were
incubated with a rabbit anti-ING5 antibody (dilution, 1:50; cat.
no. 11560-1-AP; ProteinTech Group, Inc., Chicago, IL, USA) for 15
min at 37°C, followed by incubation with an anti-rabbit secondary
antibody conjugated to horseradish peroxidase (1:100; cat no.
P0399; Dako; Agilent Technologies, Inc.) for 15 min at 37°C. All of
the incubations were performed at 37°C for 15 min in a microwave
oven as previously described (3).
Following each treatment, the slides were washed three times (1 min
each) with TBST. Bound antibodies were visualized using
3,3′-diaminobenzidine. The sections were counterstained with
Mayer's hematoxylin, dehydrated, cleared and mounted. Normal rabbit
immunoglobulin G (1:100; cat no. 500-P00; PeproTech China, Suzhou,
China) was used instead of the primary antibody as a negative
control.
Immunostaining
ING5 protein was observed under a light microscope
(BX43; Olympus Corporation, Tokyo, Japan; ×200). Initially, a
strong expression field was selected under low magnification, and
100 cells were randomly counted from five distinct and
representative fields of each section blindly by 2 independent
pathologists. The percentage of counted cells was scored as
follows: Negative, 0–10% and positive, 11–100%.
Statistical analysis
A χ2 test was performed to compare the
positive rates of different groups. SPSS 10.0 software (SPSS, Inc.,
Chicago, IL, USA) was used for analysis. P<0.05 was considered
to indicate a statistically significant difference.
Results
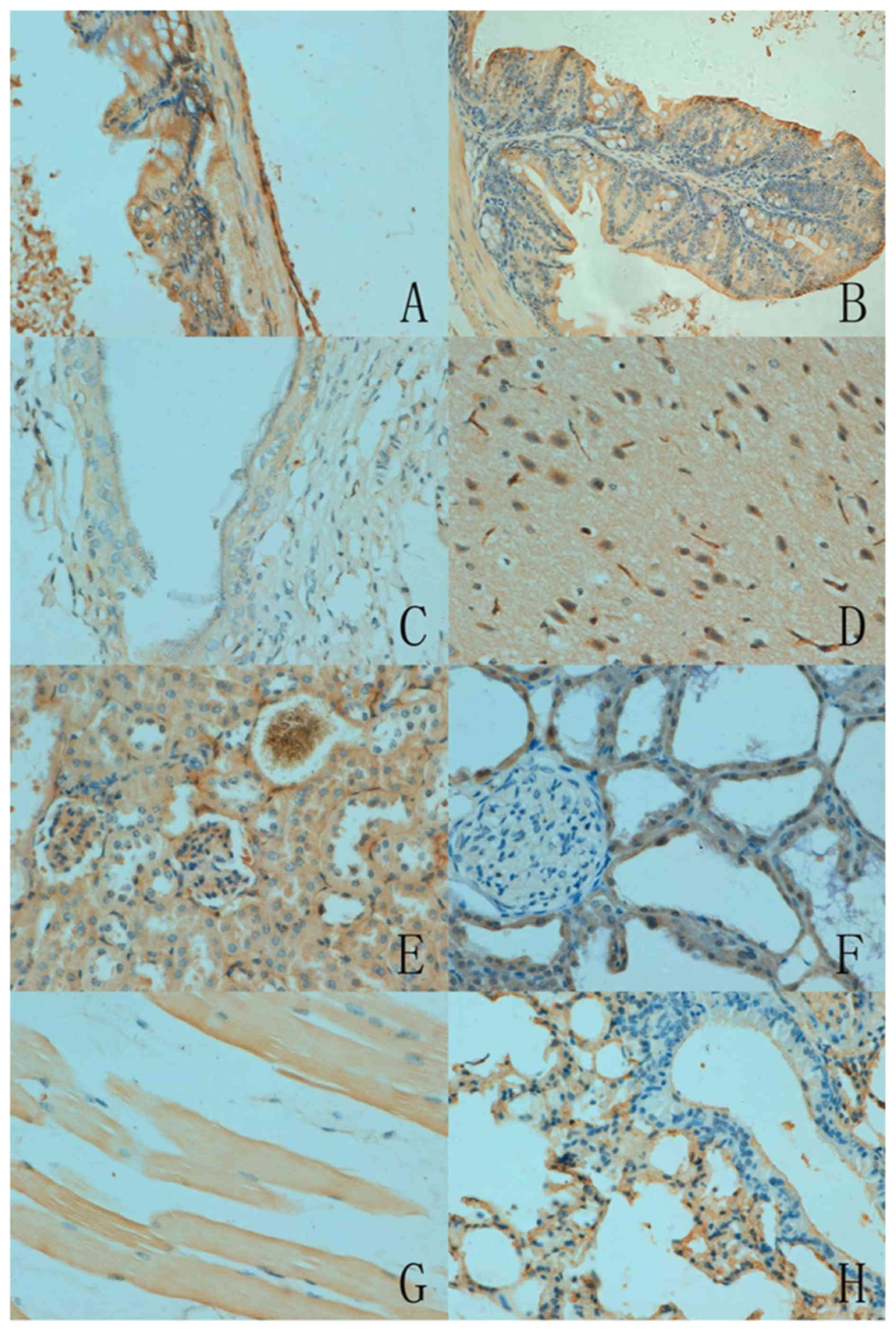
As presented in Fig.
2, the subcellular location of ING5 was cytoplasmic or
nucleo-cytoplasmic in the mouse tissues with either a sporadic or
local distribution. There was variation in ING5 expression among
the different tissues. ING5 reactivity was detectable in the
cytoplasm of neurons, the nephric tubule and glomerulus, alveolar
epithelium, glands of the stomach and intestine, squamous
epithelium of the skin and skeletal muscles. Furthermore, ING5 was
also detected in the nucleus of the breast glandular epithelium
(Table I).
 | Table I.Immunohistochemical analysis of ING5
in normal mouse tissues. |
Table I.
Immunohistochemical analysis of ING5
in normal mouse tissues.
| Organ or tissue | Cell types | Expression
pattern |
|---|
| Brain | Neuron | – |
| Heart | Sporadic | + |
| Lung | Alveolar
epithelium | – |
| Kidney | Nephric tubule and
glomerulus | – |
| Stomach | Glandular
epithelium | – |
| Intestine | Glandular
epithelium | – |
| Spleen | Sporadic | + |
| Skin | Squamous
epithelium | – |
| Muscle | Striated muscle
cell | – |
| Liver | Sporadic | + |
| Breast | Glandular
epithelium | – |
In human tissues, the ING5 protein was principally
distributed in the cytoplasm. ING5 expression was detected in the
cytoplasm and nucleus in a number of tissue types, including the
tongue, stomach, intestine, lung and breast (Fig. 3). According to the density, strong
immunoreactivity of ING5 was detected in the tongue, stomach and
skin, whereas weak immunoreactivity was observed in the cerebellum,
brain stem, thymus and skeletal muscle (Table II).
 | Table II.ING5 protein expression in normal
human tissues. |
Table II.
ING5 protein expression in normal
human tissues.
|
| ING5 expression |
|---|
|
|
|
|---|
| Tissue type | Nucleus | Cytoplasm |
|---|
| Cerebrum | – | + |
| Cerebellum | – | + |
| Brain stem | – | + |
| Thymus | – | + |
| Heart muscle | – | + |
| Aorta | – | + |
| Tongue | + | + |
| Thyroid | – | + |
| Esophagus | – | + |
| Stomach | + | + |
| Intestine | + | + |
| Liver | – | + |
| Pancreas | – | + |
| Lung | + | + |
| Trachea | – | + |
| Skin | – | + |
| Appendix | – | + |
| Smooth muscle | – | + |
| Skeletal muscle | – | + |
| Heart | – | + |
| Testis | – | + |
| Bladder | – | + |
| Prostate | – | + |
| Cervix | – | + |
| Endometrium | – | + |
| Ovary | – | + |
| Breast | + | + |
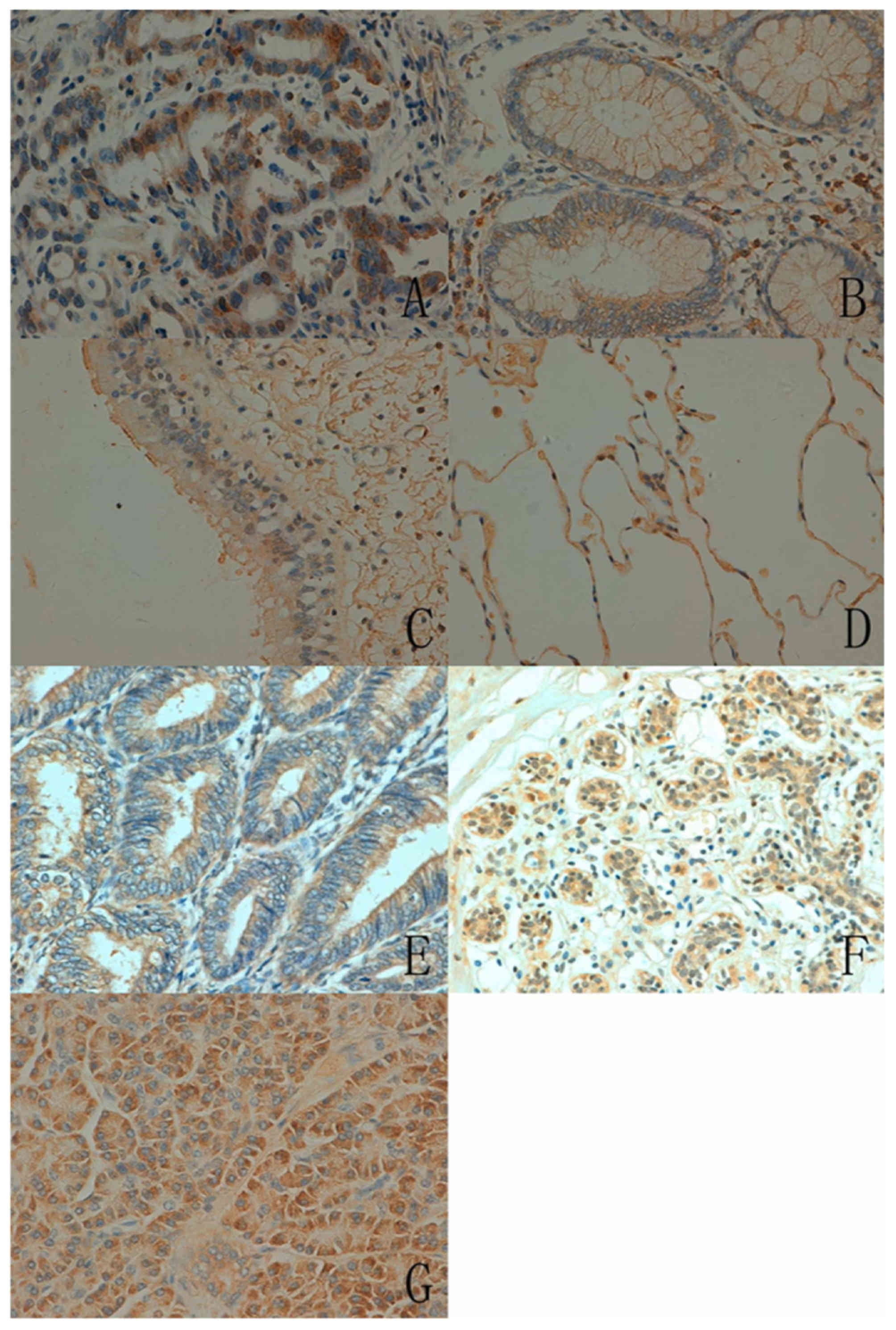
In total, ING5 expression was detected in 400/986
cancer tissues (40.6%), with a homogenous expression pattern
(Fig. 4; Table III). In the majority of cases, ING5
expression was restricted to the cytoplasm of cancer cells. ING5
expression was also detected in the nucleus and cytoplasm of
gastric, colorectal and lung cancer. Notably, ING5 was more
frequently expressed in breast (79.9%, 115/144), colorectal (56.3%,
54/96) and endometrial cancer (50.0%, 48/96) compared with
hepatocellular (14.5%, 9/62) and pancreatic cancer (22.6%, 14/62,
P<0.05).
 | Figure 4.ING5 expression in human cancer issues
(magnification, ×200). (A) Gastric, (B) colorectal, (C) esophageal,
(D) renal, (E) breast, (F) hepatocellular, (G) pancreatic, (H)
lung, (I) endometrial and (J) cervical carcinoma. ING5, inhibitor
of growth family 5. |
 | Table III.ING5 protein expression in human
cancer tissues. |
Table III.
ING5 protein expression in human
cancer tissues.
|
|
|
|
| ING5
expression |
|---|
|
|
|
|
|
|
|---|
| Type of
carcinoma | Total cases
(n) | Positive cases
(n) | Positive rate
(%) | Nucleus | Cytoplasm |
|---|
| Hepatocellular | 62 | 9 | 14.5 | – | + |
| Renal clear
cell | 62 | 20 | 32.3 | – | + |
| Pancreatic | 62 | 14 | 22.6 | – | + |
| Esophageal | 45 | 14 | 31.1 | – | + |
| Cervical | 31 | 13 | 41.9 | – | + |
| Breast | 144 | 115 | 79.9 | – | + |
| Gastric | 196 | 63 | 32.1 | + | + |
| Colorectal | 96 | 54 | 56.3 | + | + |
| Endometrial | 96 | 48 | 50.0 | – | + |
| Lung | 192 | 50 | 26.0 | + | + |
Discussion
ING5 is a member of the ING family and is involved
in DNA repair, apoptotic induction, chromatin remodeling, cellular
senescence and proliferative suppression (1–3). There is
an NLS in the C-terminal of ING5, which mediates its nuclear import
(19). In the present study, the
expression profile and cellular localization of the ING5 protein
were characterized in normal mouse and human tissues, as well as in
human cancer tissues. ING5 expression was primarily detected in the
cytoplasm of normal mouse and human tissues, and human cancer
tissues. Additionally, ING5 expression was detected in the
cytoplasm and nuclei of gastrointestinal glands, the squamous
epithelium of the skin, tongue and breast tissue. These results
indicate that the expression pattern of ING5 has cell-specific
features, and ING5 has distinct functions in different types of
cells.
The phosphorylation of ING1 by 14-3-3 family
(20) or Src (21) is able to induce the cytoplasmic
relocalization of ING1 for apoptotic induction. The degradation of
ING3 by a cytoplasmic Skp1-cullin-F-box protein complex-mediated
ubiquitin-proteasome system provides further evidence for the
cytosolic localization of ING3 protein (22). Therefore, the authors of the present
study hypothesize that the chemical modification of ING5 causes its
relocalization to the cytoplasm.
Amino acid sequence alignment indicated a high
similarity between human and mouse ING5 as they share >90%
identity of the amino acid sequence (Fig.
1) (18). The present study
identified no marked differences in the patterns of ING5 expression
between mouse and human non-cancerous tissue samples. In human
normal tissues, strong expression of ING5 was detected in in the
tongue, stomach and skin. By contrast, weak expression was detected
in the human cerebellum, brain stem, thymus and skeletal muscle
tissues. These findings suggest the functional involvement of ING5
in specific cell types. Therefore, a conditional knockdown of ING5
will be performed using a cell-specific cre mouse in order to
establish an ING5 knockout mouse model of cancer in the future.
ING5 protein has been reported to be able to inhibit
cell growth, induce apoptosis and remodel chromatin by interacting
with p53 and EP300/p300 (1–3). Therefore, ING5 overexpression was
detectable in the tongue, skin and breast, which display a higher
cell regeneration, compared with muscle and nerve tissue. ING5 is a
candidate tumor suppressor gene, and its expression is
downregulated in numerous types of tumors (14,15). In
the present study, the investigation focused on epithelial tumors
and demonstrated that there is a high incidence of ING5 expression
in gynecological cancer types, including endometrial and cervical
cancer. This indicates that ING5 protein may be involved in
estrogen production or regulated by estrogen. Notably, the highest
level of cytoplasmic ING5 expression was detected in colorectal
cancers, suggesting that cytoplasmic IN5 expression may be closely
associated with colorectal carcinogenesis.
In a previous study conducted by the authors,
nucleo-cytoplasmic translocation of ING5 occurred during colorectal
carcinogenesis. Additionally, cytoplasmic ING5 expression was
positively correlated with depth of invasion, lymphatic invasion,
and TNM staging of colorectal cancer, while the inverse was true
for nuclear ING5 expression (15). In
the present study, the positive rate of ING5 protein was 56.3% in
colorectal cancer, but hepatocellular and pancreatic cancer
exhibited lower ING5 expression at a lower positive rate (<25%).
This would significantly facilitate the identification of cancer
patients who may potentially benefit from an ING5-targeted therapy.
Together with these findings, the profiling of ING5 expression can
be helpful to clarify the role of ING5 in proliferation and
apoptosis of various types of epithelial cancer. In the present
study, ING5 protein was detected in the cytoplasm of cancer cells,
which was contrary to the results of other previous reports
(14–16). This discrepancy in the findings may be
attributed to the use of distinct sampling and fixation
methods.
In summary, the present study examined ING5
expression patterns in normal mouse and human tissues, as well as
in human cancer tissues. The differential expression and/or
subcellular location of ING5 in various types of tissues and cells
were also analyzed. ING5 expression may affect cell regeneration
and be closely associated with colorectal carcinogenesis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Liaoning
BaiQianWan Talents Program, Outstanding Scientific Fund of
Shengjing Hospital, Award for Liaoning Distinguished Professors,
Shenyang Science and Technology Grand (grant no. 18-013-0-59) and
the National Natural Scientific Foundation of China (grant no.
81472544 and 81672700).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XFY, DFS, SZ, TRR, YG, SS and JCW conducted the
experiments and analyzed the data. HCZ and HCS designed the study
and finished the organization and writing.
Ethics approval and consent to
participate
The human sample collection and animal protocols
were approved by the Ethics Committee of our hospital of Jinzhou
Medical University. Writing consent was provided by all patients
for the present research.
Patient consent for publication
The patients consented to publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gunduz M, Gunduz E, Rivera RS and
Nagatsuka H: The inhibitor of growth (ING) gene family: Potential
role in cancer therapy. Curr Cancer Drug Targets. 8:275–284. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soliman MA and Riabowol K: After a decade
of study-ING, a PHD for a versatile family of proteins. Trends
Biochem Sci. 32:509–519. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y, Wang J and Li G: Leucine
zipper-like domain is required for tumor suppressor ING2-mediated
nucleotide excision repair and apoptosis. FEBS Lett. 580:3787–3793.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Unoki M, Kumamoto K, Takenoshita S and
Harris CC: Reviewing the current classification of inhibitor of
growth family proteins. Cancer Sci. 100:1173–1179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Champagne KS, Saksouk N, Peña PV, Johnson
K, Ullah M, Yang XJ, Côté J and Kutateladze TG: The crystal
structure of the ING5 PHD finger in complex with an H3K4me3 histone
peptide. Proteins. 72:1371–1376. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ullah M, Pelletier N, Xiao L, Zhao SP,
Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, et al:
Molecular architecture of quartet MOZ/MORF histone
acetyltransferase complexes. Mol Cell Biol. 28:6828–6843. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu M, Du Y, Gao J, Liu J, Kong X, Gong Y,
Li Z, Wu H and Chen H: Aberrant expression miR-196a is associated
with abnormal apoptosis, invasion and proliferation of pancreatic
cancer cells. Pancreas. 42:1169–1181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu N, Wang J, Wang J, Wang R, Liu Z, Yu Y
and Lu H: ING5 is a Tip60 cofactor that acetylates p53 in response
to DNA damage. Cancer Res. 73:3749–3760. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Huang W, Wu Y, Hou J, Nie Y, Gu H,
Li J, Hu S and Zhang H: MicroRNA-193 pro-proliferation effects for
bone mesenchymal stem cells after low-level laser irradiation
treatment through inhibitor of growth family, member 5. Stem Cells
Dev. 21:2508–2519. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shiseki M, Nagashima M, Pedeux RM,
Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y,
Appella E, Yokota J and Harris CC: p29ING4 and p28ING5 bind to p53
and p300 and enhance p53 activity. Cancer Res. 63:2373–2378.
2003.PubMed/NCBI
|
|
11
|
Cengiz B, Gunduz M, Nagatsuka H, Beder L,
Gunduz E, Tamamura R, Mahmut N, Fukushima K, Ali MA, Naomoto Y, et
al: Fine deletion mapping of chromosome 2q21-37 shows three
preferentially deleted regions in oral cancer. Oral Oncol.
43:241–247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cengiz B, Gunduz E, Gunduz M, Beder LB,
Tamamura R, Bagci C, Yamanaka N, Shimizu K and Nagatsuka H:
Tumor-specific mutation and downregulation of ING5 detected in oral
squamous cell carcinoma. Int J Cancer. 127:2088–2094. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi L and Zhang Y: Truncation of inhibitor
of growth family protein 5 effectively induces senescence, but not
apoptosis in human tongue squamous cell carcinoma cell line. Tumour
Biol. 35:3139–3144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li X, Nishida T, Noguchi A, Zheng Y,
Takahashi H, Yang X, Masuda S and Takano Y: Decreased nuclear
expression and increased cytoplasmic expression of ING5 may be
linked to tumorigenesis and progression in human head and neck
squamous cell carcinoma. J Cancer Res Clin Oncol. 136:1573–1583.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng HC, Xia P, Xu XY, Takahashi H and
Takano Y: The nuclear to cytoplasmic shift of ING5 protein during
colorectal carcinogenesis with their distinct links to pathologic
behaviors of carcinomas. Hum Pathol. 42:424–433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xing YN, Yang X, Xu XY, Zheng Y, Xu HM,
Takano Y and Zheng HC: The altered expression of ING5 protein is
involved in gastric carcinogenesis and subsequent progression. Hum
Pathol. 42:25–35. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kumada T, Tsuneyama K, Hatta H, Ishizawa S
and Takano Y: Improved 1-h rapid immunostaining method using
intermittent microwave irradiation: Practicability based on 5 years
application in Toyama Medical and Pharmaceutical University
Hospital. Mod Pathol. 17:1141–1149. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC540017/#__sec22title
|
|
19
|
Shah S, Smith H, Feng X, Rancourt DE and
Riabowol K: ING function in apoptosis in diverse model systems.
Biochem Cell Bio. 87:117–125. 2009. View
Article : Google Scholar
|
|
20
|
Gong W, Russell M, Suzuki K and Riabowol
K: Subcellular targeting of p33ING1b by phosphorylation-dependent
14-3-3 binding regulates p21WAF1 expression. Mol Cell Biol.
26:2947–2954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu L, Thakur S, Leong-Quong RY, Suzuki K,
Pang A, Bjorge JD, Riabowol K and Fujita DJ: Src regulates the
activity of the ING1 tumor suppressor. PLoS One. 8:e609432013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen G, Wang Y, Garate M, Zhou J and Li G:
The tumor suppressor ING3 is degraded by SCF (Skp2)-mediated
ubiquitin-proteasome system. Oncogene. 29:1498–1508. 2010.
View Article : Google Scholar : PubMed/NCBI
|