Introduction
Breast cancer is the most common type of cancer and
the second leading cause of cancer-related mortality in women
globally (1). Although the mortality
rates have recently decreased due to the advancement in detection
techniques and treatment (2), the
efficacy of available breast cancer therapies remain
unsatisfactory. For instance, triple-negative breast cancer (TNBC)
and drug resistance pose a therapeutic challenge. Currently, there
are no specific targetsfor TNBC (3).
Thus, investigation into novel biomarkers that can act as drug
targets iscritical (4–6).
DNA/RNA-binding protein KIN17 (kin17) is a
constitutively expressed protein in mammalian cells that is
generally expressed at low levels in human tissues and organs
(7). Studies have shown that kin17 is
closely correlated with DNA replication, mRNA processing and cell
cycle regulation (8,9). Kin17 was previously revealed to be
upregulated in various tumors, including breast cancer, colorectal
carcinoma, hepatocellular carcinoma and non-small cell lung cancer
(10–13). Specifically, Zeng et al
(10) demonstrated that kin17 is
essential for the proliferation of breast cancer. Therefore, the
present study aimed to comprehensively investigate the role of
kin17 in breast cancer.
In the present study, the influence of kin17
knockdown in the proliferation and apoptosis of MDA-MB-231 cells,
the representative cell line of TNBC which is negative for estrogen
receptor (ER), progesterone receptor (PR) and human epidermal
growth factor receptor 2 (HER2), was investigated.
Materials and methods
Cell culture and transfection
Human breast cancer MDA-MB-231 cells were purchased
from the American Type Culture Collection (Manassas, VA, USA), and
cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum and 1% antibiotic cocktail (60 µg/ml penicillin
and 100 µg/ml streptomycin). The cultures were maintained in 5%
CO2 and 95% humidity at 37°C. Cells were seeded in
6-well plates ata density of 1×105 cells/well and
transfected with lentiviral vector against kin17
(MDA-MB-231KD cells) or NC vector
(MDA-MB-231NC cells) (Shanghai GeneChem Co., Ltd.,
Shanghai, China) using opti-MEM (Gibco; Thermo Fisher Scientific,
Inc.) and polybrene (Shanghai GeneChem Co., Ltd., Shanghai, China)
according to the manufacturer's protocol. The volume of lentiviral
vector against kin17 and NC vector were 3.5 and 4.6 µl,
respectively, which were calculated according to the manufacturer's
formula and the concentration of polybrene was 5 µg/ml as
recommended by the manufacturer. MDA-MB-231 cells without
transfection with vector (Mock MDA-MB-231 cells) were used as a
blank control. The transfected cells were cultured in the
suspension supplemented with 1.5 ug/ml of puromycin on the
pre-medium in 5% CO2 and 95% humidity at 37°C and the
subsequent experiments were conducted when the cells containing the
fluorescent vector reached 90% and the knockdown of kin17 was
verified by western blot analysis prior to any other
experiments.
Cytotoxicity assays
Cytotoxicity was measured in 96-well plates using a
Cell Counting kit-8 (CCK-8) assay kit (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) according to manufacturer's
protocols. MDA-MB-231NC and MDA-MB-231KD
cells were seeded at a density of 2,000 cells/well and allowed to
grow for 24 h. Absorbance was measured at 450 nm using amicroplate
reader (Thermo Fisher Scientific, Inc.) following incubation for
24, 48, 72, 96 and 120 h.
Clone formation test
The clone formation test was performed to measure
cellproliferation. MDA-MB-231NC and
MDA-MB-231KD cells were cultured in 6-well plates at a
density of 200 cells/well for 2 weeks. Cell clones were immobilized
with methanol and stained with crystal violet at room temperature
for 15–20 min. Colonies containing >50 cells were counted using
CKX41 Inverted MicroScope (OLYMPUS Corporation, Tokyo, Japan) at
×10 magnification and ImageQuant TL7.0 Image Analysis software was
applied to image analysis (GE Healthcare Life Sciences, Little
Chalfont, UK).
Flow cytometry
MDA-MB-231NC and MDA-MB-231KD
cells were digested with EDTA-free trypsin and harvested by
centrifugation at 400 × g for 5 min at room temperature. The
collected cells were washed twice using PBS (Gibco; Thermo Fisher
Scientific, Inc.). The final pellet was resuspended in binding
buffer and stained with Annexin V-APC and propidium iodide (PI)
(Shanghai GeneChem Co.) for 15 min in the dark at room temperature
prior to the apoptosis analysis. For cell cycle analysis, the final
pellet was fixed in 70% cold ethanol overnight at 4°C and treated
with RNaseA for 30 min in a water bath at 37°C. PI staining was
then performed according to the manufacturer's protocol. Flow
cytometry analysis (BD LSRFortessa™; BD Biosciences, San Jose, CA,
USA) was used forthe determination of the level of apoptosis and
distribution of the cell cycle. The fluorescence lifetime,
intensity and other optical data were collected from 10,000 cells
using Annexin V-APC or PI for apoptosis and cell cycle analysis,
respectively, using BD FACSDiva Software v8.0.1 (BD
Biosciences).
TUNEL assay
TUNEL assay was performed using In Situ Cell
Death Detection kit, TMR Red (Roche Applied Science, Penzberg,
Germany) according to the manufacturer's protocol. Positive and
negative controls were assessed in parallel, and the experiment was
performed in triplicate. Briefly, cells were cultured in 96-well
plates at a volume of 100 µl/well and fixed with 4%
paraformaldehyde at pH7.4 at room temperature for 60 min. Cells
were then permeabilized with 0.1% Triton X-100 in 0.1% sodium
citrate (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). PBS was
then used to wash the cells. For the experimental and positive
control groups, the prepared cells were incubated with DNase I at
room temperature for 10 min and 50 µl TUNEL reaction mixture was
added following the incubation. The negative control group was
treated with Label solution (50 µl) only, without incubation with
DNase I. The cells were then incubated with the mixture in a
humidified atmosphere for 60 min at 37°C in the dark. DAPI was
added prior to the analysis with a fluorescence microscope. Three
high-power fields were randomly evaluated from each sample at ×100
magnification.
Caspase 3/7 assay
MDA-MB-231KD and MDA-MB-231NC
cells were harvested and resuspended in binding buffer. The
suspension was added to 96-well plates in 100-µl volumes to give a
concentration of 1×104 cells/well.
Caspase-Glo® 3/7 assay (Promega Corporation, Madison,
WI, USA) was later added to detect apoptotic cells according to the
manufacturer's protocol. A blank control was set up in parallel to
eliminate background interference. Absorbance was measured at 405
nm using amicroplate reader (Thermo Fisher Scientific, Inc.)
following incubation for 2 h at 37°C. The activity of Caspase 3/7
in the MDA-MB-231KD group was calculated compared with
the NC group, and therefore the mean value of NC group was always
100%.
Western blot analysis
Protein was extracted using Cell Total Protein
Extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China).
The MDA-MB-231KD, MDA-MB-231NC and mock
MDA-MB-231 cells were digested by radioimmunoprecipitation lysis
buffer on ice and then collected in Eppendorf tubes. The suspension
was centrifuged at 20.8×103 × g at 4°C for 20 min to
collect the supernatant. Protein concentration was determined by
Bradford protein quantitation assay (Nanjing KeyGen Biotech Co.,
Ltd.), and 5XSDS was added to 0.25% volume of the collected
supernatant. The mixture was heated in boiling water for 10 min.
Following this, 50 µg protein from the MDA-MB-231KD and
MDA-MB-231NC cells was loaded on to 12% SDS-PAGE gels.
The separated proteins were transferred to polyvinylidene
difluoride membranes (Merck KGaA) and blocked with 5% non-fat milk
suspended in tris-buffered saline with 0.1% Tween-20 buffer at room
temperature for 1 h. The samples were incubated with anti-kin17
(dilution, 1:500; cat. no. sc-32769; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), anti-PARP (dilution, 1:1,000; cat. no.
sc-8007; Santa Cruz Biotechnology, Inc.), anti-cleaved PARP
(dilution, 1:1,000; cat no. #5625; Cell Signaling Technology, Inc.,
Danvers, MA, USA) and anti-GADPH (dilution, 1:500; cat no.
sc-47778; Santa Cruz Biotechnology, Inc.) monoclonalprimary
antibodies overnight at 4°C. Following the incubation, the samples
were washed with PBS and incubated with horseradish
peroxidase-conjugated secondary antibodies (donkey anti-mouse IgG,
dilution, 1:2,000, cat. no. sc-2318; Santa Cruz Biotechnology and
goat anti-rabbit IgG, dilution, 1:2,000, cat. no. #7074; Santa Curz
Biotechnology) at room temperature for 1 h. Membranes were
visualized withan enhanced chemiluminescence system (Fdbio Science,
Hangzhou, China) and observed by QuantityOne v4.62 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Data analysis was performed with GraphPad Prism v6.0
and SPSS v19.0. Data were presented as mean ± standard deviation.
Continuity correction and Student's t test was used to assess
variance among groups. All statistical tests and corresponding
P-values reported were for two-sided tests, and P<0.05 was
considered to indicate a statistically significant difference.
Results
Knockdown of kin17 reduces MDA-MB-231
cell proliferation
To investigate the role of kin17 in breast cancer,
MDA-MB-231 cells were transfected with lentiviral vector against
kin17 to knockdown its expression (Fig.
1A). Western blot assays confirmed that the protein levels of
kin17 in the MDA-MB-231KD cells were markedly decreased
compared with the levels in the MDA-MB-231NC and mock
MDA-MB-231 cells (Fig. 1B).
CCK8 assay and cell cycle analysis were performed to
assess the effects of kin17-knockdown on the proliferation of
breast cancer cells. Notably, MDA-MB-231KD cell
proliferation was significantly reduced compared with that of
MDA-MB-231NC cells (P<0.05; Fig. 1C), indicating that kin17-knockdown
altered the proliferation of breast cancer cells. Cell cycle
analysis revealed that the ratios of cells in the S and G2/M phases
were lower in the knockdown group compared with the NC group, while
the percentage of G1 cells was higher in the knockdown group
(Fig. 1D). The clone formation test
was also performed to assess the effects of kin17 on proliferation,
which revealed that the number of clones was significantly less in
the knockdown group than in the NC group (P<0.05; Fig. 1E).
Knockdown of kin17 enhances apoptosis
of MDA-MB-231 cells
To assess the role of kin17 in breast cancer cell
apoptosis, endogenous kin17 was silenced in MDA-MB-231 cells with a
lentivirus vector against kin17. The kin17-knockdown group revealed
significantly higher apoptosis rates compared with the NC group
(P<0.05) (Fig. 2A). TUNEL assay
also indicated that kin17-knockdown enhanced apoptosis in the
MDA-MB-231 cells compared with that in the control cells
(P<0.05; Fig. 2B).
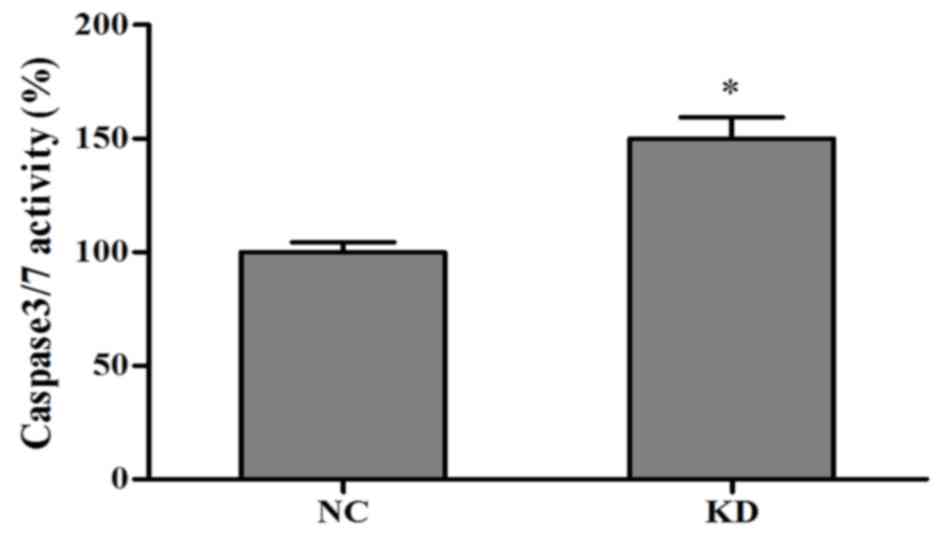
Caspase 3/7 may be involved in
kin17-knockdown-induced apoptosis
Caspase 3/7 were detected following 3 days of
culture. Notably, caspase activity was higher in the
kin17-knockdown group than in the kin17 negative control group
(P<0.05; Fig. 3).
Kin17-knockdown may induce apoptosis
of MDA-MB-231 cells via poly (adenosine diphosphate-ribose)
polymerase (PARP)
The effect of kin17-knockdown on expression levels
of PARP and cleaved PARP in MDA-MB-231 cells was investigated. The
expression levels of PARP were markedly decreased following kin17
knockdown, while the level of cleaved PARP was simultaneously
increased compared with mock and negative control cells (Fig. 4).
Discussion
TNBC is a subtype of breast cancer that does not
express ER, PR and HER2; it accounts for 15% of all breast cancer
cases (14). Patients with TNBC
always demonstrate higher rates of mortality and metastasis
compared with other subtypes due to the absence of effective target
therapies (15,16). Efforts to identify effective targets
for TNBC have thus far yielded unsatisfactory results.
To investigate the potential targets for TNBC, the
role of kin17 in a TNBC cell line was investigated. Kin17, a
constitutive gene with extremely low expression in the majority of
human tissues, has been reported to be markedly overexpressed in
multiple tumors including breast cancer (10–13). In
the present study, a lentiviral vector was used to stably knockdown
kin17. Kin17-knockdown was revealed to significantly suppress the
proliferation of MDA-MB-231 cells and prevent G1 to S phase
transition, corroborating the findings of Zeng et al
(10). Kin17 was also revealed to
facilitate the apoptosis of MDA-MB-231 cells, as investigated with
flow cytometry and TUNEL assays. Higher caspase 3/7 activity in the
knockdown group compared with the NC group also suggested that
caspase 3 and 7, which are wellknown toinduce cell apoptosis
(17,18), may be involved in the kin17-mediated
apoptosis of MDA-MB-231 cells. The underlying mechanism, however,
requires further investigation.
Hormonal therapy has been considered as the
first-line therapy for breast cancer since its emergence in 1973
(19,20). However, its applicationis limited, as
only 20–30% of the patients exhibit a HER-2-positive subtype
(21). PARPs are a family of enzymes
that areinvolved in numerous processes, including DNA damage repair
(22). PARP inhibitor, designed for
germline breast cancer susceptibility protein-mutated TNBC, was
revealed to be effective for the treatment of TNBC in a recent
breast cancer trial (23,24). In the present study, the level of PARP
was revealed to be significantly decreased by western blot analysis
following knockdown of kin17, indicating that kin17 may enhance
apoptosis of MDA-MB-231 cells by affecting the function of PARP. A
previous study also reported that kin17 upregulation was
significantly associated with p53 mutation and increased cyclin D1
and extracellular signal related kinase 1/2 expression in breast
cancer cells, including TNBC cells (10). Thus, kin17 could become a novel target
for the personalized treatment of TNBC and other breast cancer
subtypes.
Frequent adverse events and the increasing risk of
endometrial diseases, including endometrial cancer and endometrial
polyps, have not only been observed in TNBC, but also in other
types of breast cancer due to existing treatment methods (25,26).
Development of targets for evaluation or treatment is necessary for
the improvement of breast cancer therapy. Since it is unclear how
kin17 alters proliferation and apoptosis of breast cancer cells,
future investigations should focus on determining underlying
mechanisms that could facilitate the use of kin17 a target for
breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Guangdong
Planning Project of Science and Technology (grant nos.
2014A020212190 and 2016A020215118), the Knowledge Innovation
Program of Shenzhen Innovation Committee (grant no.
JCYJ20160428101420063) and the Guangzhou Planning Project of
Science and Technology (grant no. 201710010015). The funders had no
role in the study design, data collection and analysis, manuscript
preparation or decision to publish.
Availability of data and materials
The datasets used or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TZ, XG and KW conceived and designed the
experiments. XG, YZ and MZ performed the experiments. XG, ZL and HW
analyzed the data. XG and TZ wrote the paper.
Ethics approval and consent
toparticipate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jensen MB, Ejlertsen B, Mouridsen HT and
Christiansen P; Danish Breast Cancer Cooperative Group, .
Improvements in breast cancer survival between 1995 and 2012 in
Denmark: The importance of earlier diagnosis and adjuvant
treatment. Acta Oncol. 2 Suppl 55:S24–S35. 2016. View Article : Google Scholar
|
|
3
|
Lukong KE: Understanding breast cancer –
The long and winding road. BBA Clin. 7:64–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Reilly EA, Gubbins L, Sharma S, Tully R,
Guang MH, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell
M and McCann A: The fate of chemoresistance in triple negative
breast cancer (TNBC). BBA Clin. 3:257–275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miah S, Banks CA, Adams MK, Florens L,
Lukong KE and Washburn MP: Advancement of mass spectrometry-based
proteomics technologies to explore triple negative breast cancer.
Mol Biosyst. 13:42–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Denkert C, Liedtke C, Tutt A and von
Minckwitz G: Molecular alterations in triple-negative breast
cancer-the road to new treatment strategies. Lancet. 389:2430–2442.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kannouche P, Mauffrey P, Pinon-Lataillade
G, Mattei MG, Sarasin A, Daya-Grosjean L and Angulo JF: Molecular
cloning and characterization of the human KIN17 cDNA encoding a
component of the UVC response that is conserved among metazoans.
Carcinogenesis. 21:1701–1710. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miccoli L, Frouin I, Novac O, Di Paola D,
Harper F, Zannis-Hadjopoulos M, Maga G, Biard DS and Angulo JF: The
human stress-activated protein kin17 belongs to the multiprotein
DNA replication complex and associates in vivo with mammalian
replication origins. Mol Cell Biol. 25:3814–3830. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pinon-Lataillade G, Masson C,
Bernardino-Sgherri J, Henriot V, Mauffrey P, Frobert Y, Araneda S
and Angulo JF: KIN17 encodes an RNA-binding protein and is
expressed during mouse spermatogenesis. J Cell Sci. 117:3691–3702.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeng T, Gao H, Yu P, He H, Ouyang X, Deng
L and Zhang Y: Up-regulation of kin17 is essential for
proliferation of breast cancer. Plos One. 6:e253432011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu M, Zhang Z, Yu H, Xue C, Yuan K, Miao M
and Shi H: KIN enhances stem cell-like properties to promote
chemoresistance in colorectal carcinoma. Biochem Biophys Res
Commun. 448:63–69. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Huang S, Gao H, Wu K, Ouyang X,
Zhu Z, Yu X and Zeng T: Upregulation of KIN17 is associated with
non-small cell lung cancer invasiveness. Oncol Lett. 13:2274–2280.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kou WZ, Xu SL, Wang Y, Wang LW, Wang L,
Chai XY and Hua QL: Expression of Kin17 promotes the proliferation
of hepatocellular carcinoma cells in vitro and in vivo. Oncol Lett.
8:1190–1194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hammond ME, Hayes DF, Dowsett M, Allred
DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS,
Hayes M, et al: American society of clinical oncology/college of
american pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer. Arch Pathol Lab Med. 134:907–922. 2010.PubMed/NCBI
|
|
15
|
Kohler BA, Sherman RL, Howlader N, Jemal
A, Ryerson AB, Henry KA, Boscoe FP, Cronin KA, Lake A, Noone AM, et
al: Annual report to the nation on the status of cancer, 1975-2011,
featuring incidence of breast cancer subtypes by race/ethnicity,
poverty, and state. J Natl Cancer Inst. 107:djv0482015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu J, Xue X, Hu C, Xu H, Kou D, Li R and
Li M: Comparison of clinicopathological features and prognosis in
triple-negative and non-triple negative breast cancer. J Cancer.
7:167–173. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thornberry NA and Lazebnik Y: Caspases:
Enemies within. Science. 281:1312–1316. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Earnshaw WC, Martins LM and Kaufmann SH:
Mammalian caspases: Structure, activation, substrates, and
functions during apoptosis. Annu Rev Biochem. 68:383–424. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boccardo F and Rubagotti A: Switching to
aromatase inhibitors in early breast cancer. Lancet. 369:533–535.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mirkin S and Pickar JH: Selective estrogen
receptor modulators (SERMs): A review of clinical data. Maturitas.
80:52–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Montero JC, Ocaña A, Abad M, Ortiz-Ruiz
MJ, Pandiella A and Esparís-Ogando A: Expression of Erk5 in early
stage breast cancer and association with disease free survival
identifies this kinase as a potential therapeutic target. PLoS One.
4:e55652009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vyas S, Matic I, Uchima L, Rood J, Zaja R,
Hay RT, Ahel I and Chang P: Family-wide analysis of
poly(ADP-ribose) polymerase activity. Nat Commun. 5:44262014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lux MP, Janni W, Hartkopf AD, Nabieva N,
Taran FA, Overkamp F, Kolberg HC, Hadji P, Tesch H, Ettl J, et al:
Update breast cancer 2017-implementation of novel therapies.
Geburtshilfe Frauenheilkd. 77:1281–1290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Okuma HS and Yonemori K: BRCA gene
mutations and poly(ADP-Ribose) polymerase inhibitors in
triple-negative breast cancer. Adv Exp Med Biol. 1026:271–286.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hamaoka S, Kitazawa N, Nara K, Sasaki A,
Kamada A and Okabe T: Selective estrogen receptor modulator. US:
pp. 469–476. 2011
|
|
26
|
Jameera Begam A, Jubie S and Nanjan MJ:
Estrogen receptor agonists/antagonists in breast cancer therapy: A
critical review. Bioorg Chem. 71:257–274. 2017. View Article : Google Scholar : PubMed/NCBI
|