Introduction
Primary systemic therapy (PST) followed by surgery
is a standard therapy for advanced breast cancer, including
triple-negative breast cancer (TNBC) (1–3). Since
patients with TNBC, which is used as a surrogate term for
basal-like breast cancer subtypes, lack expression of estrogen
receptor (ER), progesterone receptor (PgR), and human epidermal
growth factor receptor 2 (HER2), no significant advances have yet
been made regarding therapies for treating TNBC patients.
Therefore, TNBC remains a deadly type of breast cancer, and further
study is required to fully understand it. According to
previously-reported gene expression analyses, TNBC has a distinct
profile with non-TNBC (4,5) and a heterogeneous tumor (6). This heterogeneity could be associated
with the therapeutic response of PST for TNBC; however, the
association between gene expression and PST response is still
unknown.
Cell adhesion molecule (CADM) genes encode an
immunoglobulin superfamily molecule and protect against malignant
conversion and metastasis by maintaining cell-cell adhesion in
epithelia (7,8). Expression of CADM1 located on chromosome
11q23.2 was preferentially lost in invasive lesions compared to
non-invasive lesions of lung adenocarcinoma (9). Several studies have also demonstrated
that CADM1 is frequently inactivated in various cancers, including
breast cancer (10–14). By using primary breast cancer
specimens and breast cancer cell lines, loss of CADM1 and 4.1B
expression has been reported to be associated with the development
and progression of breast cancer, especially in invasion and
metastasis (15). Therefore,
CADM1 works as a tumor suppressor gene in non-small cell
lung cancer as well as breast cancer (16). CADM4, which is located on
chromosome 19q13.31, also works as a tumor suppressor gene in renal
clear cell carcinoma, colon cancer, and breast cancer (17–19). We
recently demonstrated that CADM1 and CADM4 were less frequently
inactivated in TNBC than in non-TNBC (20). Our results that the roles of CADM1 and
CADM4 in TNBC are different than their in non-TNBC, and those genes
may therefore be associated with the therapeutic response to PST
for TNBC.
The efficacy of PST for TNBC can be improved by
further understanding of the molecular expression levels in
individual tumors. Herein, we report the evaluation of CADM1 and
CADM4 expression in TNBC patients who had received PST, and try to
define a therapeutic response based on CADM1 and CADM4 expression
levels.
Materials and methods
Patients and tissue samples
Primary invasive breast cancer samples were obtained
from patients who had undergone surgical resections with
chemotherapy prior to surgery at the Department of Breast Surgery
in Fukushima Medical University Hospital, Fukushima, Japan. This
cohort consisted of 16 patients who had been enrolled consecutively
at the time of surgery between Jan, 2006 and Dec, 2012. All 16
patients had received a core needle biopsy (CNB) at the initial
histological diagnosis, after which they received PST;
5-fluorouracil, epirubicin, and cyclophosphamide (FEC) or FEC
followed by paclitaxel (PTX) or docetaxel (DTX). The clinical
therapeutic response to PST was evaluated using the Response
Evaluation Criteria In Solid Tumors (RECIST) guidelines (21), and each case was categorized as
complete response (CR), partial response (PR), stable disease (SD),
or progressive disease (PD). After surgical resection, therapeutic
response to PST was evaluated histologically, and the patients were
divided into pathological CR (pCR) or non-pCR groups.
Detailed backgrounds of each tissue donor, including
age, sex, clinical staging, and hormone status, were collected.
Tumor histopathology was classified according to the Union for
International Cancer Control (UICC) TNM classification (the 7th
classification) (22). Written
informed consent was obtained from all patients, and the study was
approved by the Institutional Review Board of Fukushima Medical
University, Japan.
IHC staining
IHC staining for CADM1 and CADM4 was performed by
same method as previously described (20). The breast cancer tissue samples were
fixed in 10% formalin, embedded in paraffin, cut into 4-µm sections
and stained with hematoxylin and eosin (H&E) and other primary
antibodies. Rabbit polyclonal antibodies against CADM1 (1:500,
C-18, generated by the Division of Molecular Pathology, Institute
of Medical Science, The University of Tokyo) and CADM4 (1:500,
Bc-2, generated by the Division of Molecular Pathology, Institute
of Medical Science, The University of Tokyo, Tokyo, Japan) were
used as described previously (17).
The antibodies used for IHC staining were as follows: Anti-ER
(1:500, cat. no. MA5-13191; Dako; Agilent Technologies GmbH,
Waldbronn, Germany); and anti-PgR (1:500, cat. no., MA5-12581;
Dako; Agilent Technologies GmbH). For HER2 status, the
Histofine® Simple Stain HER2 mono assay kit was used
(cat. no. 427041; Nichirei Biosciences, Inc., Tokyo, Japan).
Analyses of ER, PgR, and HER2 were performed using IHC staining
according to the manufacturer's protocol. The sections were
deparaffinized in xylene, and hydrated using a graded series of
ethanol at room temperature. Subsequently, the sections were washed
three times in PBS and endogenous peroxidase was blocked with 0.3%
in methanol for 30 min at room temperature. Antigens were retrieved
by autoclaving the sections on slides in 0.01 M pH 6.0 citrate
buffer for 10 min at 121°C. Subsequent to washing in PBS, the
sections were incubated in primary antibody overnight at 4°C. A
further wash in PBS was followed by treatment with the secondary
antibody [K1491, Dako EnVision kit/horseradish peroxidase (HRP)]
for 30 min at room temperature and diaminobenzidine (K1491, Dako
EnVision kit/HRP) was used for staining detection (both from Dako:
Agilent Technologies GmbH). Finally, the sections were
counterstained with hematoxylin. Expression of these proteins was
evaluated using optical microscopy (BX43; Olympus Corporation,
Tokyo, Japan) at ×400 magnification.
Assessment of IHC staining
Stain signals of CADM1 and CADM4 protein levels were
detected in the membranes in normal mammary epithelial cells.
Cytoplasmic immunoreactivity without membrane staining was defined
as aberrant expression. Membranous staining of CADM1 or CADM4 was
evaluated by calculating the percentage of cancer cells with
membrane expression in the entire area of invasive and non-invasive
lesions. The tumors or lesions were then scored as previously
described (15); those with scores of
1 (11–30% cells with membrane expression), 2 (31–60%) or 3
(61–100%) were defined as positive staining for CADM1 or CADM4
expression, and those with a score of 0 (0–10%) were defined as
negative staining. ER, PgR and HER2 expression levels were
evaluated semi-quantitatively, with scores representing the ratio
of the number of positive staining cells compared with negative
cells, as previously described (23).
Assessment of the staining was performed blindly by two independent
investigators, including an experienced pathologist (Dr Akiteru
Goto from Akita University, who was the pathologist, and Dr
Motonobu Saito from Fukushima Medical University). Any
disagreements between the two investigators was resolved by
discussion.
Statistical analysis
Fisher's exact test was performed by GraphPad Prism
6 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Patient characteristics
The clinical characteristics of the 16 TNBC patients
are shown in Table I. All patients
were female with ages ranging from 33 to 78 years (mean 49.1
years), and had been diagnosed as having invasive ductal carcinoma.
The patients included four stage IIA, nine stage IIB, and three
stage IIIB patients. The cases received one of the following
treatments: PST as FEC only (n=2); FEC followed by PTX (n=7) or FEC
followed by DTX (n=7). The clinical tumor responses to PST as
evaluated by RECIST were CR (n=4), PR (n=10), SD (n=1), and PD
(n=1). The histological diagnoses in the surgical specimens were
pCR (n=3) and non-pCR (n=13).
 | Table I.Clinicophathological characteristics
of patients with PST triple negative breast cancer. |
Table I.
Clinicophathological characteristics
of patients with PST triple negative breast cancer.
| Characteristics | n (%) |
|---|
| Age (years) |
|
| Mean | 49 |
|
Range | 33–78 |
| Sex |
|
|
Female | 16 (100) |
| Histological
type |
|
| Invasive
carcinoma of no special type | 16 (100) |
| TNM stage at initial
diagnosis (22) |
|
| I | 0 |
| IIA | 4 (25) |
| IIB | 9 (56) |
| IIIA | 0 |
| IIIB | 3 (19) |
| PST regimens |
|
| FEC | 2 (13) |
|
FEC+PTX | 7 (44) |
|
FEC+DTX | 7 (44) |
| Clinical tumor
responsea |
|
| CR | 4
(25) |
| PR | 10 (63) |
| SD | 1 (6) |
| PD | 1 (6) |
| PST effect |
|
| pCR | 3
(19) |
|
Non-pCR | 13 (81) |
Associations between CADM1 and CADM4
expression and clinicopathological factors in TNBC
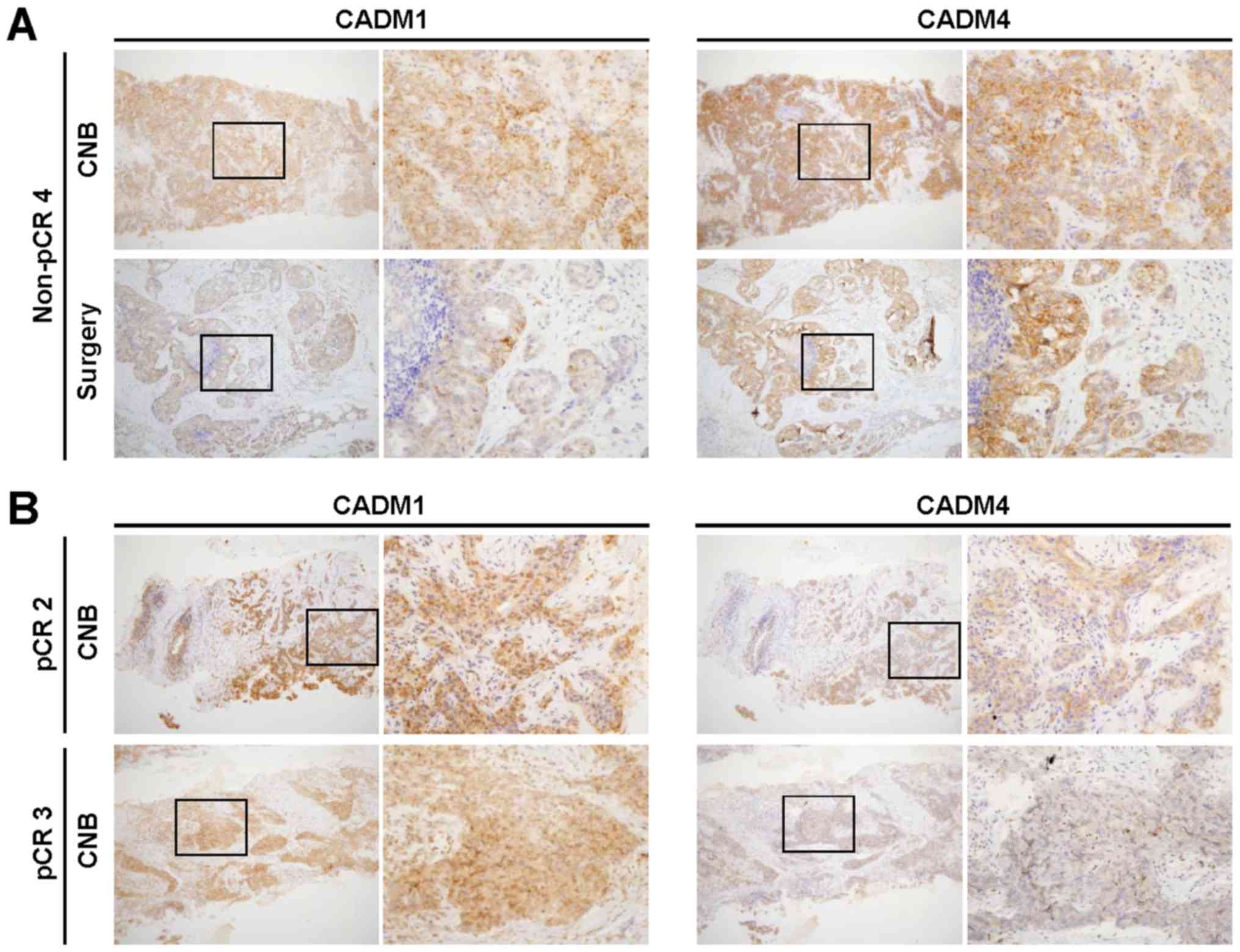
We evaluated CADM1 and CADM4 expression by IHC
staining in surgical or CNB specimens from three pCR and 11 non-pCR
TNBC patients who received PST (Table
II). While surgical specimens are usually used for IHC staining
to measure protein expression, the usefulness of IHC staining with
CNB specimens at the initial diagnosis is still unknown. Therefore,
we firstly compared CADM1 and CADM4 expression between the CNB and
surgical specimens. Among the 13 non-pCR cases in our cohort, nine
could be used in this comparison, seven of which had an identical
staining intensity score for CADM1 and CADM4 between CNB and
surgical specimens (non-pCR 3, non-pCR 4, non-pCR 5, non-pCR 6,
non-pCR7, non-pCR 12, and non-pCR 13 cases) (Fig. 1A). Two cases (non-pCR 1 and non-pCR 2
Cases) showed weaker staining intensity in the CNB specimen (score
1) than in the surgical specimen (scores 2 and 3).
 | Table II.Comparison of CADM1 and CADM4
expression at initial diagnosis or surgery in non-pCR and pCR TNBC
patients. |
Table II.
Comparison of CADM1 and CADM4
expression at initial diagnosis or surgery in non-pCR and pCR TNBC
patients.
| Case | Age (years) | Stage [initial
diagnosis, (22)] | PST | Specimen | CADM1 score | CADM4 score |
|---|
| Non-pCR 1 | 45 | IIB | FEC+DTX | CNB | 1 | 1 |
|
|
|
|
| surgery | 2 and 3 | 2 and 3 |
| Non-pCR 2 | 61 | IIA | FEC+PTX | CNB | 1 | 1 |
|
|
|
|
| surgery | 2 and 3 | 2 and 3 |
| Non-pCR 3 | 34 | IIA | FEC | CNB | 2 and 3 | 2 and 3 |
|
|
|
|
| Surgery | 2 and 3 | 2 and 3 |
| Non-pCR 4 | 44 | IIB | FEC+DTX | CNB | 2 and 3 | 2 and 3 |
|
|
|
|
| Surgery | 2 and 3 | 2 and 3 |
| Non-pCR 5 | 46 | IIA | FEC+DTX | CNB | 2 and 3 | 2 and 3 |
|
|
|
|
| Surgery | 2 and 3 | 2 and 3 |
| Non-pCR 6 | 54 | IIB | FEC+PTX | CNB | 2 and 3 | 2 and 3 |
|
|
|
|
| Surgery | 2 and 3 | 2 and 3 |
| Non-pCR 7 | 58 | IIB | FEC+PTX | CNB | 2 and 3 | 2 and 3 |
|
|
|
|
| Surgery | 2 and 3 | 2 and 3 |
| Non-pCR 8 | 44 | IIIB | FEC | CNB | NA | NA |
|
|
|
|
| Surgery | 0 | 1 |
| Non-pCR 9 | 33 | IIB | FEC+DTX | CNB | NA | NA |
|
|
|
|
| Surgery | 1 | 2 and 3 |
| Non-pCR 10 | 78 | IIIB | FEC+DTX | CNB | NA | NA |
|
|
|
|
| Surgery | 1 | 2 and 3 |
| Non-pCR 11 | 51 | IIIB | FEC+DTX | CNB | NA | NA |
|
|
|
|
| Surgery | 2 and 3 | 1 |
| Non-pCR 12 | 45 | IIA | FEC+PTX | CNB | 2 and 3 | 2 and 3 |
|
|
|
|
| Surgery | 2 and 3 | 2 and 3 |
| Non-pCR 13 | 52 | IIB | FEC+PTX | CNB | 2 and 3 | 2 and 3 |
|
|
|
|
| Surgery | 2 and 3 | 2 and 3 |
| pCR 1 | 40 | IIB | FEC+PTX | CNB | 2 and 3 | 1 |
|
|
|
|
| Surgery | NA | NA |
| pCR 2 | 50 | IIB | FEC+PTX | CNB | 2 and 3 | 2 and 3 |
|
|
|
|
| Surgery | NA | NA |
| pCR 3 | 51 | IIB | FEC+DTX | CNB | 2 and 3 | 0 |
|
|
|
|
| Surgery | NA | NA |
Next, we assessed the impact of CADM1 and CADM4
expression on therapeutic responses. Among the 13 non-pCR cases,
one exhibited negative CADM1 expression and two showed weak
positive CADM1 expression (score 1) in the surgical specimens,
while two cases showed weak positive CADM4 expression (score 1)
(Table II). Since there were no
available resected tumor specimens for IHC staining in three pCR
cases, the evaluation of therapeutic responses according to CADM1
and CADM4 expression levels between non-pCR and pCR cases was
impossible. Therefore, the CNB specimens were used for this
comparison (Fig. 1B). While all three
pCR cases showed strong positive CADM1 expression (score 2 and 3),
two cases showed weak positive CADM1 expression (score 1) and 7
cases showed strong CADM1 expression (score 2 and 3) in non-pCR
cases (P=1) (Table III). On the
other hand, while one weak positive (score 1) and one strong
positive (score 2 and 3) CADM4 expression in pCR cases, two cases
showed weak positive CADM1 expression (score 1) and 7 cases showed
strong CADM4 expression (score 2 and 3) in non-pCR cases (P=0.49).
Although we have expected that loss of or weak positive CADM1 or
CADM4 expression may affect therapeutic response, our results lead
to no common tendencies.
 | Table III.Comparison of CADM1 and CADM4
expression at initial diagnosis in non-pCR and pCR TNBC
patients. |
Table III.
Comparison of CADM1 and CADM4
expression at initial diagnosis in non-pCR and pCR TNBC
patients.
|
| CADM1 score in CNB
specimen |
| CADM4 score in CNB
specimen |
|---|
|
|
|
|
|
|---|
| Case | 1 | 2 and 3 |
P-valuea | Case | 1 | 2 and 3 |
P-valuea |
|---|
| non-pCR (n=9) | 2 | 7 | 1.00 | non-pCR (n=9) | 2 | 7 | 0.49 |
| pCR (n=3) | 0 | 3 |
| pCR (n=2) | 1 | 1 |
|
Discussion
Inactivated expression of CADM1 and CADM4 has been
correlated with local invasion, lymph node metastasis,
lymphovascular invasion (15) and
poor prognosis in breast cancer (18,24).
Therefore, it is considered that CADM1 and CADM4 suppress tumor
development in cases of breast cancer. However, we previously
evaluated CADM1 and CADM4 expression in breast cancer and found
that those expression levels were less frequently decreased in TNBC
cases than in non-TNBC cases (20).
In the present study, we investigated further into CADM1 and CADM4
expression levels in both CNB and surgical specimens in TNBC
patients who received PST. This analysis allowed us to predict
therapeutic efficacy using the pair samples of pre- and post-PST.
As a result, we revealed that loss or weak positive expression of
CADM1 was frequently observed in non-pCR patients. That loss of
CADM1 and CADM4 was less associated with tumorigenesis in the TNBC
cases than with that in the non-TNBC cases suggests that TNBC is a
unique subtype of breast cancer, which requires further
examination.
Approximately 80% of TNBC is classified as
basal-like breast cancer, which is one of the intrinsic subtypes as
categorized by the gene expression profile (4,5).
Basal-like breast cancer is different from other intrinsic
subtypes, such as luminal A, luminal B, and HER2 overexpressing
breast cancer. Therefore, breast cancer should be considered to be
a heterogeneous disease, and TNBC should also be considered to be a
unique subtype of breast cancer. In addition, TNBC exhibits a high
level of genomic instability, resulting in a high level of
intratumor heterogeneity (6), and is
categorized into several distinct subtypes (25,26).
Recently, next-generation sequencing (NGS) has progressed
significantly and, by using the NGS method, we have revealed gene
aberration profiles of breast tumors in adolescent and young adult
females (27). Noteworthy, the NGS
has also revealed intratumor heterogeneity and accelerated the
understanding of clonal evolution in cancer (28,29).
Phylogenic trees, which were constructed based on clonal (truncal)
and subclonal (branched) mutation analyses, uncovered highly
important issues regarding malignant tumors. For example, genomic
sequencing from different regions of a tumor may sometimes lead to
different mutation profiles. When this situation occurs in driver
genes, it results in different treatment strategy selection
(30). Protein expression may also
have the same issue, suggesting that it is not possible to analyze
data without considering intratumoral heterogeneity. Therefore, we
compared CADM1 and CADM4 expression levels between CNB and surgical
specimens. Ultrasound-guided CNB was performed to obtain breast
cancer tissue samples for pathological diagnosis at the initial
diagnosis. After PST, the tumor tissue was resected and compared
with a CNB specimen. In our study, both CADM1 and CADM4 expression
levels in the CNB specimens were almost consistent with those in
the surgical specimens.
The present study has several limitations. First, it
is a retrospective study with a small sample size. Second, although
we suggest that CADM1 has an anti-PST role in TNBC, no biological
functions are proposed. Further large scale and functional studies
are required.
Acknowledgements
Not applicable.
Funding
This work was supported by a research grant from
Japanese Society of Strategies for Cancer Research and Therapy
(JSCT) and the JSPS KAKENHI grant no. 17k10556.
Availability of data and materials
All data generated or analyzed during this study are
included within the manuscript.
Authors' contributions
YK, KK and MS designed the study. YK, MS, KS, AG,
and YM performed immunohistochemical staining and analyzed the
data. NA and TO collected clinical information. YK, MS and KK wrote
the manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients, and the Institutional Review Board of Fukushima Medical
University, Japan approved the present study. Each patient provided
written informed consent for the publication of any data and
associated images.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Berruti A, Generali D, Bertaglia V, Brizzi
MP, Mele T, Dogliotti L, Bruzzi P and Bottini A: Intermediate
endpoints of primary systemic therapy in breast cancer patients. J
Natl Cancer Inst Monogr. 2011:142–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Berruti A, Generali D, Kaufmann M, Puztai
L, Curigliano G, Aglietta M, Gianni L, Miller WR, Untch M, Sotiriou
C, et al: International expert consensus on primary systemic
therapy in the management of early breast cancer: Highlights of the
Fourth Symposium on Primary Systemic Therapy in the Management of
Operable Breast Cancer, Cremona, Italy (2010). J Natl Cancer Inst
Monogr. 2011:147–151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gralow JR, Burstein HJ, Wood W, Hortobagyi
GN, Gianni L, von Minckwitz G, Buzdar AU, Smith IE, Symmans WF,
Singh B and Winer EP: Preoperative therapy in invasive breast
cancer: Pathologic assessment and systemic therapy issues in
operable disease. J Clin Oncol. 26:814–819. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weigelt B, Baehner FL and Reis-Filho JS:
The contribution of gene expression profiling to breast cancer
classification, prognostication and prediction: A retrospective of
the last decade. J Pathol. 220:263–280. 2010.PubMed/NCBI
|
|
5
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Turner NC and Reis-Filho JS: Tackling the
diversity of triple-negative breast cancer. Clin Cancer Res.
19:6380–6388. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ito A, Ichiyanagi N, Ikeda Y, Hagiyama M,
Inoue T, Kimura KB, Sakurai MA, Hamaguchi K and Murakami Y:
Adhesion molecule CADM1 contributes to gap junctional communication
among pancreatic islet α-cells and prevents their excessive
secretion of glucagon. Islets. 4:49–55. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Murakami Y: Functional cloning of a tumor
suppressor gene, TSLC1, in human non-small cell lung cancer.
Oncogene. 21:6936–6948. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goto A, Niki T, Chi-Pin L, Matsubara D,
Murakami Y, Funata N and Fukayama M: Loss of TSLC1 expression in
lung adenocarcinoma: Relationships with histological subtypes, sex
and prognostic significance. Cancer Sci. 96:480–486. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hui AB, Lo KW, Kwong J, Lam EC, Chan SY,
Chow LS, Chan AS, Teo PM and Huang DP: Epigenetic inactivation of
TSLC1 gene in nasopharyngeal carcinoma. Mol Carcinog. 38:170–178.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Honda T, Tamura G, Waki T, Jin Z, Sato K,
Motoyama T, Kawata S, Kimura W, Nishizuka S and Murakami Y:
Hypermethylation of the TSLC1 gene promoter in primary gastric
cancers and gastric cancer cell lines. Jpn J Cancer Res.
93:857–860. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jansen M, Fukushima N, Rosty C, Walter K,
Altink R, Heek TV, Hruban R, Offerhaus JG and Goggins M: Aberrant
methylation of the 5′ CpG island of TSLC1 is common in pancreatic
ductal adenocarcinoma and is first manifest in high-grade PanlNs.
Cancer Biol Ther. 1:293–296. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fukuhara H, Kuramochi M, Fukami T,
Kasahara K, Furuhata M, Nobukuni T, Maruyama T, Isogai K, Sekiya T,
Shuin T, et al: Promoter methylation of TSLC1 and tumor suppression
by its gene product in human prostate cancer. Jpn J Cancer Res.
93:605–609. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Steenbergen RD, Kramer D, Braakhuis BJ,
Stern PL, Verheijen RH, Meijer CJ and Snijders PJ: TSLC1 gene
silencing in cervical cancer cell lines and cervical neoplasia. J
Natl Cancer Inst. 96:294–305. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takahashi Y, Iwai M, Kawai T, Arakawa A,
Ito T, Sakurai-Yageta M, Ito A, Goto A, Saito M, Kasumi F and
Murakami Y: Aberrant expression of tumor suppressors CADM1 and 4.1B
in invasive lesions of primary breast cancer. Breast Cancer.
19:242–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kuramochi M, Fukuhara H, Nobukuni T, Kanbe
T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura
T, et al: TSLC1 is a tumor-suppressor gene in human non-small-cell
lung cancer. Nat Genet. 27:427–430. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nagata M, Sakurai-Yageta M, Yamada D, Goto
A, Ito A, Fukuhara H, Kume H, Morikawa T, Fukayama M, Homma Y and
Murakami Y: Aberrations of a cell adhesion molecule CADM4 in renal
clear cell carcinoma. Int J Cancer. 130:1329–1337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jang SM, Sim J, Han H, Ahn HI, Kim H, Yi
K, Jun YJ, Rehman A, Chung MS, Jang K and Paik SS:
Clinicopathological significance of CADM4 expression in invasive
ductal carcinoma of the breast. J Clin Pathol. 66:681–686. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jang SM, Han H, Jun YJ, Jang SH, Min KW,
Sim J, Ahn HI, Lee KH, Jang KS and Paik SS: Clinicopathological
significance of CADM4 expression, and its correlation with
expression of E-cadherin and Ki-67 in colorectal adenocarcinomas. J
Clin Pathol. 65:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saito M, Goto A, Abe N, Saito K, Maeda D,
Ohtake T, Murakami Y and Takenoshita S: Decreased expression of
CADM1 and CADM4 are associated with advanced stage breast cancer.
Oncol Lett. 15:2401–2406. 2018.PubMed/NCBI
|
|
21
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sobin LH, Gospodarowicz M and Wittekind
Ch: International Union Against Cancer (UICC) TNM Classification of
Malignant Tumors. 7th edition. Wiley-Blackwell; Oxford, UK:
2010
|
|
23
|
Saito M, Matsuzaki M, Sakuma T, Katagata
N, Watanabe F, Yamaguchi Y, Schetter AJ, Takenoshita S and Nomizu
T: Clinicopathological study of non-palpable familial breast cancer
detected by screening mammography and diagnosed as DCIS. Breast
Cancer. 21:140–145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wikman H, Westphal L, Schmid F, Pollari S,
Kropidlowski J, Sielaff-Frimpong B, Glatzel M, Matschke J, Westphal
M, Iljin K, et al: Loss of CADM1 expression is associated with poor
prognosis and brain metastasis in breast cancer patients.
Oncotarget. 5:3076–3087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abramson VG, Lehmann BD, Ballinger TJ and
Pietenpol JA: Subtyping of triple-negative breast cancer:
Implications for therapy. Cancer. 121:8–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Abramson VG and Mayer IA: Molecular
heterogeneity of triple negative breast cancer. Curr Breast Cancer
Rep. 6:154–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanke Y, Shimomura A, Saito M, Honda T,
Shiraishi K, Shimada Y, Watanabe R, Yoshida H, Yoshida M, Shimizu
C, et al: Gene aberration profile of tumors of adolescent and young
adult females. Oncotarget. 9:6228–6237. 2017.PubMed/NCBI
|
|
28
|
Greaves M and Maley CC: Clonal evolution
in cancer. Nature. 481:306–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang J, Fujimoto J, Zhang J, Wedge DC,
Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, et al: Intratumor
heterogeneity in localized lung adenocarcinomas delineated by
multiregion sequencing. Science. 346:256–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
de Bruin EC, McGranahan N, Mitter R, Salm
M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan
AJ, et al: Spatial and temporal diversity in genomic instability
processes defines lung cancer evolution. Science. 346:251–256.
2014. View Article : Google Scholar : PubMed/NCBI
|