Introduction
Lung cancer is an extremely malignant human tumor
and has a high cancer-associated morbidity worldwide (1). Non-small cell lung cancer (NSCLC)
accounts for >80% of lung cancer globally, including small
squamous cell carcinoma (SCC), adenocarcinoma (AD) and large cell
carcinoma (LCC), in 2015 (2). Despite
significant advances in the presently available treatment methods
including surgery combined with radiotherapy and/or chemotherapy,
due to local recurrence and metastasis in the disease, the 5-year
survival rate is poor (1,3). Therefore, it is necessary to investigate
biological markers with high specificity and sensitivity for early
diagnosis and novel therapeutic targets for NSCLC.
Accumulating evidence have demonstrated that long
non-coding RNAs (lncRNAs) function as regulators in a variety of
diseases, including cancer, neurological disorders, and fragile X
syndrome (4). Abnormal alterations in
the function of lncRNAs in certain tumors were identified to either
promote or suppress tumor formation, development, progression and
metastasis (5). A number of lncRNAs,
including H19 and Homeobox transcript antisense RNA, have potential
clinical applications as prognostic markers or therapeutic targets
in some tumors (6).
MIR31HG is a lncRNA that has been identified as
>2,166 nucleotides in length (7).
A previous study reported that inhibition of MIR31HG promoted a
strong p16 (INK4A)-dependent senescence phenotype (7). Downregulation of lncRNA MIR31HG was
associated with a shorter survival time in patients with gastric
cancer and promoted cell proliferation (8). In bladder cancer, MIR31HG expression
levels were revealed to be remarkably reduced and was negatively
associated with Tumor-node metastasis (TNM) stage (9). The lncRNA MIR31HG exhibited oncogenic
potential in pancreatic ductal adenocarcinoma and was negatively
regulated by miR-193b in order to promote tumor progression
(10). Recent studies showed that
increased MIR31HG lncRNA expression increased gefitinib resistance
in NSCLC lines through the EGFR/PI3K/AKT signaling pathway
(11). However, the clinical role and
biological function of MIR31HG involved in NSCLC was has not been
completely investigated.
The present study demonstrated that MIR31HG was
significantly increased in tumor tissues, and patients with higher
a MIR31HG expression, were predicted a shorter overall survival
(OS) time. Using in vitro assays, the downregulation of
MIR31HG expression was demonstrated to significantly inhibit cell
proliferation, invasion and the epithelial-mesenchymal transition
(EMT) phenotype in NSCLC cells. Furthermore, downregulated MIR31HG
inhibited the Wnt/β-catenin signaling pathway. Taken together,
these data demonstrated that MIR31HG could be identified as a poor
prognostic biomarker and a novel therapeutic target for patients
with NSCLC.
Materials and methods
Human tissue samples
Human NSCLC tissue and paired normal tissue samples
were collected from 88 patients (49 males and 39 females; mean age,
50 years; range, 32–76 years) who underwent radical surgery at The
First Affiliated Hospital and College of Clinical Medicine of Henan
University of Science and Technology (Luoyang, China) between March
2007 and July 2012. No patient had received radiotherapy or
chemotherapy prior to surgery. All tissue samples were immediately
frozen in liquid nitrogen and stored at −80°C until subsequent
experimentation. The experimental protocol was conducted according
to the principles of the Declaration of Helsinki and was approved
by the Ethics Committee of The First Affiliated Hospital and
College of Clinical Medicine of Henan University of Science and
Technology (Luoyang, China). Written, informed consent was obtained
from all patients. The TNM staging followed NSCLC TNM staging
criteria of American Joint Committee on Cancer 2003 edition
(12).
Cell culture
Three human NSCLC cell lines A549, H1299 and NCIH460
and a normal human bronchial epithelial cell line 16HBE were
purchased from Cell Bank at Chinese Academy of Sciences (Shanghai,
China). Cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and were supplemented
with 10% fetal bovine serum (FBS) and 100 U/ml penicillin and 100
µg/ml streptomycin (Thermo Fisher Scientific, Inc.). The cell lines
were maintained at 37°C in a humidified atmosphere of 5%
CO2.
Cell transfection
The small interfering (si)-negative control (NC),
si-MIR31HG-1 and si-miR31HG-2 used in this study were synthesized
by Ribobio (Guangzhou RiboBio Co., Ltd., Guangzhou, China). The
following sequences were used: si-MIR31HG-1, sense,
5′-AAGAAUGUGUUGUGGACACAA-3′, and anti-sense,
5′-UUGUGUCCACAACACAUUCUU-3′. si-miR31HG-2, sense,
5′-AAUGGAGCACAAAUAGUUU-3′, and anti-sense,
5′-AAACUAUUUGUGCUCCAUU-3′. si-NC, sense,
5′-UUCUCCGAACGUGUCACGUTT-3′, and anti-sense,
5′-ACGUGACACGUUCGGAGAATT-3′. The cells were transfected with
si-MIR31HG-1, si-miR31HG-2 or si-NC (100 nM, respectively)
according to the manufacturer's protocol. Cells transfection was
conducted using Lipofectamine 2000 reagents (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Cells
were harvested following transfection at 48 h.
Cell proliferation, cell colony
formation and cell migration assays
A Cell Counting kit-8 (CCK-8) assay was performed to
evaluate NCIH460 or A549 cell proliferation by using the CCK-8
assay kit (Beyotime Institute of Biotechnology, Haimen, China).
Briefly, 5,000 cells/well were cultured on a 96-well plate. The
si-NC, si-MIR31HG-1 and si-MIR31HG-2 were transfected into the
cells using Lipofectamine 2000 according to the manufacturer's
protocol, as described previously. After 1, 2, 3, 4 and 5 days of
transfection with RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) containing sterile CCK-8 dye (10 µl) was added to
each well, after which the cells were incubated at 37°C for a
further 4 h and the absorbance at 450 nm was measured in a
microtiter plate reader (Molecular Devices, LLC, Sunnyvale, CA,
USA). For the cell colony formation assay, a total of 100
cells/well transfected with si-NC, si-MIR31HG-1 and si-MIR 31HG-2
were seeded into a 12-well plate and cultured for 2 weeks. Cells
were then fixed with 4% paraformaldehyde for 10 min at room
temperature and stained with 0.1% crystal violet for 10 min at room
temperature. The cells were observed and calculated with an
inverted microscope (IX71; Olympus Corporation, Tokyo, Japan,
magnification, ×200). For the cell migration assay, the cells
invasive ability was measured using Transwell insert with 8.0 µm
pore polycarbonate membrane coated with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA). Briefly, 1×105 cells/well
transfected with si-NC, si-MIR31HG-1 and si-MIR31HG-2 were plated
onto the upper chambers of the transwell coated with Matrigel in
serum-free RPMI-1640 medium and the lower chambers of the transwell
were added with RPMI-1640 supplemented with 10% FBS. Following a 48
h incubation at room temperature, cells on the upper surface of the
filter were removed using a cotton swab, then cells were fixed with
4% paraformaldehyde for 20 min at room temperature and stained with
0.1% crystal violet for 20 min at room temperature. The cells were
observed and measured in 10 randomly selected fields using an
inverted microscope (magnification ×200; CKX41; Olympus
Corporation; magnification, ×200).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
Total RNA from tissues and NSCLC and 16HBE cells was
purified by the TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol. RNA
quality was determined by a Nanodrop spectrophotometer. The RNA was
reverse-transcribed into cDNA using RevertAid First Strand cDNA
Synthesis kit (catalog no. K1622; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The mRNA expression was
detected using SYBR Green PCR Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol, on ABI7500 system (ABI7500; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were as follows: 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec.
Relative mRNA expression was calculated via 2−ΔΔCq
methods (13). RT-qPCR experiments
were replicated at least three times. The PCR primer sequences were
as follows: MIR31HG forward, 5′-CGCTTCTGTCCTCCTACTCG-3′, and
MIR31HG reverse, 5′-ACAAGCAGACCCTTGGAATG-3′. GAPDH forward,
5′-ATGGGGAAGGTGAAGGTCG-3′, and GAPDH reverse,
5′-GGGTCATTGATGGCAACAATATC-3′. Vimentin forward,
5′-AGGAATGGCTCGTCACCTTCGTGAATA-3′ and Vimentin reverse,
5′-GGAGTGTCGGTTGTTAAGAACTAGAGCGGA0-3′. Twist1 forward,
5′-CATGTCCGCGTCCCACTAG-3′, and Twist1, reverse
5′-TGTCCATTTTCTCCTTCTCTCG-3′. E-cadherin forward,
5′-TAGAGGGTCACCGCGTCTAT-3′, and E-cadherin reverse,
5′-CGTACCGCTGATTGGCTGAG-3′.
Western blot assays
NCIH460 and A549 cells were lysed in RIPA buffer
(Beyotime Institute of Biotechnology, Shanghai, China) and
concentrations of supernatant protein were determined using the BCA
protein assay kit (Beyotime Institute of Biotechnology).
Supernatant samples containing 40 µg total proteins were separated
by 10% sodium dodecylsulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), and then were transferred to nitrocellulose membrane
(Merck KGaA, Darmstadt, Germany). After blocking with 5% fat-free
milk at room temperature for 1 h, the membrane was incubated with
anti-E-cadherin antibody (cat. no. sc-21791; 1:1,000), anti-Twist1
antibody (cat. no. sc-6070; 1:2,000), anti-Vimentin antibody (cat.
no. sc-80975; 1:1,000), anti-glycogen synthase kinase 3β (GSK3β;
cat. no. sc-53931; 1:2,000), anti-β-catenin (cat. no. sc-1496;
1:2,000), anti-p-GSK3β (cat. no. sc-373800; 1:2,000) and anti-GAPDH
antibody (cat. no. 166574; 1:1,000 dilution) (all from Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight and then
incubated with appropriate horseradish peroxidase-conjugated
secondary antibodies (goat anti-rabbit; dilution, 1:5,000; cat. no.
ab7832; Abcam, Cambridge, MA, USA) at room temperature for 2 h and
visualized using Pierce Enhanced Chemiluminescent Western Blotting
Substrate (Pierce; Thermo Fisher Scientific, Inc.). The intensity
of the bands was quantified using Image Lab™ Software (version
4.6.9; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
The data are presented as mean ± standard deviation
from at least three experiments. Statistical analysis was performed
using SPSS version 11.0 for Windows (SPSS, Inc., Chicago, IL, USA).
Comparisons of quantitative data were analyzed by Student's t-test
between two groups (two-tailed). One-way analysis of variance
(ANOVA) for comparisons of multiple groups and Student Newman-Keuls
was used as a post hoc test following ANOVA. P<0.05 was
considered to indicate a statistically significant difference.
Results
MIR31HG has a higher expression in
NSCLC tissues and cells
RT-qPCR was applied to determine the expression
levels of MIR31HG in 88 cases of tumor tissues and adjacent normal
tissues in patients with NSCLC. As demonstrated in Fig. 1, MIR31HG was highly expressed in tumor
tissues, when compared with adjacent normal tissues (P<0.05).
Furthermore, the present study evaluated whether MIR31HG expression
was correlated with clinicopathological factors in patients. The
patients with NSCLC were classified into two groups, comprised of
the higher expression and lower expression group, according to the
median value (2.68) of relative MIR31HG expression. The results
demonstrated that the expression of MIR31HG was closely associated
with histological differentiation grade (P=0.024), lymph node
metastasis (P=0.021) and TNM stage (P=0.002) (Table I). Furthermore, Kaplan-Meier curves
and log-rank tests were performed to analyze the prognostic value
of MIR31HG expression for the OS time in patients with NSCLC. The
results demonstrated that patients with higher MIR31HG expression
have a poor OS time (P<0.05, Fig.
1B). In addition, MIR31HG expression levels were detected in
the three human NSCLC cells A549, H1299 and NCIH460, and a normal
human bronchial epithelial cell line 16HBE, the results confirmed
that MIR31HG expression levels were higher in NSCLC cells compared
with that in 16HBE cells (Fig.
1C).
 | Figure 1.MIR31HG has higher expression in NSCLC
tissues and cells. (A) RT-qPCR analysis of MIR31HG expression in 88
cases of NSCLC tissues and adjacent normal tissues. The expression
of MIR31HG was normalized to GAPDH expression. (B) Kaplan-Meier
survival analysis was performed to detect the association between
MIR31HG expression and clinicopathologic parameters in 88 cases of
NSCLC patients. (C) RT-qPCR analysis of MIR31HG expression in the
three human NSCLC cell lines A549, H1299 and NCIH460 and the normal
human bronchial epithelial cell line 16HBE. The expression of
MIR31HG was normalized to GAPDH expression. RT-qPCR analysis of
MIR31HG expression following transfection with si-NC, si-MIR31HG-1,
si-MIR31HG-2 into (D) NCIH460 or (E) A549 cells, The expression of
MIR31HG was normalized to GAPDH expression. **P<0.05 vs.
non-tumor, 16HBE, si-NC. Every independent experiment was performed
three times. NSCLC, non-small cell lung cancer; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; si, small
interfering; NC, negative control. |
 | Table I.Associations between MIR31HG
expression and clinicopathological parameters in 88 cases of NSCLC
patients. |
Table I.
Associations between MIR31HG
expression and clinicopathological parameters in 88 cases of NSCLC
patients.
|
| MIR31HG expression
levels |
|---|
|
|
|
|---|
| Clinicopathologic
parameters | Patient no. | Lower (n=41) | Higher (47) | P-value |
|---|
| Age (years) |
|
|
| 0.594 |
| ≤60 | 52 | 23 | 29 |
|
|
>60 | 36 | 18 | 18 |
|
| Sex |
|
|
| 0.224 |
| Male | 49 | 20 | 29 |
|
|
Female | 39 | 21 | 18 |
|
| Tumor size |
|
|
| 0.876 |
| <3
cm | 48 | 22 | 26 |
|
| >3
cm | 40 | 19 | 21 |
|
| Smoking |
|
|
| 0.285 |
| No | 44 | 23 | 21 |
|
| Yes | 44 | 18 | 26 |
|
| Histological
differentiation grade |
|
|
| 0.024a |
|
Moderate-well | 32 | 20 | 12 |
|
|
Poorly | 56 | 21 | 35 |
|
| Histology |
|
|
| 0.294 |
| SCC | 55 | 28 | 27 |
|
| AC | 33 | 13 | 20 |
|
| Lymph node
metastasis |
|
|
| 0.021a |
|
Negative | 40 | 24 | 16 |
|
|
Positive | 48 | 17 | 31 |
|
| TNM stage (12) |
|
|
| 0.002a |
|
I–II | 49 | 30 | 19 |
|
|
III | 39 | 11 | 28 |
|
Decreased MIR31HG expression
suppresses cell proliferation and migration ability in NSCLC
cells
To evaluate the effects of MIR31HG expression on
NSCLC cell proliferation and migration ability, two siRNA
oligonucleotides against MIR31HG were used to knockdown MIR31HG in
NCIH460 and A549 cells. The results revealed that MIR31HG was
efficiently knocked-down following transfection with
siRNA-MIR31HG-1 or siRNA-MIR31HG-2 into NCIH460 or A549 cells
(Fig. 1D and E). CCK-8 cell
proliferation assays demonstrated that the cell proliferation
ability was inhibited following MIR31HG silencing in NCIH460 or
A549 cells, compared with the si-NC group (Fig. 2A and B). The cell colony formation
number was significantly decreased following MIR31HG silencing in
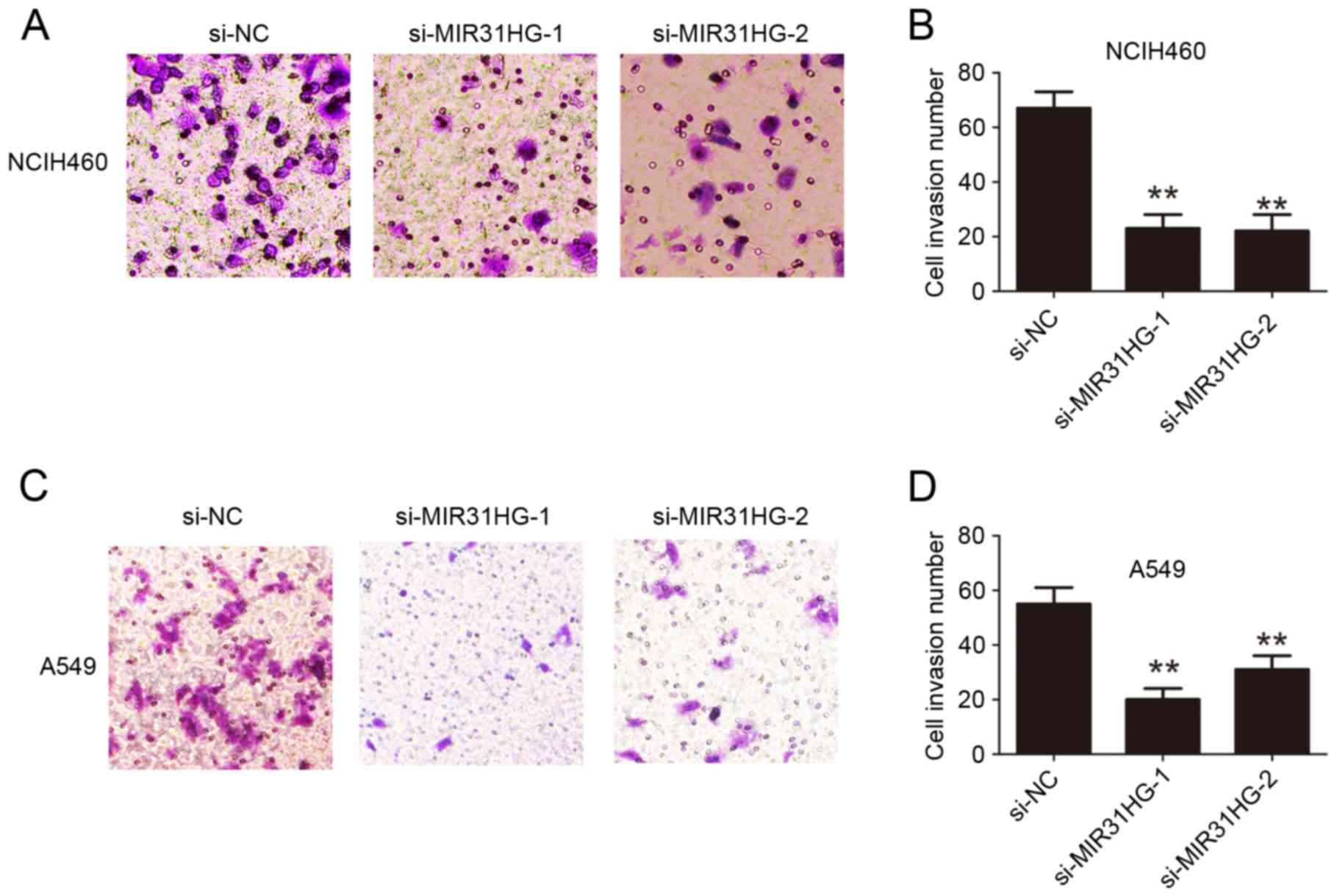
NCIH460 or A549 cells compared with the si-NC group (Fig. 2C-F). Transwell cell invasion assays
analysis demonstrated that cell invasion ability in NCIH460 or A549
cells was also inhibited following MIR31HG silencing compared with
the si-NC group (Fig. 3). Taken
together, these results confirmed that MIR31HG promoted cell
proliferation and invasion ability in NSCLC.
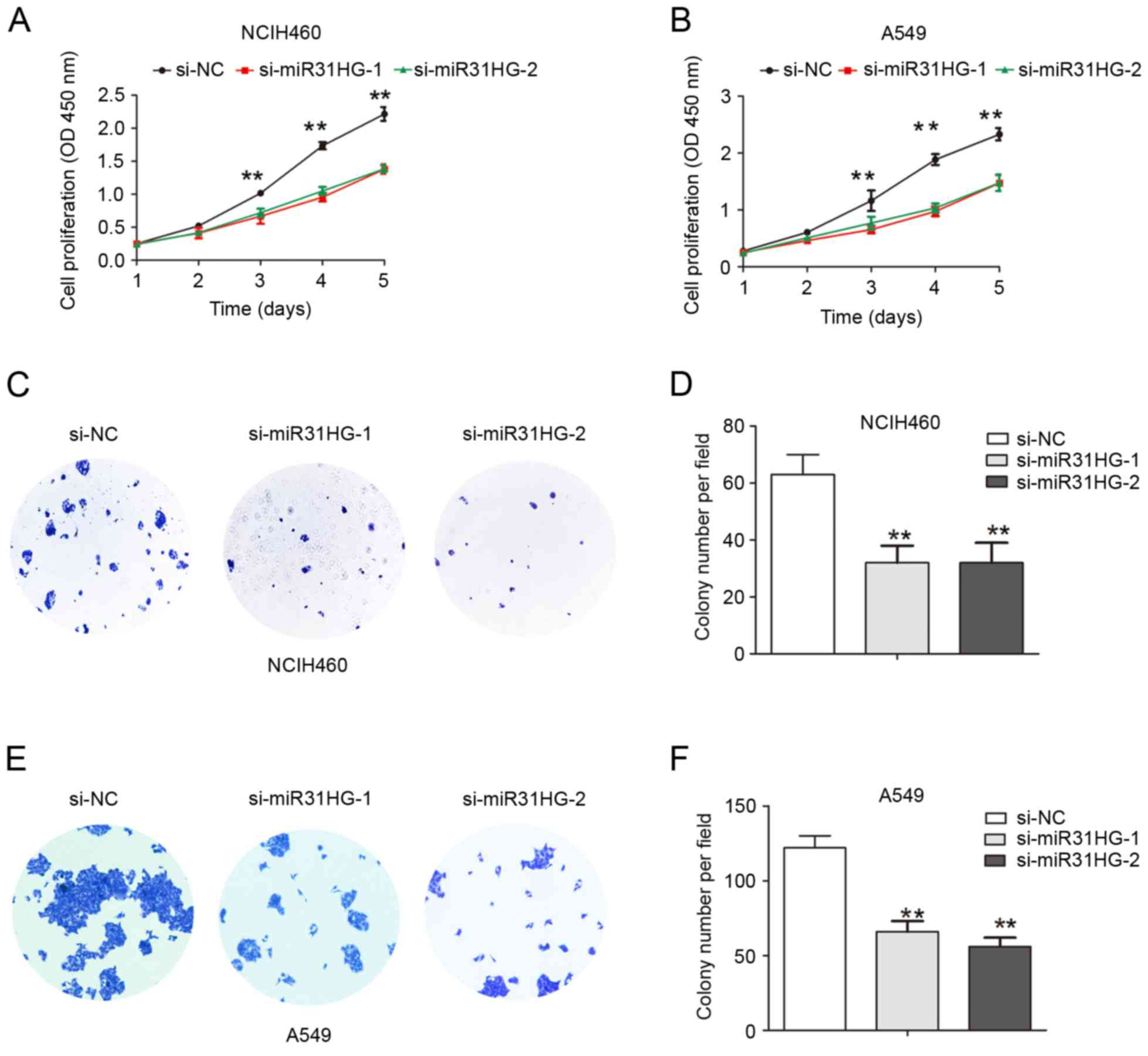 | Figure 2.Knockdown of MIR31HG suppressed cell
proliferation in NSCLC cells. (A) NCIH460 or (B) A549 cells were
transfected with si-NC, si-MIR31HG-1, si-MIR31HG-2 and cell
proliferation was determined using CCK8 assays. (C) NCIH460 cells
were transfected with si-NC, si-MIR31HG-1, si-MIR31HG-2 and cell
colony formation assay was performed, and (D) quantified. (E) A549
cells were transfected with si-NC, si-MIR31HG-1, si-MIR31HG-2 and
cell colony formation assay was performed, and (F) quantified.
**P<0.05 vs. si-NC. Every independent experiment was performed
three times. NSCLC, non-small cell lung cancer; si, small
interfering; NC, negative control; OD, optical density. |
Knockdown of MIR31HG inhibits cell EMT
and Wnt/β-catenin signaling pathway in NSCLC cells
Cell invasion was associated with tumor EMT process,
to explore whether MIR31HG affected cell EMT in NSCLC, the RT-qPCR
and Western blotting assays were performed. The results showed that
the mRNA expression levels of EMT related transcription factor
Twist1 and mesenchymal marker Vimentin were significantly
decreased, but epithelial marker E-cadherin was dramatically
increased after MIR31HG silencing in NCIH460 or A549 cells
(Fig. 4A and B). Furthermore, we
detected their protein expression and the results demonstrated that
protein expression of Twist1 and Vimentin were upregulated, but
E-cadherin was downregulated after MIR31HG silencing in NCIH460 or
A549 cells (Fig. 4C and D).
Wnt/β-catenin signaling is associated with cell invasion and EMT
process (14). Furthermore, the
present study demonstrated that downregulated MIR31HG inhibited the
Wnt/β-catenin signaling pathway by decreasing the expression of
GSK3β and β-catenin, but increasing the expression of p-GSK3β in
NCIH460 or A549 cells (Fig. 5A and
B). Therefore, these results indicated that MIR31HG promoted
EMT by regulating the Wnt/β-catenin signaling pathway in NSCLC
cells.
 | Figure 4.Knockdown of MIR31HG inhibited EMT in
NSCLC cells. RT-qPCR analysis of Twist1, E-cadherin and Vimentin
expression levels following transfection with si-NC, si-MIR31HG-1,
si-MIR31HG-2 into (A) NCIH460 or (B) A549 cells, The expression of
mRNA was normalized to GAPDH expression. Western blot analysis of
Twist1, E-cadherin and Vimentin expression levels after transfected
with si-NC, si-MIR31HG-1, si-MIR31HG-2 into (C) NCIH460 or (D) A549
cells. The numbers present the protein expression levels normalized
to GAPDH expression. **P<0.05 vs. si-NC. Every independent
experiment was performed three times. EMT, epithelial-mesenchymal
transition; NSCLC, non-small cell lung cancer; si, small
interfering; NC, negative control. |
Discussion
The precise molecular mechanism involved in lung
carcinogenesis, at present, has not been completely elucidated.
Recent studies provided the evidence that lncRNAs acted as
prognostic predictor in lung cancer (15,16),
including increased expression of the long non-coding RNA ANRIL
promoting lung cancer cell metastasis and was significantly
correlated with poor prognosis (15).
A previous study reported that growth arrest specific 5 (GAS5)
expression was decreased in NSCLC plasma and the circulating long
non-coding RNA GAS5 was a novel biomarker for the diagnosis of
NSCLC (16). The expression level of
tumor suppressor candidate 7 (TUSC7) was lower in NSCLC tissues and
low expression of TUSC7 was an independent poor prognostic
indicator for patients with NSCLC (17). Increased serum XIST and HIF1A-AS1
could be used as a predictive biomarker for NSCLC screening, and
combination of XIST and HIF1A-AS1 showed a higher positive
diagnostic efficiency of NSCLC (18).
In the present study, it was revealed that MIR31HG was
significantly increased in NSCLC tissues, when compared with
adjacent normal tissues. Compared with lower MIR31HG in patients,
higher MIR31HG predicted a poor prognosis for patients with
NSCLC.
EMT is a cellular process that is involved in
embryonic development and is classically characterized by the
dedifferentiation from an epithelial to mesenchymal phenotype
(19). The EMT phenotype is marked by
the downregulated expression of E-cadherin and upregulated
expression of N-cadherin, vimentin and culminates in the higher
expression of the transcription factors Snail, Zeb, Twist (20,21).
HNF1A-AS1 was revealed to promote tumor proliferation and
metastasis, in vitro and in vivo, by regulating the
expression of cyclin D1, E-cadherin, N-cadherin and β-catenin
(22). A previous study demonstrated
that the loss of the lncRNA FOXF1-AS1 regulated EMT, stemness and
metastasis of NSCLC cells (23).
Downregulation of lncRNA BRAF activated non-coding RNA and was
associated with poor prognosis for NSCLC and promoted metastasis by
affecting EMT (24). LncRNA BC087858
induced non-T790M mutation and acquired resistance to EGFR-TKIs by
activating PI3K/AKT and MEK/ERK pathways and EMT in NSCLC (25). In the present study, it was
demonstrated that the knockdown of MIR31HG inhibited the cell
proliferation, migration and EMT in NSCLC cells by downregulating
the expression of Twist1 and Vimentin and upregulating the
expression of E-cadherin.
Wnt/β-catenin signaling has been reported to be
involved in NSCLC invasion and metastatic abilities (14). For example, Astrocyte elevated gene-1
(AEG-1) induced EMT in lung cancer through activating Wnt/β-catenin
signaling (26). Cucurbitacin B
inhibits the stemness and metastatic abilities of NSCLC via
downregulation of the Wnt/β-catenin signaling axis (27). In the present study, downregulated
MIR31HG was demonstrated to inhibit the Wnt/β-catenin signaling
pathway by decreasing the expression of GSK3β and β-catenin, but
increasing the p-GSK3β expression in NSCLC cells. The limitation of
the present study is that it was only demonstrated that
downregulated MIR31HG inhibited the Wnt/β-catenin signaling
pathway. The effects of MIR31HG expression on other signaling
pathways requires further investigated. The investigation of
molecular mechanism underlying NSCLC cell proliferation and
invasion for MIR31HG may provide a potential target for NSCLC
therapy in the future.
To conclude, the results of the present study
demonstrated that MIR31HG was significantly increased in NSCLC
tissues and increased MIR31HG predicted a poor survival time in
patients with NSCLC. Furthermore, the downregulation of MIR31HG
significantly inhibited NSCLC cell proliferation, cell invasion and
EMT phenotype. Additionally, downregulated MIR31HG inhibited the
Wnt/β-catenin signaling pathway. These results demonstrated that
MIR31HG could be identified as a poor prognostic biomarker and a
therapeutic target for patients with NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SZ and XZ designed the study. SZ and XZ performed
the experiments. XW and JL analyzed the data. All authors
collaborated to interpret results and develop the manuscript. The
final version of the manuscript has been read and approved by all
authors.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The First Affiliated Hospital and College of Clinical Medicine of
Henan University of Science and Technology (Luoyang, China).
Written, informed consent was obtained from all patients.
Patient consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Crinò L, Weder W, van Meerbeeck J and
Felip E; ESMO Guidelines Working Group, . Early stage and locally
advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical
Practice Guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 21 Suppl 5:v103–v115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hackner K, Errhalt P, Mueller MR, Speiser
M, Marzluf BA, Schulheim A, Schenk P, Bilek J and Doll T: Canine
scent detection for the diagnosis of lung cancer in a
screening-like situation. J Breath Res. 10:0460032016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boffa DJ, Allen MS, Grab JD, Gaissert HA,
Harpole DH and Wright CD: Data from The Society of Thoracic
Surgeons General Thoracic Surgery database: The surgical management
of primary lung tumors. J Thorac Cardiovasc Surg. 135:247–254.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen J, Wang R, Zhang K and Chen LB: Long
non-coding RNAs in non-small cell lung cancer as biomarkers and
therapeutic targets. J Cell Mol Med. 18:2425–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Montes M, Nielsen MM, Maglieri G, Jacobsen
A, Højfeldt J, Agrawal-Singh S, Hansen K, Helin K, van de Werken
HJ, Pedersen JS and Lund AH: The lncRNA MIR31HG regulates
p16(INK4A) expression to modulate senescence. Nat Commun.
6:69672015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nie FQ, Ma S, Xie M, Liu YW, De W and Liu
XH: Decreased long noncoding RNA MIR31HG is correlated with poor
prognosis and contributes to cell proliferation in gastric cancer.
Tumour Biol. 37:7693–7701. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He A, Chen Z, Mei H and Liu Y: Decreased
expression of LncRNA MIR31HG in human bladder cancer. Cancer
Biomark. 17:231–236. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang H, Liu P, Zhang J, Peng X, Lu Z, Yu
S, Meng Y, Tong WM and Chen J: Long noncoding RNA MIR31HG exhibits
oncogenic property in pancreatic ductal adenocarcinoma and is
negatively regulated by miR-193b. Oncogene. 35:3647–3657. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang B, Jiang H, Wang L, Chen X, Wu K,
Zhang S, Ma S and Xia B: Increased MIR31HG lncRNA expression
increases gefitinib resistance in non-small cell lung cancer cell
lines through the EGFR/PI3K/AKT signaling pathway. Oncol Lett.
13:3494–3500. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kaur H, Sehgal IS, Bal A, Gupta N, Behera
D, Das A and Singh N: Evolving epidemiology of lung cancer in
India: Reducing non-small cell lung cancer-not otherwise specified
and quantifying tobacco smoke exposure are the key. Indian J
Cancer. 54:285–290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak and Schmittgen: Analysis of relative
gene expression data using real-time quantitative PCR and the
2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kahlert UD, Nikkhah G and Maciaczyk J:
Epithelial-to- mesenchymal(-like) transition as a relevant
molecular event in malignant gliomas. Cancer Lett. 331:131–138.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin L, Gu ZT, Chen WH and Cao KJ:
Increased expression of the long non-coding RNA ANRIL promotes lung
cancer cell metastasis and correlates with poor prognosis. Diagn
Pathol. 10:142015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang W, Lv T, Shi X, Liu H, Zhu Q, Zeng
J, Yang W, Yin J and Song Y: Circulating long noncoding RNA GAS5 is
a novel biomarker for the diagnosis of nonsmall cell lung cancer.
Medicine (Baltimore). 95:e46082016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Jin Y, Ren H, Ma X, Wang B and
Wang Y: Downregulation of the long non-coding RNA TUSC7 promotes
NSCLC cell proliferation and correlates with poor prognosis. Am J
Transl Res. 8:680–687. 2016.PubMed/NCBI
|
|
18
|
Tantai J, Hu D, Yang Y and Geng J:
Combined identification of long non-coding RNA XIST and HIF1A-AS1
in serum as an effective screening for non-small cell lung cancer.
Int J Clin Exp Pathol. 8:7887–7895. 2015.PubMed/NCBI
|
|
19
|
Micalizzi DS and Ford HL:
Epithelial-mesenchymal transition in development and cancer. Future
Oncol. 5:1129–1143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Felipe Lima J, Nofech-Mozes S, Bayani J
and Bartlett JM: EMT in breast carcinoma-A review. J Clin Med.
5(pii): E652016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prat A and Perou CM: Deconstructing the
molecular portraits of breast cancer. Mol Oncol. 5:5–23. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Y, Liu H, Shi X, Yao Y, Yang W and Song
Y: The long non-coding RNA HNF1A-AS1 regulates proliferation and
metastasis in lung adenocarcinoma. Oncotarget. 6:9160–9172.
2015.PubMed/NCBI
|
|
23
|
Miao L, Huang Z, Zengli Z, Li H, Chen Q,
Yao C, Cai H, Xiao Y, Xia H and Wang Y: Loss of long noncoding RNA
FOXF1-AS1 regulates epithelial-mesenchymal transition, stemness and
metastasis of non-small cell lung cancer cells. Oncotarget.
7:68339–68349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun M, Liu XH, Wang KM, Nie FQ, Kong R,
Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, et al: Downregulation of
BRAF activated non-coding RNA is associated with poor prognosis for
non-small cell lung cancer and promotes metastasis by affecting
epithelial-mesenchymal transition. Mol Cancer. 13:682014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pan H, Jiang T, Cheng N, Wang Q, Ren S, Li
X, Zhao C, Zhang L, Cai W and Zhou C: Long non-coding RNA BC087858
induces non-T790M mutation acquired resistance to EGFR-TKIs by
activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell
lung cancer. Oncotarget. 7:49948–49960. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He W, He S, Wang Z, Shen H, Fang W, Zhang
Y, Qian W, Lin M, Yuan J, Wang J, et al: Astrocyte elevated gene-1
(AEG-1) induces epithelial-mesenchymal transition in lung cancer
through activating Wnt/β-catenin signaling. BMC Cancer. 15:1072015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shukla S, Sinha S, Khan S, Kumar S, Singh
K, Mitra K, Maurya R and Meeran SM: Cucurbitacin B inhibits the
stemness and metastatic abilities of NSCLC via downregulation of
canonical Wnt/β-catenin signaling axis. Sci Rep. 6:218602016.
View Article : Google Scholar : PubMed/NCBI
|