Introduction
Gastric cancer is one of the most common types of
malignancy globally (1). Improvements
in diagnosis and treatment have resulted in greater long-term
survival for patients with early gastric cancer; however, the
prognosis for patients with advanced gastric cancer remains
relatively poor, with locally advanced and widespread metastatic
disease associated with a high mortality rate (2). Despite the need for an improved
understanding on how gastric cancer metastasizes, the mechanisms
underlying gastric cancer progression remain unclear.
Gastric cancer has the ability to spread locally via
direct invasion through the gastric wall into adjacent tissue; it
then has the ability to metastasize to regional lymph nodes and
distant organs through the lymphatic and venous vessels. Recent
evidence has emerged suggesting that acquisition of invasiveness in
cancer cells is accompanied by the loss of epithelial features and
the gain of a mesenchymal phenotype, in a process termed the
epithelial-mesenchymal transition (EMT) (3).
A variety of molecules have been identified in tumor
samples as EMT-associated markers, including the epithelial marker
E-cadherin (4) and the mesenchymal
marker Vimentin (5).
Transcriptional factors, including the zinc-finger protein Snail
(6), the basic helix-loop-helix
protein Twist (7) and the
E-box-binding protein ZEB1/2 (8,9), are also
involved in regulating EMT-associated changes. It has been
previously reported that matrix metalloproteinases serve key roles
in the invasion and metastasis of gastric cancer (10–13).
Expression of the actin-binding protein L-plastin in
SW480 cells was recently demonstrated to cause a downregulation of
E-cadherin and an increase in invasiveness (14). Furthermore, it was previously reported
that the Plastin3 (PLS3) gene, which is known to code for an
actin bundling protein that inhibits cofilin-mediated
depolymerization of actin, may be used as a marker for circulating
tumor cells; PLS3 has been demonstrated to serve an
important role in the EMT of colorectal carcinoma cells (15). However, there are few reports to data
regarding the clinical significance of PLS3 in epithelial
malignancies.
In the present study, the clinical significance of
PLS3 in gastric cancer was identified as an independent
prognostic factor and the association between the EMT markers
Cadherin1 (CDH1), Vimentin and PLS3 was indicated.
Furthermore, it was demonstrated in an in vitro study that
PLS3 serves a key role in invasion and migration in gastric
cancer.
Materials and methods
Clinical samples and cell lines
Primary gastric carcinoma and adjacent normal
gastric epithelial tissues were obtained from a total of 163
patients who underwent gastric resection, without preoperative
treatment, at The Oita Prefectural Hospital (Oita, Japan) or The
Kyushu University Beppu Hospital (Beppu, Japan) between 1993 April
and 2003 December. All of the obtained tissue samples were cut and
immediately placed into RNAlater® (Takara Bio, Inc.,
Otsu, Japan), then frozen in liquid nitrogen and kept at −80°C
until RNA extraction. Written informed consent was obtained from
all of the patients and the protocol of the study was approved by
the Ethics Committee of Kyushu University (Fukuoka, Japan).
Clinicopathological information, including age (range, 40–87
years), sex, pathology, differentiation and tumor-node metastasis
classification, was available for all patients. The tumor size was
calculated under the microscope by pathologists. The median overall
survival (OS) time was 32 months and ranged from 1–144 months.
Human gastric cancer cell lines NUGC3, NUGC4, MKN7,
MKN45, MKN74 and AGS were provided by the Cell Resource Centre for
Biomedical Research Institute of Development, Aging and Cancer,
Tohoku University (Sendai, Japan). All cells were grown in
RPMI-1640 medium (Cambrex Corporation, East Rutherford, NJ, USA),
supplemented with 10% foetal bovine serum (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany), 2 mmol/l glutamine, 100 U/ml penicillin
and 100 µg/ml streptomycin (Cambrex Corporation); this was followed
by incubation at 37°C in a humidified chamber with 5%
CO2.
Immunohistochemistry
Immunohistochemical studies of PLS3 were performed
on formalin-fixed paraffin-embedded surgical sections (5-µm thick)
obtained from 43 patients with gastric cancer treated at the Kyushu
University Beppu Hospital (Beppu, Japan). Tissue sections were
deparaffinised and boiled in 0.01 mol/l sodium citrate buffer (pH
9.0; 0.01 ml of 1 M disodium citrate and 99.99 ml of 1 M trisodium
citrate) in a microwave for 10 min at 500 W for antigen retrieval
and then washed in phosphate-buffered saline (PBS). The primary
antibody used was goat anti-PLS3 (C15; cat no. sc-16555; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), which was diluted to a ratio
of 1:100 and incubated with the slides for 15 min at room
temperature. All tissue sections were immunohistochemically stained
with the avidin-biotin-peroxidase LASB2 kit (cat. no. K0675; Dako;
Agilent Technologies, Inc., Santa Clara, CA, USA) according to the
protocol of the manufacturer, and were counterstained with
haematoxylin at room temperature for 3 min. Representative
photomicrographs were captured with a Nikon Eclipse E800 microscope
equipped with a Nikon DXM1200 digital camera (Nikon instruments,
Melville, NY, USA). Slides were observed under light microscopy at
×40 magnification, and five regions were selected for every slide
at random. The degree of differentiation was independently
evaluated by two of the authors (Dr Junji Kurashige and Dr Kosuke
Mima) using a blinded protocol design, with the observers having no
knowledge of the clinical outcome or any other clinicopathological
data. For evaluation of the PLS3 expression, the staining intensity
was scored as 0 (negative), 1 (weak), 2 (medium) or 3 (strong) as
previously indicated (16).
Total RNA isolation and first-strand
cDNA synthesis
Total RNA was isolated from frozen tissue samples
and cultured cell lines using a modified
acid-guanidine-phenol-chloroform method as described previously
(15). The purity and concentration
of all RNA samples were evaluated using an absorbance ratio of
260/280 nm with a NanoDrop™ ND-1000 spectrophotometer
(NanoDrop Technologies; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Total RNA was reverse transcribed with M-MLV RT,
according to the manufacturer's protocol (Invitrogen; Thermo Fisher
Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The expression levels of PLS3, CDH1 and
Vimentin were determined using a LightCycler® 480
Probes Master kit (Roche Molecular Diagnostics, Pleasanton, CA,
USA), according to the manufacturer's instructions. The primer
sequences used in RT-qPCR were as follows: PLS3 forward
5′-GAAACTTACACCCTTCATCATTCAG-3′, reverse
5′-TTCTGCACCAATGTTCACAAC-3′ and universal probe no. 29; CDH1
forward 5′-GGCCAGGAAATCACATCCTA-3′, reverse
5′-GGCAGTGTCTCTCCAAATCC-3′ and universal probe no. 36;
Vimentin forward 5′-TACAGGAAGCTGCTGGAAGG-3′, reverse
5′-ACCAGAGGGAGTGAATCCAG-3′ and universal probe no. 13; GAPDH
forward 5′-AGCCACATGCTCAGACAC-3′, reverse
5′-AATACGACCAAATCCGTTGACT-3′ and universal probe no. 60. All
RT-qPCR reactions were run in a LightCycler® 480 system
II (Roche Molecular Diagnostics). The relative amounts of PLS3,
CDH1 and Vimentin were measured using the
2−∆∆Cq method (17) and
normalized to GAPDH. All RT-qPCR reactions were performed in
triplicate.
PLS3 RNA interference
Small interfering RNAs (siRNA) against PLS3
no. 1 (5′-CAGCAACGGAUUCAUUUGUTT-3′) and 2
(5′-CUGCUUAGAUGGGCAAACUTT-3′), along with control non-targeting
siRNA were obtained from Ambion (Thermo Fisher Scientific, Inc.).
The non-silencing control siRNA, which has no sequence homology to
any known human gene sequence, was used as a control with
non-specific effects for all experiments. The siRNA oligomer was
diluted with Opti-MEM I without serum (Invitrogen; Thermo Fisher
Scientific, Inc.). The diluted siRNA oligomer was mixed with
diluted Lipofectamine® RNAiMAX (Invitrogen; Thermo
Fisher Scientific, Inc.), and incubated for 15 min at room
temperature to allow siRNA-Lipofectamine® RNAiMAX
complexes to form. The optimal amount of siRNA used for
transfection was determined to be 20 nmol/l. Diluted logarithmic
growth phase NUGC3 and AGS cells without antibiotics were seeded at
a density of 2×105 cells/well in a final volume of 2.5
ml into 6-well flat-bottom microtiter plates. The cells were
incubated in a humidified atmosphere (37°C and 5% CO2).
The assay was performed after a 72-h incubation.
Migration and invasion assay
Cell migration was assessed using The BD
Falcon™ FluoroBlok™ 24 Multiwell Insert
system (BD Bioscience, San Jose, CA, USA). For a period of 24 h
prior to the assay, cells were transfected with PLS3 siRNA
or a negative control siRNA. The NUGC3 and AGS cells
(1.0×104) were placed in the upper chamber of a 24-well
plate with serum-free RPMI-1640 medium. The lower chamber was
filled with 750 µl medium with 10% foetal bovine serum, in which
the serum acted as a chemoattractant, and the cell migration plate
was incubated in a humidified atmosphere (37°C and 5%
CO2). After a 48 h incubation, the upper chamber was
transferred into a second 24-well plate containing 500 µl/well of 4
µg/ml calcein AM in HBSS (Invitrogen; Thermo Fisher Scientific,
Inc.) and incubated for 1 h (37°C and 5% CO2). Invasive
cells that migrated through the membrane were evaluated using a
fluorescence plate reader at excitation/emission wavelengths of
485/535 nm. Each independent experiment was performed three times.
NUGC3 cell invasion was assessed using the BD BioCoat™
Tumor Invasion system 24-Multiwell (BD Bioscience, San Jose, CA,
USA) and the same procedure as that used for migration.
Statistical analysis
All experiments were performed ≥3. Continuous
variables were expressed as the mean ± standard deviation. The
association between the expression of PLS3 and the patient
clinicopathological characteristics was analysed using a Student's
t-test or χ2 analysis. The OS time curves were plotted
according to the Kaplan-Meier method and the generalized log-rank
test was applied to compare the survival curves. P<0.05 was
considered to indicate a statistically significant difference. All
tests were performed using JMP® software 8th edition
(SAS Institute, Inc., Cary, NC, USA).
Results
Immunohistochemistry of PLS3
expression
Expression of the PLS3 protein was evaluated by
immunohistochemical analysis of resected gastric cancer specimens
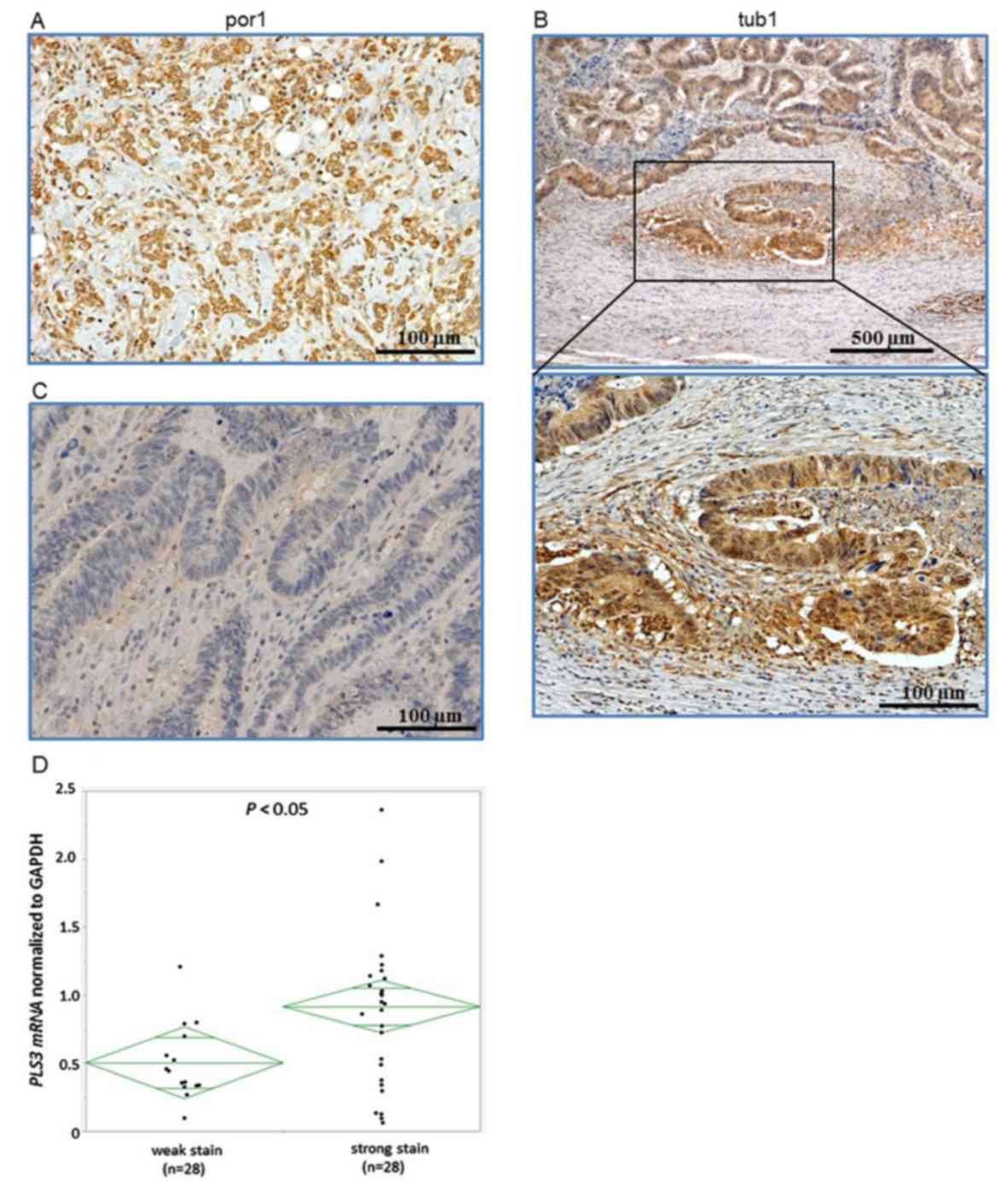
using the anti-PLS3 antibody. PLS3 staining was notably strong in
poorly differentiated gastric cancer tissues (Fig. 1A) and in the budding foci in well
differentiated gastric cancer (Fig.
1B). On the other hand, PLS3 staining was weak in
differentiated gastric cancer tissues (Fig. 1C). Overall, the staining was primarily
located within the cytoplasm.
PLS3 mRNA expression and
clinicopathological characteristics
Expression levels of PLS3 were examined in
163 gastric cancer clinical samples using RT-qPCR, with quantified
values used to calculate PLS3/GAPDH ratios. In the specimens
with strong staining, the mRNA expression levels of PLS3
were significantly higher compared with that in the weak staining
group (P<0.05; Fig. 1D). The 163
patients with gastric cancer were divided into two groups according
to the median PLS3 mRNA expression level: 82 of the cases
were placed in the high PLS3 expression group, and the
remaining 81 cases were placed into the low PLS3 expression
group. The association between patient clinicopathological
characteristics and PLS3 expression is summarized in
Table I. PLS3 expression was
significantly associated with cancer differentiation (well and
moderately differentiated vs. poorly differentiated and others;
P=0.010), depth of tumour invasion (T1 and T2 vs. T3 and T4;
P=0.029), distant metastasis (present vs. absent; P=0.010) and
cancer staging (stage I and II vs. III and IV; P=0.002). By
contrast, there was no significant association between PLS3
expression and age (P=0.143), sex (P=0.953), lymph node metastasis
(present vs. absent; P=0.825), venous invasion (present vs. absent;
P=0.788) and peritoneal metastasis (present vs. absent;
P=0.900).
 | Table I.PLS3 mRNA expression and
clinicopathological features. |
Table I.
PLS3 mRNA expression and
clinicopathological features.
|
|
| PLS3 mRNA
expression |
|
|---|
|
|
|
|
|
|---|
| Features | Total (n=163) | High (n=82)
(%) | Low (n=81) (%) | P-value |
|---|
| Age (years) |
|
|
| 0.143 |
| Mean ±
SD | 65.9±11.2 | 64.6±10.9 | 67.2±11.9 |
|
| Sex |
|
|
| 0.953 |
|
Male | 105 | 53 (64.6) | 52 (64.2) |
|
|
Female | 58 | 29 (35.4) | 29 (35.8) |
|
|
Differentiation |
|
|
| 0.010a |
|
Well/moderately | 76 | 30 (36.6) | 46 (56.8) |
|
|
Poorly/others | 87 | 52 (63.4) | 35 (43.2) |
|
| Depth of tumor
invasion |
|
|
| 0.029a |
|
T1-2 | 59 | 23 (28.0) | 36 (44.4) |
|
|
T3-4 | 104 | 59 (72.0) | 45 (55.6) |
|
| Lymph node
metastasis |
|
|
| 0.825 |
|
Absent | 57 | 28 (34.1) | 29 (35.8) |
|
|
Present | 106 | 54 (65.9) | 52 (64.2) |
|
| Lymphatic
invasion |
|
|
| 0.788 |
|
Absent | 58 | 30 (36.6) | 28 (34.6) |
|
|
Present | 105 | 52 (63.4) | 53 (65.4) |
|
| Venous
invasion |
|
|
| 0.900 |
|
Absent | 120 | 60 (73.2) | 60 (74.1) |
|
|
Present | 43 | 22 (26.8) | 21 (25.9) |
|
| Peritoneal
metastasis |
|
|
| 0.806 |
|
Absent | 138 | 69 (84.1) | 67 (82.7) |
|
|
Present | 27 | 13 (15.9) | 14 (17.3) |
|
| Distant
metastasis |
|
|
| 0.010a |
|
Absent | 150 | 71 (86.6) | 79 (97.5) |
|
|
Present | 13 | 11 (13.4) | 2 (2.5) |
|
| Stage |
|
|
| 0.002a |
|
I–II | 93 | 37 (45.1) | 56 (69.1) |
|
|
III–IV | 70 | 45 (54.9) | 25 (30.9) |
|
Survival analysis for the expression
of PLS3
Analysis of the 5-year OS revealed that the high
PLS3 expression group had a significantly poorer prognosis
compared with the low expression group (P=0.012; Fig. 2). In a univariate Cox regression
analysis, the high PLS3 expression group displayed
significantly higher overall mortality [hazard ratio (HR), 2.211;
95% confidence interval (CI), 1.299–3.764; P=0.012; Table II] compared with the low PLS3
expression group. Variables with P<0.05 from univariate analysis
were selected for multivariate analysis with the Cox's proportional
hazard model. In a multivariate Cox regression analysis for OS
time, including tumour depth, tumour size, lymph node metastasis,
venous invasion, peritoneal metastasis and PLS3 expression,
high PLS3 expression was indicated to be an independent
prognostic factor (HR, 1.770; 95% CI, 1.020–3.074; P=0.042;
Table II).
 | Table II.Univariate and multivariate analysis
of clinicopathological features for 5-year OS (Cox's proportional
regression model). |
Table II.
Univariate and multivariate analysis
of clinicopathological features for 5-year OS (Cox's proportional
regression model).
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Features | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age years
(>65/65≥) | 0.765 | 0.460–1.272 | 0.301 |
|
|
|
| Sex
(male/female) | 0.893 | 0.513–1.553 | 0.688 |
|
|
|
| Differentiation
(well, moderate/poor, other) | 1.375 | 0.820–2.305 | 0.228 |
|
|
|
| T (1,2/3,4) | 3.239 | 1.681–3.514 |
<0.001a | 1.301 | 0.575–2.944 | 0.577 |
| Tumor size
(<50/≥50 mm) | 2.068 | 1.225–3.490 | 0.007a | 0.835 | 0.445–1.570 | 0.527 |
| Lymph node
metastasis (absent/present) | 4.387 | 2.149–8.958 |
<0.001a | 2.876 | 1.318–6.274 | 0.008a |
| Venous invasion
(absent/present) | 3.386 | 2.013–5.696 |
<0.001a | 1.734 | 0.975–3.086 | 0.061a |
| Lymphatic invasion
(absent/present) | 3.748 | 1.835–7.656 |
<0.001a |
|
|
|
| Peritoneal
metastasis (absent/present) | 6.644 | 3.706–11.912 |
<0.001a | 3.553 | 1.873–6.740 |
<0.001a |
| Stage (I, II/III,
IV) | 5.460 | 3.096–9.629 |
<0.001a |
|
|
|
| PLS3
expression (low/high) | 2.211 | 1.299–3.764 | 0.012a | 1.770 | 1.020–3.074 | 0.042a |
Inhibition of PLS3 gene expression
with PLS3 siRNA in human gastric cancer cells
Knockdown of PLS3 mRNA was performed in NUGC3
and AGS gastric cancer cells, which have the highest PLS3
mRNA expression levels among eight different gastric cancer cell
lines examined (NUGC3, NUGC4, MKN7, MKN45, MKN74, AGS) (data not
shown). Evaluation of the expression levels following PLS3
siRNA transfection confirmed that PLS3 mRNA expression in
cells transfected with PLS3 siRNA no. 1 and 2 was lower
compared with that in cells transfected with the negative control
siRNA (Fig. 3A). The transfected
cells were used to investigate whether PLS3 knockdown
altered migration and invasion. NUGC3 and AGS cells transfected
with PLS3 siRNA exhibited reduced migratory abilities
(P<0.05; Fig. 3B), and NUGC cells
were significantly less invasive (P<0.05; Fig. 3C), compared with those cells
transfected with the negative control siRNA. These results indicate
that knockdown of PLS3 suppressed the migration and invasion
of gastric cancer cells in vitro.
Association between CDH1 and Vimentin
mRNA expression, and PLS3 mRNA expression in gastric cancer
The association between the expression of
PLS3, and that of CDH1 and Vimentin in
patients with gastric cancer was investigated. The 163 gastric
cancer cases were classified into high and low expression groups,
according to the median PLS3 mRNA expression levels
determined by RT-qPCR. As presented in Fig. 4A, Vimentin mRNA expression
levels were significantly increased in the high PLS3
expression group (n=82), compared within the low PLS3
expression group (n=81; P<0.01); however, there was no
significant difference in CDH1 mRNA expression between the
high and low PLS3 expression groups. Similarly,
Vimentin mRNA was significantly decreased in NUGC3 cells
transfected with siRNA no. 1 and 2, compared with the negative
control (Fig. 4B); however,
CDH1 mRNA expression was not significantly different in
NUGC3 cells transfected with siRNA compared with the negative
control group (Fig. 4B).
Discussion
In the present study, it was revealed that the
patients with a high PLS3 expression had a significantly
poorer prognosis compared with those with a low PLS3
expression. In addition, PLS3 expression was associated with
cancer differentiation, depth of tumour invasion and distant
metastasis. Furthermore, it was demonstrated that there was a
significant positive association between PLS3 and
Vimentin expression in gastric cancer. It was concluded that
downregulation of PLS3 in gastric cancer cells was
associated with the inhibition of cellular invasion and
migration.
The PLS3 gene, located on chromosome Xq23,
encodes a protein that functions to polymerize actin fibers through
the inhibition of cofilin-mediated actin depolymerization (18); however, there is limited knowledge
regarding the biology of PLS3 and its relevance to solid
cancer development and progression. A previous study demonstrated
that colorectal carcinoma cell lines overexpressing PLS3
acquire invasiveness through the downregulation of E-cadherin
(19). Furthermore, supportive
evidence that PLS3 may be involved in EMT via TGFB1 stimulation was
revealed. In addition, the results of the present study
demonstrated that PLS3 is associated with colorectal cancer
metastasis and cancer stemness (15).
In the present study, it was determined that Vimentin
expression is significantly higher in the group with high
PLS3 expression than in the low expression group, and the
downregulation of PLS3 caused a decrease in the expression
of Vimentin mRNA; however, CDH1 exhibited no
significant changes in expression levels between the high and low
PLS3 expression groups. Additionally, it was revealed that
there was a strong association between PLS3 expression and
distant metastasis in gastric cancer. The findings of the present
study propose that PLS3 is a key factor in EMT, which in
turn leads to regulation of Vimentin. Plastin are a recently
described family of actin-binding proteins, which have been
implicated in invasion and metastasis (18). The present study provides the first
evidence for the clinical significance of PLS3 in primary
gastric cancer. Conversely, the present study identified no
significant associations between PLS3 expression and
peritoneal dissemination; however, a strong association between
PLS3 expression and distant metastasis was determined.
Therefore, the results suggest that PLS3 expression may be
used in predicting hematogenous metastasis. Furthermore,
PLS3 was demonstrated to regulate the invasion and migration
in gastric cancer, PLS3-targeted therapy may be more
effective in blocking distant metastasis of gastric cancer.
It was reported that the presence of PLS3 in
peripheral blood in colorectal cancer cases was significantly
associated with disease-free survival and OS time, and
PLS3-expressing cells were detected in the peripheral blood
of ~33% of Japanese patients with colorectal cancer (15). It was expected that the high
PLS3 expression levels in the peripheral blood and bone
marrow of patients with advanced gastric cancer could be detected,
and therefore the expression of PLS3 in preoperative
peripheral blood and bone marrow from the patients with gastric
cancer could be investigated; however, there was no significant
association between PLS3 expression and cancer staging in
the peripheral blood and bone marrow, and there was no significant
difference between gastric cancer and healthy volunteers (data not
presented). The results in the current study revealed that
PLS3 serves an important role in the invasion of gastric
cancer cells in primary tumors; however, there was no
clinicopathological significance to the presence of the PLS3
gene in the circulation in the gastric cancer cases. This
discrepancy may have indicated the differences in hematogenous
pathways of cancer progression between colorectal cells and gastric
cancer cells. PLS3 is not a useful circulating tumour cell
marker in patients with gastric cancer; therefore, the expression
of PLS3 in gastric cancer may be lower than in colorectal
cancer. A conclusive marker to detect circulating tumor cells has
not yet been established for gastric cancer and further studies are
required to identify other novel circulating tumour markers of
gastric cancer.
In conclusion, the present study demonstrated that
high expression levels of PLS3 are independently associated
with poor prognosis, and that PLS3 serves an important role
in EMT through the inhibition of cellular migration and invasion,
and the regulation of Vimentin expression in gastric
cancer.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EMT
|
epithelial-mesenchymal transition
|
|
PLS3
|
plastin3
|
|
CDH1
|
Cadherin1
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
siRNA
|
small interfering RNA
|
|
OS
|
overall survival
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. Cancer J Clin. 61:69–90.
2011. View Article : Google Scholar
|
|
2
|
Lordick F and Siewert JR: Recent advances
in multimodal treatment for gastric cancer: A Review. Gastric
Cancer. 8:78–85. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Graziano F, Mandolesi A, Ruzzo A, Bearzi
I, Testa E, Arduini F, Silva R, Muretto P, Mari D, Berardi R, et
al: Predictive and prognostic role of E-cadherin protein expression
in patients with advanced gastric carcinomas treated with
palliative chemotherapy. Tumour Biol. 25:106–1010. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Otsuki S, Inokuchi M, Enjoji M, Ishikawa
T, Takagi Y, Kato K, Yamada H, Kojima K and Sugihara K: Vimentin
expression is associated with decreased survival in gastric cancer.
Oncol Rep. 25:1235–1242. 2011.PubMed/NCBI
|
|
6
|
Shin NR, Jeong EH, Choi CI, Moon HJ, Kwon
CH, Chu IS, Kim GH, Jeon TY, Kim DH, Lee JH and Park DY:
Overexpression of snail is associated with lymph node metastasis
and poor prognosis in patients with gastric cancer. BMC Cancer.
12:5212012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu AN, Zhu Z-HH, Chang SJ and Hang XS:
Twist expression associated with the epithelial-mesenchymal
transition in gastric cancer. Mol Cell Biochem. 367:195–203. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okugawa Y, Inoue Y, Tanaka K, Kawamura M,
Saigusa S, Toiyama Y, Ohi M, Uchida K, Mohri Y and Kusunoki M: Smad
interacting protein 1 (SIP1) is associated with peritoneal
carcinomatosis in intestinal type gastric cancer. Clin Exp
Metastasis. 30:417–429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Okugawa Y, Toiyama Y, Tanaka K, Matsusita
K, Fujikawa H, Saigusa S, Ohi M, Inoue Y, Mohri Y, Uchida K and
Kusunoki M: Clinical significance of zinc finger E-box binding
homeobox 1 (ZEB1) in human gastric cancer. J Surg Oncol.
106:280–285. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kosaka Y, Mimori K, Fukagawa T, Ishikawa
K, Etoh T, Katai H, Sano T, Watanabe M, Sasako M and Mori M:
Clinical significance of molecular detection of matrix
metalloproteinase-1 in bone marrow and peripheral blood in patients
with gastric cancer. Ann Surg Oncol. 19 Suppl:S430–S437. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mimori K, Fukagawa T, Kosaka Y, Ishikawa
K, Iwatsuki M, Yokobori T, Hirasaki S, Takatsuno Y, Sakashita H,
Ishii H, et al: A large-scale study of MT1-MMP as a marker for
isolated tumor cells in peripheral blood and bone marrow in gastric
cancer cases. Ann Surg Oncol. 15:2934–2942. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mori M, Mimori K, Shiraishi T, Fujie T,
Baba K, Kusumoto H, Haraguchi M, Ueo H and Akiyoshi T: Analysis of
MT1-MMP and MMP2 expression in human gastric cancers. Int J Cancer.
74:316–321. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mimori K, Mori M, Shiraishi T, Fujie T,
Baba K, Haraguchi M, Abe R, Ueo H and Akiyoshi T: Clinical
significance of tissue inhibitor of metalloproteinase expression in
gastric carcinoma. Br J Cancer. 76:531–536. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Foran E, McWilliam P, Kelleher D, Croke DT
and Long A: The leukocyte protein L-plastin induces proliferation,
invasion and loss of E-cadherin expression in colon cancer cells.
Int J Cancer. 118:2098–20104. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yokobori T, Iinuma H, Shimamura T, Imoto
S, Sugimachi K, Ishii H, Iwatsuki M, Ota D, Ohkuma M, Iwaya T, et
al: Plastin3 is a novel marker for circulating tumor cells
undergoing the epithelial-mesenchymal transition and is associated
with colorectal cancer prognosis. Cancer Res. 73:2059–2069. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kurashige J, Watanabe M, Iwatsuki M,
Kinoshita K, Saito S, Nagai Y, Ishimoto T, Baba Y, Mimori K and
Baba H: RPN2 expression predicts response to docetaxel in
oesophageal squamous cell carcinoma. Br J Cancer. 107:1233–1238.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Delanote V, Vandekerckhove J and Gettemans
J: Plastins: Versatile modulators of actin organization in
(patho)physiological cellular processes. Acta Pharmacol Sin.
26:769–779. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Willis ND, Cox TR, Rahman-Casañs SF, Smits
K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M,
Wilson RG, de Bruïne A and Hutchison CJ: Lamin A/C is a risk
biomarker in colorectal cancer. PLoS One. 3:e29882008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edge SB and Compton CC: The american joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|