Introduction
Cervical cancer is the second most common cause of
cancer-related deaths in women worldwide (1). Each year, approximately 500,000 women
are diagnosed with cervical cancer, and 270,000 will die from it
(1). Patients with advanced cervical
cancer are treated with surgery, radiation therapy, chemotherapy,
or a combination of these strategies. However, the prognosis of
advanced cervical cancer remains poor, with no significant
improvements in the overall treatment outcome over the last three
decades (2). Cervical cancer is
caused by high-risk human papillomavirus (HPV) infection (3–5).
Specifically, persistent infection of the basal epithelial cells of
the cervix by high-risk HPV causes the integration of the HPV
genome into the host chromosome, which leads to the expression of
the HPV-E6 oncoprotein that inactivates the tumor suppressor gene
p53, resulting in tumor formation (6,7). Thus,
suppressing the expression of E6 may lead to the treatment of
cervical cancer. Furthermore, the expression of E6 is limited to
cervical lesions, with no expression in healthy tissue, including
the cervix (8). Therefore, specific
targeting of E6 expression is likely a safe strategy that would
spare normal tissue.
CRISPR-Cas9 is one of the genome editing
technologies that has gained much attention in recent years. It
induces double stranded breaks (DSBs) at specific target DNA
locations by the combined action of a single guide RNA (sgRNA) that
recognizes a specific DNA sequence and the Cas9 nuclease that
induces DSBs (9). During the repair
of these DSBs by non-homologous end joining, gene mutations often
occur in the form of base insertions or deletions. Expression of a
gene can be knocked out if these mutations occur in the coding
region of the gene (10). Thus,
CRISPR-Cas9 can be used to induce the specific knockout of E6
expression in cervical cancer cells. However, application of such a
strategy in clinical practice requires an effective vector that
enables gene transfer into targeted cervical cancer cells.
Adeno-associated virus (AAV) vector is an attractive
option for gene transfer as it is derived from a non-pathogenic
virus and can induce gene transfer in non-dividing cells (11). Previous studies have demonstrated
prolonged transgene expression and clinical benefits following the
direct administration of the AAV vector into humans (12). Several serotypes of AAV vectors with
varying gene transfer efficacy for specific tissues and organs have
been reported. For example, AAV serotypes 1 and 7, 2 and 3, 5, and
8 are effective in gene transfer in the skeletal muscles, nerves,
and liver, respectively (13). We
previously reported that the AAV type 2 vector was the most
effective in inducing gene transfer into cervical cancer cells
(14).
There are no treatment strategies for cervical
cancer that target high-risk HPV E6. In the present study, we
performed in vitro and in vivo experiments to develop
a CRISPR-Cas9-based, effective, and highly specific therapy
targeting high-risk HPV E6 for cervical cancer.
Materials and methods
Cell culture
HPV 18-positive human cervical cancer cell lines
(HeLa, HCS-2, SKG-I), and human immortal cell line 293 were
purchased from the Japanese Collection of Research Bioresources
Cell Bank (JCRB, Osaka, Japan). These cells were cultured in
Dulbecco's Modified Eagle Medium/F12 (DMEM/F12; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10%
inactivated fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and 1% penicillin/streptomycin (Thermo Fisher
Scientific, Inc.) at 37°C under 5% CO2.
Construction of the plasmid
vector
The part of the CMV promotor and Cas9 which was
tagged with human influenza hemagglutinin (HA) were cut out from
the SpeI and XbaI site of pRGEN-Cas9-CMV (Toolgen, Seoul, South
Korea) and inserted into the SpeI and XbaI sites of pCMV-IRES-bsr
(15) to produce the Cas9-expression
vector (pCMV-Cas9-HA-IRES-bsr).
Optimized CRISPR Design (crispr.mit.edu/) was used to search for the HPV18 E6
sequence to be targeted by CRISPR. The sequence with the highest
score was selected. Two DNA oligonucleotides,
5′-CACCGGAGCTTGTAGGGTCGCCGTGT-3′ and
5′-AAACACACGGCGACCCTACAAGCTC-3′, were annealed and inserted at the
BsaI site of pRGEN-U6-sgRNA (Toolgen) to produce a vector
expressing an sgRNA targeting E6 (sgE6) (pRGEN-U6-sgE6). Two
inverted terminal repeats (ITRs) cleaved from pWlacZ (16) were then added to the vector to produce
pW-U6-sgE6. Two ITRs cleaved from pWlacZ were also added to
pRGEN-U6-sgRNA to produce pW-U6-sgRNA, which was used as a control
vector.
Establishment of Cas9-expressing
cervical cancer cell lines
HeLa, HCS-2, and SKG-I cell lines were transfected
with pCMV-Cas9-HA-IRES-bsr using Lipofectamine LTX and Plus Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Transfected cells
were selected by culturing in cell culture media containing 10
µg/ml of Blasticidin S Hydrochloride (Funakoshi, Tokyo, Japan) to
collect single colonies.
Cell growth curve
Tumor cells were plated onto 96-well plates (500
cells/well), and 10 µl of Premix WST-1 Cell Proliferation Assay
System (Takara Bio Inc., Tokyo, Japan) was added to each well every
24 h. Absorption was measured at 450 nm 24 h after premix added
using SpectraMax 190 (Molecular Devices, LLC, Sunnyvale, CA, USA)
to produce a cell growth curve.
AAV vector preparation
sgE6-containing and control AAV vectors were
prepared by transfecting the 293 cells with 3 plasmids, including
pW-U6-sgE6, or pW-U6-sgRNA, adenovirus helper plasmid (16), and AAV type 2 helper plasmid (17,18) via
calcium phosphate transfection, and cells were collected 72 h
later. Cells were then exposed to three freeze-thaw cycles to
obtain the recombinant AAV vectors. The solution containing the
vectors was purified by cesium chloride density gradient
centrifugation, and the vector titer was measured by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) as
described previously (19).
Detection of mutations in the E6
genome
Cas9-expressing cells were seeded onto 6-well plates
at a concentration of 5×104 cells/well, and were
transduced 24 h later with AAV-sgE6 (1×105 viral genomes
(vg)/cell). Cells were collected by trypsin 48 h later, and DNA was
extracted according to the protocol using the QIAamp®
DNA Mini kit (Qiagen GmbH, Hilden, Germany). The extracted DNA was
used as a template to perform PCR using TaKaRa Ex Taq Hot Start
version (Takara Bio Inc.) and PTC-100 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) to amplify the E6 genome. The following primers
were used for the reaction: Forward: 5′-GGGAGTGACCGAAAACGGTC-3′,
reverse: 5′-GTGTTTCTCTGCGTGTTGT-3′. PCR was carried out using 40
cycles of heating at 95°C for 30 sec, 56°C for 30 sec, and 75°C for
30 sec. The PCR product was cloned according to the protocol using
the Mighty TA-cloning kit (Takara Bio Inc.), and Sanger sequencing
was performed using the Applied Biosystems 3730×l DNA Analyzer
(Thermo Fisher Scientific, Inc.).
T7 Endonuclease 1 (T7E1) assay
Tumor cells were seeded onto 6-well plates at
5×104 cells/well, incubated for 24 h, and transduced
with either AAV-sgE6 or AAV-sgNC at 1×105 vg/cell. Cells
were collected by trypsin 48 h later, and DNA was extracted
according to the protocol using the QIAamp® DNA Mini
kit. The extracted DNA was used as a template to amplify the E6
genome. Using the thermal cycler PTC-100, PCR products were
denatured for 2 min at 95°C, then cooled to 30°C over 10 min, and
annealed. Double-strand DNA was reacted with T7 Endonuclease
(M0302S; New England BioLabs, Inc., Ipswich, MA, USA) at 37°C for
20 min. Electrophoresis was performed with a 0.8% agarose gel, and
imaged using the BioDoc-It® Imaging System (UVP, Inc.,
Upland, CA, USA).
RT-qPCR
Tumor cells were seeded onto 6-well plates at
5×104 cells/well, incubated for 24 h, and transduced
with either AAV-sgE6 or AAV-sgNC at 1×105 vg/cell. Cells
were collected by trypsin 48 h later, and DNA was extracted
according to the protocol for the QIAamp® DNA mini kit.
qPCR was performed according to the protocol using the Thermal
Cycler Dice Real Time System II (Takara Bio Inc.). The PCR was
carried out using 40 cycles of heating at 95°C for 15 s, 58°C for
15 s, and 72°C for 20 s.
Mutations in the E6 genome were measured by qPCR
using a protocol adopted from a previous report (20). Primers with the following sequences
were used: Mut primer forward: 5′-TTTGAGGATCCAACACGGCGA-3′. Mut
primer reverse: 5′-GTCTTGCAGTGAAGTGCTCAG-3′. CTL primer forward:
5′-GTGCCRGAAACCGTTGAATCC-3′. CTL primer reverse:
5′-CCAGCTATGTTGTGAAATCGTCG-3′. To assess the validity of this
assay, plasmid vectors containing non-mutated and mutated E6, as
identified by Sanger sequencing, were used as templates to perform
qPCR using two pairs of primers. Furthermore, the plasmid vectors
mixed at varying ratios (10:0, 9:1, 8:2, 5:5, 2:8, 1:9, 0:10) were
used as templates for qPCR to evaluate the quantitative ability of
the assay. qPCR results were analyzed using the relative
quantification (RQ) value (21), and
the mutation rate was calculated as (1-RQ) ×100 (%).
Western blot analysis
Tumor cells were seeded onto 6-well plates at
5×104 cells/well, incubated for 24 h, and transduced
with either AAV-sgE6 or AAV-sgNC at 1×105 vg/cell. Cells
were lysed 48 h later using lysis buffer (1% NP-40, 150 mM NaCl, 50
mM Tris-HCl, pH 8.0), and extracted proteins were mixed with 1%
sodium dodecyl sulfate (SDS) sample buffer (10 mM Tris-HCl, pH 7.5,
150 mM NaCl, 1% SDS, EDTA-free Protease inhibitor cocktail (Roche,
Basel, Switzerland), separated by electrophoresis using 5% (for HA,
Rb) or 12.5% (for E6, p53, actin) polyacrylamide gels, and
transferred onto polyvinylidene fluoride (PVDF) membranes (Merck
KGaA, Darmstadt, Germany). Membranes were left at room temperature
for 1 h in PVDF Blocking Reagent for Can Get Signal®
(Toyobo Life Science, Osaka, Japan), washed three times using
Tris-buffered saline-Tween-20 (TBS-T), and incubated overnight with
the following antibodies at room temperature in Can Get
Signal® Immunoreaction Enhance Solution 1 (Toyobo Life
Science): Anti-HA-probe antibody (cat. no. sc-7392; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), anti-HPV18E6 monoclonal
antibody (cat. no. sc-365089; Santa Cruz Biotechnology, Inc.),
anti-p53 monoclonal antibody (cat. no. sc-126; Santa Cruz
Biotechnology, Inc.), anti-Rb monoclonal antibody (cat. no. 9313;
Cell Signaling Technology, Inc., Danvers, MA, USA), and anti-actin
polyclonal antibody (cat. no. A2066; Sigma-Aldrich; Merck KGaA).
After the reaction, membranes were washed three times with TBS-T,
and incubated with peroxidase-labeled anti-mouse or anti-rabbit
antibodies (GE Healthcare Japan, Tokyo, Japan) in Can Get
Signal® Immunoreaction Enhance Solution 2 (Toyobo Life
Science) at room temperature for 1 h. Membranes were then washed
three times with TBS-T, incubated with ECL prime western blotting
detection reagent (GE Healthcare Japan), and imaged using a cooled
CCD system (LAS-4000mini: GE Healthcare Japan).
Apoptosis
Cells were seeded onto 12-well plates at
5×104 cells/well, incubated for 24 h, and transduced
with either AAV-sgE6 or AAV-sgNC at 1×105 vg/cell. The
Apoptotic/Necrotic cells detection kit (PromoKine, Heidelberg,
Germany) was used as per its protocol 24 h after the transduction,
and FITC-Annexin V-positive cells were imaged using an inverted
microscope (IX73l; Olympus Corporation, Tokyo, Japan). The number
of FITC-positive cells was counted per high power field (HPF).
In vitro cell growth
Cells were seeded onto 96-well plates at 50
cells/well, and were transduced with either AAV-sgE6 or AAV-sgNC at
0–1×107 vg/cell. Premix WST-1 cell proliferation assay
system (Takara Bio Inc.) was added at 10 µl/well 96 h after the
transduction, and the absorbance was measured at 450 nm 24 h later
using SpectraMax 190 (Molecular Devices, LLC).
In vivo experiments
Six to seven-week-old (15–20 g) Balb/c nude mice
(CLEA Japan, Inc., Tokyo, Japan) were used for the in vivo
experiments (n=8). SKG-I was selected among the three cervical
cancer cell lines as it has been reported to induce tumors in nude
mice. A total of 5×106 Cas9-expressing SKG-I cells
(SKG-I/Cas9) were injected under the dorsal skin of nude mice.
Simultaneously, 2×1012 vg AAV-sgE6 or AAV-sgNC were
injected once in the same location. Tumor size was measured with
calipers twice a week, and the tumor volume was calculated as
length × width2 ×1/2. All mice were sacrificed by
cervical dislocation when the diameter of tumors in the AAV-sgNC
group reached over 20 mm. Changes in body weight were measured, and
gross observation of the injection site was performed at the
experimental endpoint. DNA was extracted from tumors in both groups
using the QIAamp DNA mini kit (Qiagen GmbH), and the E6 mutation
rate was measured by qPCR as described above. Multiple tumors were
not observed in the present study.
Mice were grown under specific pathogen-free
conditions. The experimental protocol was approved by the Jichi
Medical University Ethics Committee (Tochigi, Japan), and strictly
followed the National and Institutional Guidelines for animal
experiments.
Statistical analysis
Statistical analysis was performed using SPSS v22
(IBM Corp., Armonk, NY, USA). Student's t-test was used to compare
two groups. One-way analysis of variance with Bonferroni post-hoc
test was performed to compare multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Development of Cas9-expressing
cervical cancer cell lines
In order to confirm that the Cas9 is introduced, we
performed western blots for transfected cells using an anti-HA
antibody. Cas9 protein was expressed only in those cells that
received the Cas9 gene (Fig. 1A),
confirming that Cas9-expressing cervical cancer cell lines
HeLa/Cas9, HCS-2/Cas9, and SKG-I/Cas9 were established.
Cell growth curve
In order to assess the effect of Cas9 on cell growth
we compared the growth of wild type and Cas9-expressing cervical
cancer cell lines in vitro. As shown in Fig. 1B-D, there was no significant
difference in cell growth between wild type and Cas9-expressing
cells, indicating that Cas9 expression had no impact on cell growth
in vitro.
Detection of E6 gene mutations
The E6 gene was sequenced by Sanger sequencing for
HeLa/Cas9 cells 48 h after AAV-sgE6 transduction. Several base
insertions and deletions were identified mostly around the targeted
region (Fig. 2A), indicating that
genome editing was achieved for E6 in vitro by
CRISPR-Cas9.
 | Figure 2.Detection of mutations in the E6
gene. (A) Detection of mutations in the E6 gene in Cas9-expressing
HeLa cells by Sanger sequencing performed 48 h following AAV-sgE6
transduction. Detection of mutations in the E6 gene in
Cas9-expressing cervical cancer cells by the T7 Endonuclease 1
assay performed 48 h following AAV-sgE6 transduction. Horizontal
lines indicate base deletions. (B) HeLa/Cas9, (C) HCS-2/Cas9 and
(D) SKG-I/Cas9. White arrows indicate non-cleaved products; black
arrows indicate cleaved products. Validation was performed using
qPCR to measure the CRISPR-Cas9-induced E6 mutation rate. The E6
mutation rate was measured in Cas9-expressing cervical cancer cells
48 h following AAV-sgE6 transduction. (E) Mutation rates were
calculated based on the RQ. The mutation rate was calculated as:
(1-RQ) ×100 (%). (F) Wild type and mutated plasmid vectors were
mixed at varying ratios (10:0, 9:1, 8:2, 5:5, 2:8, 1:9 and 0:10)
and used as templates. Mutation rates were calculated based on the
RQ for different mixtures of plasmid vectors. DNA extracted from
the Cas9-expressing cervical cancer cells 48 h following AAV-sgE6
transduction was used as a template for qPCR. The rate of E6
mutations in (G) HeLa/Cas9. The rate of E6 mutations in (H)
HCS-2/Cas9 and (I) SKG-I/Cas9. Results are expressed as the mean ±
standard deviation. *P<0.05, as indicated. PAM,
protospacer-adjacent motif; sgE6, E6 target sequence; wt,
wild-type; qPCR, quantitative polymerase chain reaction; 2 bp ins,
E6 gene with a 2 bp insertion; 13 bp del, E6 gene with a 13 bp
deletion; RQ, relative quantification; AAV, adeno-associated virus;
Cas9, CRISPR associated protein 9; sg, single guide; PAM,
protospacer adjacent motif; NC, negative control. |
T7E1 assay
Products cleaved by T7E1 were detected only in cells
transduced with AAV-sgE6 (Fig. 2B-D).
Thus, gene mutations were introduced into the E6 gene by
E6-targeting CRISPR-Cas9.
Measurement of mutation rates by
qPCR
qPCR was performed using mutated plasmid vectors as
templates, and demonstrated a significant increase in mutation
rates (Fig. 2E). In addition,
mutation rates increased with an increasing proportion of mutated
plasmid vector, and this rate was equivalent to the proportion of
the mutated vectors in the plasmid mixture (Fig. 2F).
The rate of E6 mutation was measured using the same
method. Mutation rates were estimated to be 82% for HeLa/Cas9, 77%
for HCS-2/Cas9, and 87% for SKG-I/Cas9 (Fig. 2G-I). Mutations were not found in
untransduced cells or cells transduced with AAV-sgNC (Fig. 2G-I).
Western blot
E6 expression was detected in untransduced cells or
cells transduced with AAV-sgNC. These cells did not express
p53. On the other hand, cells transduced with AAV-sgE6 had
significantly decreased expression of E6, and significantly
increased expression of p53 (Fig.
3A). These results indicated that E6 was knocked out
effectively by CRISPR-Cas9 targeting of E6, consequently increasing
the expression of p53 at the protein level.
Apoptosis
Cells transduced with AAV-sgE6 had a significantly
higher number of FITC-Annexin V-positive cells (46.0±5.0/HPF) than
untransduced cells (1.0±1.4/HPF) and cells transduced with AAV-sgNC
(0.8±1.3/HPF) (Fig. 3B and C). Thus,
E6-targeting CRISPR-Cas9 induced apoptosis in tumor cells in
vitro.
In vitro cell growth
For all cell lines, the number of live cells
decreased with an increase in AAV-sgE6 vector dose (Fig. 3D). Thus, E6-targeting CRISPR-Cas9
suppressed tumor cell growth in a dose-dependent manner in
vitro.
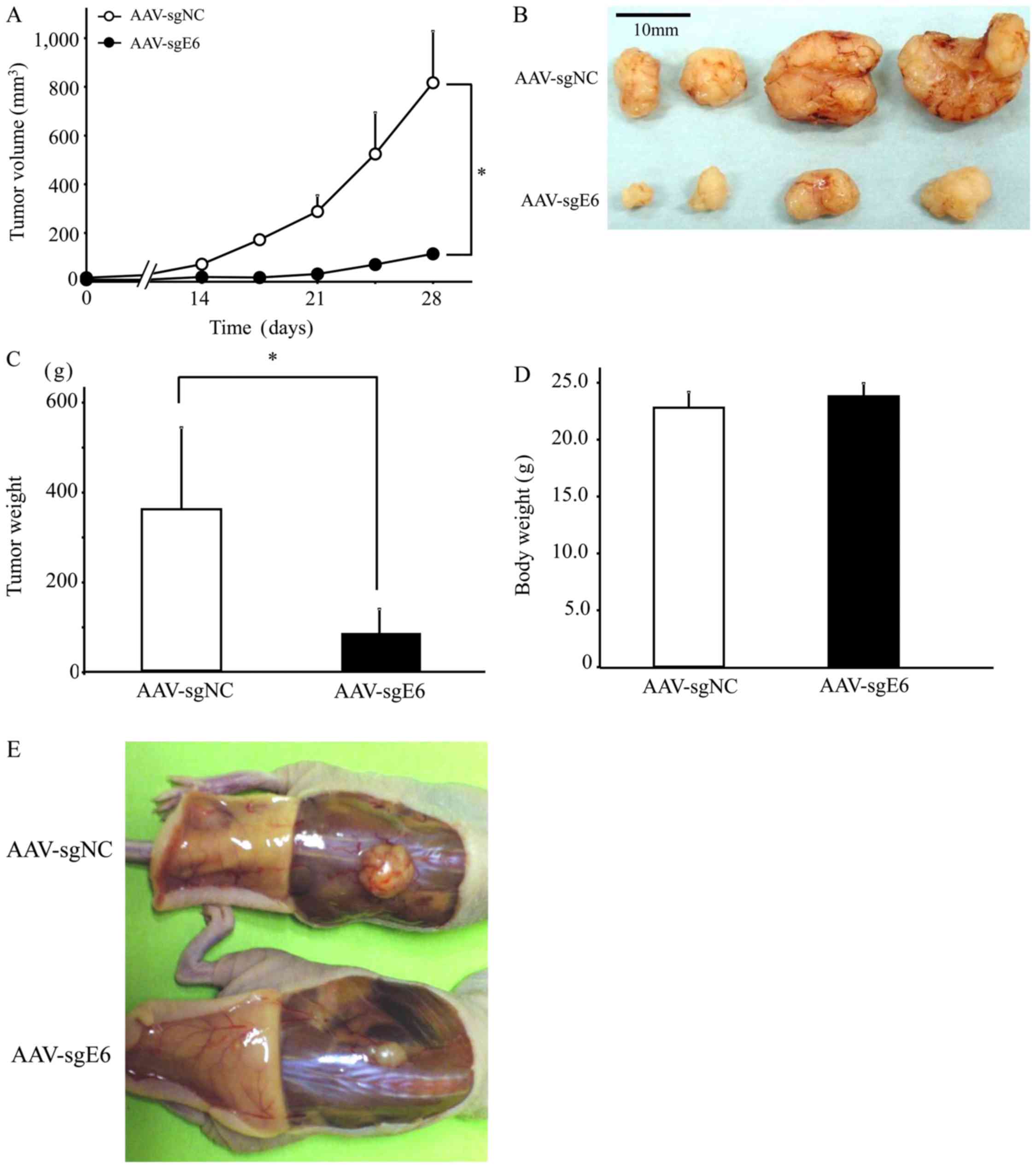
In vivo experiments
Tumor growth was suppressed significantly in mice
injected with AAV-sgE6 as compared with those injected with
AAV-sgNC (Fig. 4A). At the
experimental endpoint of 42 days after the injection, tumors in the
AAV-sgE6 group were significantly smaller than those in the
AAV-sgNC group, at 114±60 and 817±114 mm3, respectively
(n=4/group, P<0.05; Fig. 4B and
C). Thus, E6-targeting CRISPR-Cas9 suppressed tumor growth
in vivo. In addition, mutations in the E6 gene were not
identified in the remaining tumors (data not shown). There was no
significant difference in the body weights of mice measured prior
to sacrifice (Fig. 4D), there were no
abnormal findings in the subcutaneous tumor area (Fig. 4E).
Discussion
In the present study, we sought to develop an
effective and highly specific therapeutic approach for cervical
cancer by targeting high-risk HPV E6 using CRISPR-Cas9 and the AAV
vector. We previously performed screening to identify AAV vector
serotypes that have the highest efficiency for gene transfer into
cervical cancer cells, and demonstrated that the AAV serotype 2
vector was the most effective (14).
Based on this data, we selected AAV type 2 as the vector in the
present study. We first constructed the Cas9-expressing AAV vector
and observed that Cas9 was not transferred into cervical cancer
cells (data not shown). The length of genomic sequence that can be
packaged into the AAV vector is limited to approximately 5 kb
(22). Vector production efficiency
significantly decreases if longer sequences are utilized. The
length of Cas9 used in this study was approximately 4.2 kb and
exceeded 5 kb when the promoter and PolyA sequences were included;
thus, it may not have been transferred effectively using the AAV
vector. Therefore, as an alternative to using the AAV vector to
introduce Cas9, we established cervical cancer cell lines that
constitutively express Cas9 to perform our study. We demonstrated
that the growth rate of Cas9-expressing cells was not different
from that of wild type cells, confirming that Cas9 expression does
not impact the growth of cervical cancer cells. As Cas9 knock-in
mice that express Cas9 ubiquitously did not show any phenotypic
changes (23), the expression of Cas9
might not have any influence on mammalian cells.
We then constructed the sgRNA-expressing AAV vector
to target E6. We used an online service to select the target E6
sequence, and constructed the AAV vector (AAV-sgE6) that expresses
the sgRNA containing the target sequence driven by the U6 promoter.
Transduction of AAV-sgE6 into Cas9-expressing cervical cancer cells
resulted in multiple mutations in E6 as detected by Sanger
sequencing. This suggested that E6-targeting by CRISPR-Cas9 may
induce mutations in the E6 gene in cervical cancer cells.
The T7E1 assay is the most commonly used method for
the detection of mutations induced by CRISPR-Cas9. Using this
assay, we observed a new band indicating the presence of mutations
in AAV-sgE6-infected cells. That said, the T7E1 assay is complex to
set up as it involves multiple steps, and is not quantitative.
Thus, qPCR was used to detect the mutation rate (20) by mismatch PCR. A primer pair (Mut
primers) of which a part of the 3′ end of one primer corresponds to
a part of the target E6 sequence was constructed. In addition,
another primer pair (CTL primers) was constructed such that it
corresponded to another sequence distant from the target E6
sequence. When the Mut primers are used for qPCR, amplification
efficiency is significantly reduced due to mismatches caused by the
mutations in the sequence induced by CRISPR-Cas9. On the other
hand, amplification efficiency is not affected by the presence of
mutations when CTL primers are used. Thus, by comparing the RQ
value of qPCR using Mut and CTL primers, it is possible to quantify
the mutation rate as well as detect mutations in the target
sequence. We used the plasmid vector carrying the E6 sequence with
mutations identified by Sanger sequencing to validate this method.
Our data demonstrated a significant reduction in RQ in the Mut
primer group when qPCR was performed using the mutated plasmid
vector as a template. Furthermore, we performed qPCR using a
mixture of plasmid vectors as a template to examine whether the
assay is quantitative. The template was produced by mixing vectors
that carry non-mutated and mutated E6 sequences at different
ratios. Our data revealed that the rate of RQ reduction in the Mut
primer group relative to the CTL group was equivalent to the
proportion of the mutated vectors in the plasmid mixture. This
finding validated the qPCR method as a quantitative method to
detect CRISPR-Cas9-induced mutations in E6 and to quantify the rate
of mutations. We used this method to detect mutations in E6 and
determined the mutation rate in Cas9-expressing cells transduced
with AAV-sgE6. E6 mutations were identified in all cell lines, and
the frequency of these mutations was significantly higher than that
expected with the T7E1 assay.
Next, we examined the expression of E6 and related
factors using western blotting, and found decreased expression of
E6, and increased expression of p53 in all three cervical cancer
cell lines with AAV-sgE6. Furthermore, using Annexin V as a target,
we observed that apoptosis was induced in these cells. In high-risk
HPV-positive cervical cancer, p53 has no mutations (24,25), and
is normally degraded by E6 (6,7) resulting
in the inhibition of apoptosis. Our data suggest that the
suppression of E6 expression by Cas9 and AAV-sgE6 led to the
alteration of this pathway, resulting in the induction of
apoptosis.
We subsequently examined the effects of E6-targeting
CRISPR-Cas9 on the growth of cervical cancer cells in vitro,
and demonstrated that AAV-sgE6 suppressed tumor cell growth in a
dose-dependent manner. Based on this in vitro observation,
we examined the effects of E6-targeting CRISPR-Cas9 in an in
vivo model of cervical cancer, and found that tumor growth was
suppressed significantly in mice injected with AAV-sgE6 as compared
with those with AAV-sgNC. Using the qPCR method described above, we
found no mutations in E6 in DNA extracted from tumors in the
AAV-sgE6 group. This suggests that the remaining tumors may not
have been transduced by the vector, or may have been formed by
tumor cells that did not undergo genome editing following gene
delivery. Thus, in order to enhance the efficacy of this approach,
it may be necessary to increase the amount of vectors or its
frequency of injection, and/or package multiple sgRNAs targeting
several E6 sequences. A recent study revealed the presence of
KIAA0319L, a receptor essential for AAV transduction of human cells
(26). Use of this receptor may
improve the transduction efficiency of AAV vectors.
Previous in vitro and ex vivo studies
reported that E6 expression can be knocked out effectively in
cervical cancer by genome editing techniques (27–30). In
addition, in vivo experiments, the effect that E6 knockout
enhances sensitivity to an anticancer drug has been reported
(31). In the present study, we
further demonstrated its high tumor-suppressing effects in
vivo using a combination of CRISPR-Cas9, a relatively simple
and effective genome editing technique, and the AAV type 2 vector,
which has high transduction efficiency for cervical cancer
cells.
With respect to side effects, weight loss was not
observed in mice injected with AAV-sgE6 and there were no abnormal
findings in the tissue around the injection site. The lack of
apparent side effects may be because 1) AAV vectors are not
pathogenic, and 2) E6-targeting CRISPR-Cas9 did not affect the
normal tissue as E6 is only expressed in cervical cancer cells.
Thus, our findings indicate that E6-targeting CRISPR-Cas9 is a safe
therapeutic approach that does not affect normal tissue.
In order to translate the E6-targeting CRISPR-Cas9
approach into clinical practice, its effects may be tested
initially in patients with precancerous lesions, such as CIN 2/3,
or in those with early stage invasive cancers. Currently, surgical
approaches, including cervical conization and total hysterectomy,
are the only options to treat patients in these early stages of
cancer development (32,33). Cervical conization is often selected
for younger patients to preserve their fertility (34). However, compared with healthy
individuals, the risk of cervical cancer is still higher by 4-times
after cervical conization (35).
Thus, there are concerns associated with the low curability of
cervical conization. Furthermore, patients who undergo cervical
conization often develop reproductive or perinatal complications,
such as miscarriage and premature birth, due to the removal of a
portion of the endocervical canal and consequent infections in the
uterine cavity (34). Thus, there is
a need to develop an alternative, effective approach that is less
invasive. As these precancerous and early stage lesions are
localized, our approach may be the most effective strategy.
Moreover, as young individuals and those who desire to have
children are most affected by cervical cancer, treatment must be
safe for these individuals. Although we did not observe any
apparent side effects, further investigation is needed to ensure
that the CRISPR-Cas9 approach can be safely performed for future
clinical translation.
The effects of the CRISPR-Cas9 approach are
localized, and therefore unlikely to be curative for advanced
cervical cancer patients with widespread invasion and metastasis.
However, it may be effective in shrinking bulky tumors, and thus
may be used as an alternative to neoadjuvant chemotherapy to
facilitate surgery and to increase the efficacy of radiation
therapy. Furthermore, this approach may be applied to some
recurrent tumors that are localized.
We demonstrated the successful transfer of
E6-targeting sgRNA into cervical cancer cells by the AAV vector.
Cas9 derived from Streptococcus pyogenes (SpCas9) was used
in this study because it has been used the most for genome editing
since its first report in 2013. As the protospacer adjacent motif
(PAM) sequences for SpCas9 (NGG) that determine the target sequence
are relatively short, it allows for selection of many target
sequences. However, as described above, packaging of SpCas9 into
the AAV vector is challenging due to its low packaging capacity. A
recent study reported that Cas9 from Staphylococcus aureus
(SaCas9) can be packaged effectively into the AAV vector due to its
shorter length (3.3 kb) (36). This
suggests that it is possible to package both SaCas9 and sgRNA into
the AAV vector, which, in theory, should enable genome editing and
E6-targeted therapy for cervical cancer. We are currently
constructing an AAV vector containing SaCas9 and E6-targeting sgRNA
to test its effects on cervical cancer cells.
In conclusion, we report that the CRISPR-Cas9
approach targeting high-risk HPV E6 may be a highly selective and
effective therapeutic strategy for cervical cancer.
Acknowledgements
The authors would like to thank Mrs. Miyoko Mitsu,
Mrs. Satomi Fujiwara, Mrs. Akemi Takada (Division of Genetic
Therapeutics, Center for Molecular Medicine, School of Medicine,
Jichi Medical University, Tochigi, Japan) and Mrs. Michiko Ohashi
(Department of Obstetrics and Gynecology, School of Medicine, Jichi
Medical University, Tochigi, Japan) for providing excellent
technical assistance.
Funding
The present study was partly supported by the JMU
Graduate Student Research Award.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TY performed experiments, analyzed the data and
wrote the manuscript. YS conceived and designed the study,
performed experiments and the analyzed data. MU and RU analyzed the
experimental data and provided guidance during the present study.
SM and HF contributed to the interpretation of the data and revised
the manuscript. HM conceived and designed the study, and critically
revised the manuscript for important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol was approved by the Jichi
Medical University Ethics Committee (Tochigi, Japan), and strictly
followed the National and Institutional Guidelines for animal
experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Woodman CB, Collins SI and Young LS: The
natural history of cervical HPV infection: Unresolved issues. Nat
Rev Cance. 7:11–22. 2007. View
Article : Google Scholar
|
|
4
|
Boshart M, Gissmann L, Ikenberg H,
Kleinheinz A, Scheurlen W and zur Hausen H: A new type of
papillomavirus DNA, its presence in genital cancer biopsies and in
cell lines derived from cervical cancer. EMBO J. 3:1151–1157. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Durst M, Gissmann L, Ikenberg H and zur
Hausen H: A papillomavirus DNA from a cervical carcinoma and its
prevalence in cancer biopsy samples from different geographic
regions. Proc Natl Acad Sci USA. 80:3812–3815. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Narisawa-Saito M and Kiyono T: Basic
mechanisms of high-risk human papillomavirus-induced
carcinogenesis: Roles of E6 and E7 proteins. Cancer Sci.
98:1505–1511. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moody CA and Laimins LA: Human
papillomavirus oncoproteins: Pathways to transformation. Nat Rev
Cancer. 10:550–560. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Da Silva DM, Eiben GL, Fausch SC,
Wakabayashi MT, Rudolf MP, Velders MP and Kast WM: Cervical cancer
vaccines: Emerging concepts and developments. J Cell Physiol.
186:169–182. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jinek M, Chylinski K, Fonfara I, Hauer M,
Doudna JA and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hsu PD, Lander ES and Zhang F: Development
and applications of CRISPR-Cas9 for genome engineering. Cell.
157:1262–1278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mueller C and Flotte TR: Clinical gene
therapy using recombinant adeno-associated virus vectors. Gene
Ther. 15:858–863. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grieger JC and Samulski RJ:
Adeno-associated virus vectorology, manufacturing, and clinical
applications. Methods Enzymol. 507:229–254. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Z, Asokan A and Samulski RJ:
Adeno-associated virus serotypes: Vector toolkit for human gene
therapy. Mol Ther. 14:316–327. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sato N, Saga Y, Uchibori R, Tsukahara T,
Urabe M, Kume A, Fujiwara H, Suzuki M, Ozawa K and Mizukami H:
Eradication of cervical cancer in vivo by an AAV vector that
encodes shRNA targeting human papillomavirus type 16 E6/E7. Int J
Oncol. 52:687–696. 2018.PubMed/NCBI
|
|
15
|
Urabe M, Hasumi Y, Ogasawara Y, Matsushita
T, Kamoshita N, Nomoto A, Colosi P, Kurtzman GJ, Tobita K and Ozawa
K: A novel dicistronic AAV vector using a short IRES segment
derived from hepatitis C virus genome. Gene. 200:157–162. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsushita T, Elliger S, Elliger C,
Podsakoff G, Villarreal L, Kurtzman GJ, Iwaki Y and Colosi P:
Adeno-associated virus vectors can be efficiently produced without
helper virus. Gene Ther. 5:938–945. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mochizuki S, Mizukami H, Kume A, Muramatsu
S, Takeuchi K, Matsushita T, Okada T, Kobayashi E, Hoshika A and
Ozawa K: Adenoassociated virus (AAV) vector-mediated liver- and
muscle-directed transgene expression using various kinds of
promoters and serotypes. Gene Ther Mol Biol. 8:9–18. 2004.
|
|
18
|
Xiao W, Chirmule N, Berta SC, McCullough
B, Gao G and Wilson JM: Gene therapy vectors based on
adeno-associated virus type 1. J Virol. 73:3994–4003.
1999.PubMed/NCBI
|
|
19
|
Ishiwata A, Mimuro J, Mizukami H,
Kashiwakura Y, Takano K, Ohmori T, Madoiwa S, Ozawa K and Sakata Y:
Liver-restricted expression of the canine factor VIII gene
facilitates prevention of inhibitor formation in factor
VIII-deficient mice. J Gene Med. 11:1020–1029. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu C, Zhang Y, Yao S and Wei Y: A PCR
based protocol for detecting indel mutations induced by TALENs and
CRISPR/Cas9 in zebrafish. PLoS One. 9:e982822014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dong JY, Fan PD and Frizzell RA:
Quantitative analysis of the packaging capacity of recombinant
adeno-associated virus. Hum Gene Ther. 7:2101–2112. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech
L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et
al: CRISPR-Cas9 knockin mice for genome editing and cancer
modeling. Cell. 159:440–455. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Crook T, Wrede D, Tidy JA, Mason WP, Evans
DJ and Vousden KH: Clonal p53 mutation in primary cervical cancer:
Association with human-papillomavirus-negative tumours. Lancet.
339:1070–1073. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Paquette RL, Lee YY, Wilczynski SP,
Karmakar A, Kizaki M, Miller CW and Koeffler HP: Mutations of p53
and human papillomavirus infection in cervical carcinoma. Cancer.
72:1272–1280. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pillay S, Meyer NL, Puschnik AS, Davulcu
O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS
and Carette JE: An essential receptor for adeno-associated virus
infection. Nature. 530:108–112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu Z, Yu L, Zhu D, Ding W, Wang X, Zhang
C, Wang L, Jiang X, Shen H, He D, et al: Disruption of HPV16-E7 by
CRISPR/Cas system induces apoptosis and growth inhibition in HPV16
positive human cervical cancer cells. Biomed Res Int.
2014:6128232014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu L, Wang X, Zhu D, Ding W, Wang L, Zhang
C, Jiang X, Shen H, Liao S, Ma D, et al: Disruption of human
papillomavirus 16 E6 gene by clustered regularly interspaced short
palindromic repeat/Cas system in human cervical cancer cells. Onco
Targets Ther. 8:37–44. 2014.PubMed/NCBI
|
|
29
|
Zhen S, Hua L, Takahashi Y, Narita S, Liu
YH and Li Y: In vitro and in vivo growth suppression of human
papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9.
Biochem Biophys Res Commun. 450:1422–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kennedy EM, Kornepati AV, Goldstein M,
Bogerd HP, Poling BC, Whisnant AW, Kastan MB and Cullen BR:
Inactivation of the human papillomavirus E6 or E7 gene in cervical
carcinoma cells by using a bacterial CRISPR/Cas RNA-guided
endonuclease. J Virol. 88:11965–11972. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhen S, Lu JJ, Wang LJ, Sun XM, Zhang JQ,
Li X, Luo WJ and Zhao L: In vitro and in vivo synergistic
therapeutic effect of cisplatin with human papillomavirus16 E6/E7
CRISPR/Cas9 on cervical cancer cell line. Transl Oncol. 9:498–504.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sevin BU, Nadji M, Averette HE, Hilsenbeck
S, Smith D and Lampe B: Microinvasive carcinoma of the cervix.
Cancer. 70:2121–2128. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Massad LS, Einstein MH, Huh WK, Katki HA,
Kinney WK, Schiffman M, Solomon D, Wentzensen N and Lawson HW: 2012
ASCCP Consensus Guidelines Conference: 2012 updated consensus
guidelines for the management of abnormal cervical cancer screening
tests and cancer precursors. Obstet Gynecol. 121:829–846. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bevis KS and Biggio JR: Cervical
conization and the risk of preterm delivery. Am J Obstet Gynecol.
205:19–27. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Melnikow J, McGahan C, Sawaya GF, Ehlen T
and Coldman A: Cervical intraepithelial neoplasia outcomes after
treatment: Long-term follow-up from the British Columbia Cohort
Study. J Natl Cancer Inst. 101:721–728. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ran FA, Cong L, Yan WX, Scott DA,
Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et
al: In vivo genome editing using Staphylococcus aureus Cas9.
Nature. 520:186–191. 2015. View Article : Google Scholar : PubMed/NCBI
|