Introduction
Hepatocellular carcinoma (HCC) is the sixth most
prevalent malignant tumor worldwide, with rising incidence in
recent decades (1,2). The development of surgical techniques
has improved the prognosis of patients with HCC. However, due to
high mortality (3), the overall
survival rate of patients with HCC is still poor (2,4).
Notably, research has revealed that a variety of molecules play key
roles in the initiation and progression of HCC (5). Thus, finding reliable biomarkers to
diagnose HCC and developing novel strategies are required for the
effective treatment of patients with HCC.
Increasingly, evidence has indicated that different
transcription factor (TF) classes play critical roles for tumor
development (6). For example, the
member of forkhead box P (FoxP) family, FoxP1, FoxP2 and FoxP3,
have been revealed to play an important function in HCC (7–12). The
FoxP subfamily which consists of four members (FoxP1-4) belongs to
the Fox superfamily, it has a highly conserved ‘winged-helix’ or
‘fork-head’ (13,14). Fox proteins are involved in cell
cycle progression, apoptosis, proliferation as well as in
senescence and metabolism (15).
However, the function of FoxP4, the other member of the FoxP
family, has yet to be revealed.
Epithelial-mesenchymal transition (EMT) is a complex
process which is closely associated with the metastasis of cancer
cells (16). The main characteristic
of EMT is the transition of cell phenotype, epithelial cells change
their phenotype from epithelial to mesenchymal (16). Previous studies have identified that
several transcription factors are involved in EMT, including Slug,
Snail and Twist1 (17,18). A recent study demonstrated that the
member of the Fox family plays a crucial role in EMT (19).
The aim of the present study was to reveal the role
of FoxP4 in HCC. Its functions in HCCLM3 cells were investigated
with colony formation, CCK-8, wound healing and Transwell invasion
assays. Moreover, ChIP and qChIP as well as a luciferase reporter
assay demonstrated that Slug was a downstream target of FoxP4. In
conclusion, our study revealed that FoxP4 plays a key role in
HCC.
Materials and methods
Cell culture
Human HL-7702 normal liver cell line and human
HCCLM3 cells were purchased from the Shanghai Institute of
Biochemistry and Cell Biology, Shanghai Institutes for Biological
Sciences, Chinese Academy of Sciences (Shanghai, China). Cell lines
were cultured in DMEM (HyClone; GE Healthcare Life Sciences, Logan,
UT, USA) supplemented with 1% penicillin and 1% streptomycin as
well as 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life
Sciences,) and cells were maintained at 37°C in a 5% CO2
atmosphere.
Patients and tissue samples
Human HCC and their adjacent normal liver tissues
(110 pairs) were collected from HCC patients undergoing liver
resections at the Department of Liver Diseases, Rizhao Hospital of
Traditional Chinese Medicine from April 2005 to December 2017. All
tissues samples were collected from patients who had not received
any antitumor therapy and tissue samples were stored in liquid
nitrogen before use. The study was approved by the Ethics Committee
of Rizhao Hospital of Traditional Chinese Medicine and each patient
was well informed and signed informed consent forms.
Cell transfection
Vector (pcDNA3.1) and pcDNA3.1-FoxP4 were purchased
from Vigene Biosciences (Shandong, China). Scramble siRNA (SCR) and
FoxP4 siRNA (siFoxP4) were designed and synthesized by GenePharma
(Shanghai, China). For cell transfection, the cells were
transfected with 2.5 µg plasmid or 50 nM siRNA using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. After transfection for 48
h, the cells were collected and used for the subsequent
experiments.
Real-Time quantitative polymerase
chain reaction (qRT-qPCR) analysis
Total RNA was extracted from tissue samples and
cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to manufacturer's instructions. RNA (2 µg) was
reverse-transcribed into cDNA using the PrimeScript™ RT Reagent
kit. qPCR was performed using SYBR Green (Roche Diagnostics,
Mannheim, Germany). The thermocycling conditions were as follows:
95°C for 10 min, followed by 35 cycles at 95°C for 60 sec
(denaturation), 56°C for 60 sec (annealing), and 72°C for 2 min
(extension), followed by 72°C for 6 min. GAPDH served as an
internal control. Relative gene expression was quantified using
2−ΔΔCq method (20).
Western blot analysis
Approximately 2×106 HCCLM3 cells were
collected by centrifugation at 800 × g for 5 min at 4°C, and lysed
in RIPA buffer (10% NP-40, 10% sodium deoxycholate, 100 mM NaCl, 20
mM Tris-HCl pH 7.4 and 100 mM EDTA) at 4°C for 30 min. Samples were
subsequently centrifuged at 13,000 × g for 15 min at 4°C and the
supernatants were collected. The concentration of the proteins was
assessed using a BCA protein assay (Pierce; Thermo Fisher
Scientific, Inc.) according to manufacturer's protocol. Equal
amounts of samples (40 µg) were separated by 10% SDS-PAGE and
transferred onto PVDF membranes. After blocking with 5% non-fat
milk for 1 h at room temperature, the membranes were incubated with
the indicated antibodies overnight at 4°C. Following washing with
TBST solution three times, the membranes were incubated with
HRP-conjugated secondary antibodies for 1 h at room temperature.
The blots were visualized using enhanced chemiluminescence (Pierce;
Thermo Fisher Scientific, Inc.). The antibodies used were as
follows: rabbit anti-E-cadherin (1:1,000), rabbit anti-N-cadherin
(1:1,000) and mouse anti-β-actin (1:1,000) antibodies (all from
Abcam; Cambridge, MA, USA); rabbit/mouse secondary (1:5,000)
antibodies (Proteintech; Wuhan Sanying Biotechnology, Wuhan,
China).
CCK-8 assay
A CCK-8 assay was performed to establish the effect
of FoxP4 on cellular proliferation. Briefly, ~2×103
transfected cells were placed into 96-well plates with 200 µl DMEM
supplemented with 10% FBS, and 20 µl CCK-8 solution (Beyotime
Institute of Biotechnology, Shanghai, China) was added to each well
after culturing for 0, 24, 48 and 72 h. Following incubation for
another 30 min at 37°C, the absorbance value was assessed at 450 nm
using a microplate reader (Bio-Tek Instruments, Winooski, VT, USA).
Each independent experiment was replicated at least three
times.
Colony formation assay
Briefly, ~5×103 transfected cells were
cultured in 6-well plates for 12 days. The medium was replaced
every three days. After 12 days, the colonies were fixed with
methanol at room temperature for 10 min, and then stained with 0.1%
crystal violet (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at
room temperature for 15 min. The number of colonies was counted
under a light microscope. Each independent experiment was
replicated at least three times.
Wound healing assay
A wound healing assay was performed to determine the
effect of FoxP4 on the migratory capabilities of HCC cells. In
brief, ~4×105 transfected HCCLM3 cells were plated in
12-well plates and cultured in DMEM supplemented with 1% penicillin
and 1% streptomycin as well as 10% FBS at 37°C with 5%
CO2 until 90–100% confluence. A 20-µl pipette tip was
utilized to scratch a line wound. The detached cells were removed
using ice-cold PBS. Subsequently, the cells were cultured in
serum-free DMEM at 37°C with 5% CO2. The scratch wounds
were observed at 0 and 48 h under a light microscope and
photomicrographs were captured. The migration of cells was analyzed
using ImageJ 1.48 software (National Institutes of Health,
Bethesda, MD, USA). The wound healing rate was calculated according
to the following formula: Wound healing rate=[(scratch width at 0
h)-scratch width at 48 h]/(scratch width at 0 h)] ×100%. Each
independent experiment was repeated at least three times.
Transwell invasion analysis
Transwell invasion analysis was used to determine
cell migration and invasion capacities. Briefly, Transwell chambers
with 8-µm pore sizes (Corning, Cambridge, MA, USA) were coated with
80 µl 1:16-diluted Matrigel-coated (BD Biosciences, Franklin Lakes,
NJ, USA) and incubated at 37°C for 1 h. After transfection for 24
h, approximately 2.5×104 cells were placed into the
upper chamber with 300 µl serum-free DMEM medium, while 500 µl DMEM
supplemented with 10% FBS was added to the lower chamber. Following
incubation for 48 h, the cells in the upper chamber were removed by
a cotton swab and the cells in the lower chamber were then stained
with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 10 min at
room temperature. Finally, the invaded cells were counted and
photographed under a light microscope. Each independent experiment
was replicated at least three times.
Dual luciferase reporter assay
For the dual luciferase reporter assay, the promoter
region (−2,000 to +200) of Slug was cloned into pGL3-basic plasmid,
and 293T cells were placed in 12-well plates and co-transfected
with pGL3-Slug, Renilla, FoxP4, FoxP4 siRNA or the
corresponding negative control. After transcription for 24 h, the
cells were collected. The luciferase activity was assessed using a
dual luciferase reporter assay system (Promega Corporation,
Madison, WI, USA) according to manufacturer's instructions. The
firefly luciferase activity was normalized to Renilla
luciferase activity. Each independent experiment was replicated at
least three times.
Statistical analysis
Each independent experiment was repeated at least
three times. Statistical analysis was performed using SPSS
software, version 17.0 (SPSS, Inc., Chicago, IL, USA), and all data
were presented as the mean ± SD. A student's t-test was used to
determine the statistical significance of the differences between
two groups, and one-way analysis of variance followed by Tukey's
test was used to analyze the statistical significance of the
differences between multiple groups. The Kaplan-Meier method
followed by log-rank test was used to plot the survival curves
based on FoxP4 relative expression and overall survival. The
relationship of FoxP4 expression and pathological characteristics
of patients was analyzed by χ2 test. The correlations
between the expression of EMT indicator proteins and FoxP4 were
analyzed using Spearman's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
The expression of FoxP4 is elevated in
HCC tissues and cell lines
To investigate the functions of FoxP4 in HCC, we
first determined the expression of FoxP4 in HCC tissues and their
adjacent normal tissues using RT-qPCR assay, revealing that the
expression of FoxP4 was elevated in tumor tissues, compared with
adjacent normal tissues (Fig. 1A).
In addition, the expression of FoxP4 in HCC cell line HCCLM3 was
detected using RT-qPCR and western blotting assays, respectively.
The human HL-7702 normal hepatocellular cell line was used as a
control. The present results revealed that both mRNA and protein
levels of FoxP4 were higher in the HCC cell line than that in
normal hepatocytes (Fig. 1B and C).
Subsequently, the correlation between the expression of FoxP4 and
the clinical information of HCC patients was determined. The mean
value of FoxP4 mRNA content in tumor cells was used as the
standard. Thus, higher values than the standard value were defined
as high expression, and lower values than the standard value were
defined as low expression. As revealed in Table I, high expression of FoxP4 was
closely associated with tumor size, TNM stage and metastasis
(Table I), indicating that FoxP4 may
play a key role in HCC development. Additionally, analysis of
survival data using the Kaplan-Meier method revealed that patients
with high expression of FoxP4 had a poor prognosis (Fig. 1D).
 | Table I.Clinicopathological variables in 110
HCC patients. |
Table I.
Clinicopathological variables in 110
HCC patients.
|
|
| FoxP4 protein
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. (n=110) | Low (n=36) | High (n=74) | P-value |
|---|
| Sex |
|
|
|
|
| Male | 64 | 18 | 46 | 0.225 |
|
Female | 46 | 18 | 28 |
|
| Age |
|
|
|
|
| ≥40 | 78 | 25 | 53 | 0.814 |
|
<40 | 32 | 11 | 21 |
|
| Tumor size |
|
|
|
|
| Large (≥2
cm) | 62 | 14 | 48 | 0.010 |
| Small
(<2 cm) | 48 | 22 | 26 |
|
| Pathological
grade |
|
|
|
|
|
I–II | 57 | 24 | 33 | 0.030 |
|
III–IV | 53 | 12 | 41 |
|
| Lymph node
metastasis |
|
|
|
|
|
Yes | 58 | 14 | 44 | 0.043 |
| No | 52 | 22 | 30 |
|
Elevated expression of FoxP4 in HCC
cells promotes their proliferation
As revealed in Table
I, high expression of FoxP4 was closely associated with larger
tumor size, thus, we assumed that FoxP4 may play a role in cellular
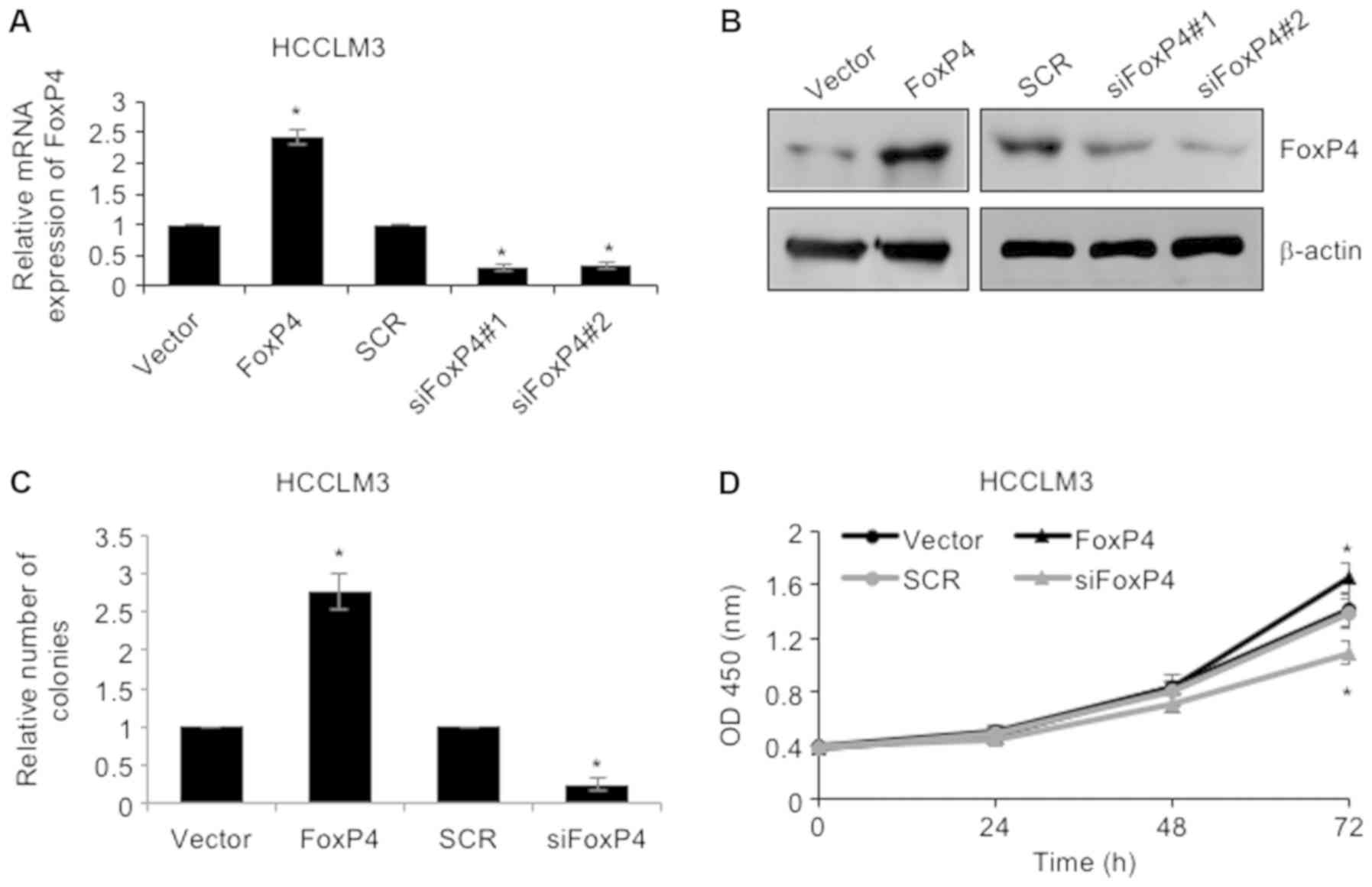
proliferation. To investigate the roles of FoxP4 in HCC, FoxP4 was
overexpressed or knocked down in HCCLM3 cells using pcDNA3.1-FoxP4
plasmid or siRNA-FoxP4, respectively. RT-qPCR and western blotting
assays were utilized to determine the success of the transfection
(Fig. 2A and B). Subsequently,
colony formation as well as CCK-8 assays were performed to
determine the effect of FoxP4 on cellular proliferation. The
results of the colony formation assay revealed that upregulation of
the expression of FoxP4 resulted in an elevated number of colonies
in HCCLM3 cells compared to the vector group, whereas knockdown of
FoxP4 notably reduced the number of colonies compared to the SCR
group (Fig. 2C). Additionally, CCK-8
analysis also demonstrated that ectopic expression of FoxP4
improved the cellular proliferation rate and knockdown of FoxP4
impaired the cellular proliferation rate in HCCLM3 cells (Fig. 2D). Collectively, the results
indicated that elevated expression of FoxP4 in HCC cells affected
cell proliferation.
Upregulation of FoxP4 promotes the
migration and invasion of HCC cells
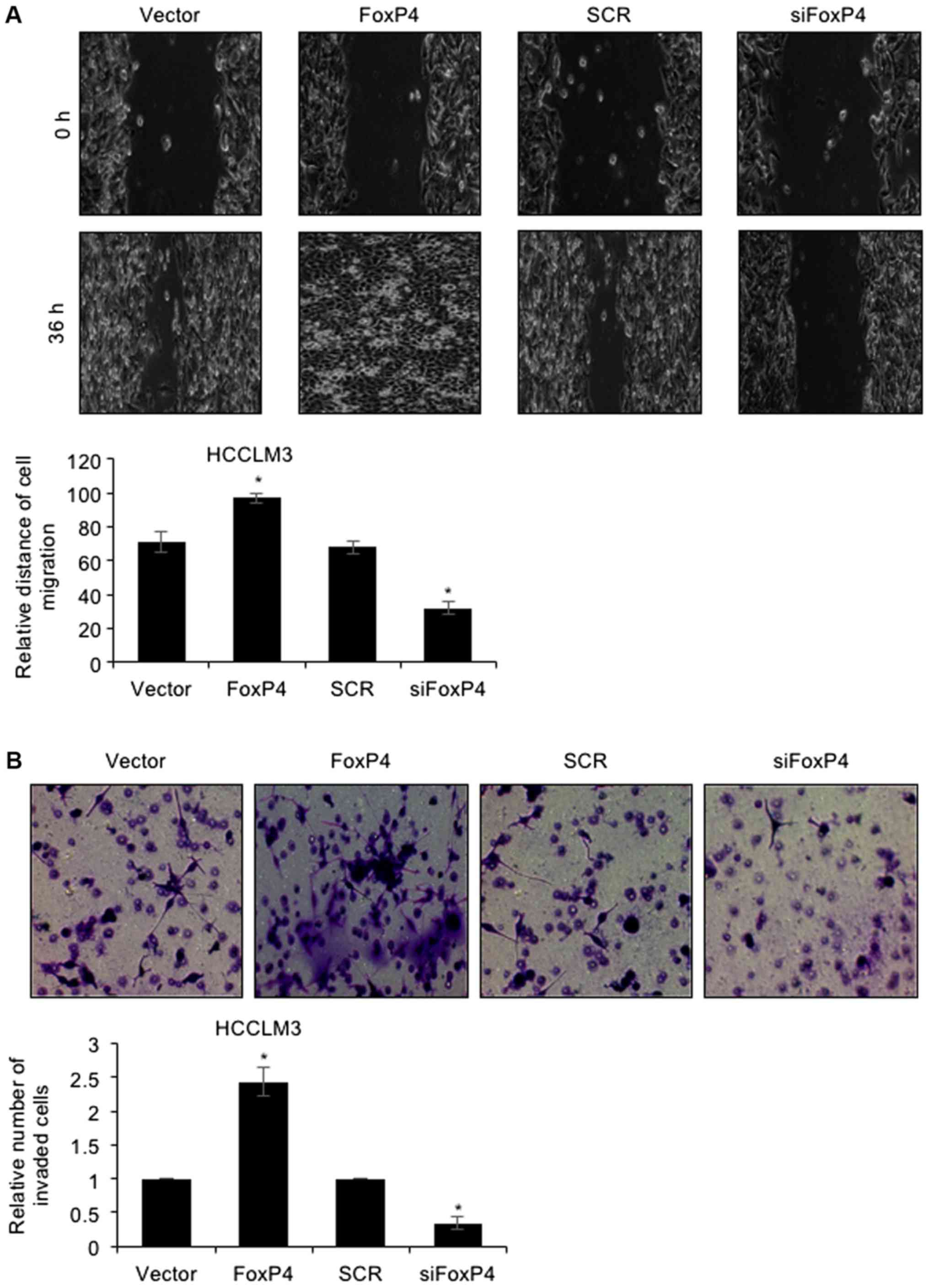
To further decipher the detailed mechanism of FoxP4
in HCC, a wound healing assay and Tranwell invasion assay were
performed. The results of the wound healing assay revealed that
ectopic expression of FoxP4 potentiated the healing of scratch
wounds in HCCLM3 cells, whereas FoxP4 inhibition resulted in the
slower healing of scratch wounds in HCCLM3 cells (Fig. 3A), indicating that the migratory
capabilities of HCC cells were enhanced following FoxP4
overexpression, whereas they were impaired following FoxP4
silencing. For the Transwell invasion assay, the present results
revealed that following FoxP4 overexpression, the number of invaded
cells was significantly increased compared with the vector cells
(Fig. 3B). Conversely, invasive
capabilities of siFoxP4-transfected cells were significantly
impaired compared with SCR-transfected cells (Fig. 3B). These observations indicated that
the upregulation of FoxP4 promoted the migration and invasion of
HCC cells.
FoxP4 promotes EMT in HCC cells
through regulation of Slug
Multiple studies have revealed that EMT can increase
the incidence of cancer metastasis (21–23).
Therefore, we ascertained whether FoxP4 promoted the migration and
invasion of HCC cells through regulation of EMT. As revealed in
Fig. 4A and B, the results of the
RT-qPCR assay revealed that following ectopic expression of FoxP4,
the mRNA and protein levels of E-cadherin were significantly
downregulated and the mRNA and protein levels of N-cadherin were
significantly upregulated compared with the vector group (Fig. 4A and B). Conversely, following
inhibition of FoxP4, the mRNA and protein levels of E-cadherin were
significantly upregulated and the mRNA and protein levels of
N-cadherin were significantly downregulated compared with the SCR
group (Fig. 4A and B). The
aforementioned results indicated that FoxP4 promoted EMT in HCC
cells. Subsequently, whether FoxP4 promoted EMT in HCC cells
through regulation of the EMT-associated transcription factors,
including Snail, Slug and Twist1, was determined. The present
results demonstrated that ectopic expression of FoxP4 resulted in
an elevated expression of Slug, whereas inhibition of FoxP4
significantly decreased the expression of Slug; however, neither
overexpression nor knockdown of FoxP4 had an effect on Snail and
Twist1 expression (Fig. 4C and D).
These observations indicated that FoxP4 promoted EMT in HCC cells
through regulation of Slug.
Slug is transcriptionally regulated by
FoxP4 in HCC cells
Since FoxP4 is a transcription factor, it was
hypothesized that FoxP4 may transcriptionally regulate Slug. To
ascertain our hypothesis, ChIP and qChIP assays were performed,
revealing that FoxP4 could directly bind to the promoter region of
Slug (Fig. 5A). Subsequently,
the promoter region of Slug was cloned into the pGL3-basic
plasmid (pGL3-Slug). 293T cells were co-transfected with pGL3-Slug
and vector or FoxP4 plasmid, SCR or siFoxP4. The results of the
dual luciferase reporter assay demonstrated that ectopic expression
of FoxP4 resulted in an elevated relative luciferase activity
compared with the vector group, and that in the siFOXP4-transfected
cells, the relative luciferase activity was significantly decreased
compared with the SCR group (Fig.
5B). Notably, E-cadherin, N-cadherin as well as Slug expression
were observed to be correlated with FoxP4 expression in HCC tissues
(Table II). Collectively, our
results indicated that Slug was transcriptionally regulated by
FoxP4 in HCC cells.
 | Table II.Spearman's correlation analysis
between the expression of FoxP4 and EMT indicator proteins in
HCC. |
Table II.
Spearman's correlation analysis
between the expression of FoxP4 and EMT indicator proteins in
HCC.
|
|
| FoxP4
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. (n=110) | Low | High | P-value |
|---|
| E-cadherin
expression |
|
|
| 0.03 |
|
High | 57 | 24 | 33 |
|
|
Low | 53 | 12 | 41 |
|
| N-cadherin
expression |
|
|
| 0.003 |
|
High | 56 | 11 | 45 |
|
|
Low | 54 | 25 | 29 |
|
| Slug
expression |
|
|
| 0.042 |
|
High | 55 | 13 | 42 |
|
|
Low | 55 | 23 | 32 |
|
Discussion
In malignant tumors, >90% of patients with cancer
succumbed to tumor metastasis in 2015 (24). Hepatocellular carcinoma (HCC) is a
fast-growing type of cancer that is characterized by highly
invasive and metastatic capabilities (25,26).
Unfortunately, almost 60–80% of patients with HCC are diagnosed at
an advanced stage, thereby losing the opportunity for surgical
treatment (27).
In the present study, FoxP4 was identified to be
upregulated in HCC tissues and cells compared with adjacent normal
tissues and normal hepatocytes, respectively. Notably, FoxP4
expression was closely associated with tumor size, TNM stage and
lymph node metastasis in patients with HCC. Epithelial-mesenchymal
transition (EMT) has been revealed to promote cellular invasion and
metastasis of cancer (28–30), and the results of our present study
revealed that upregulation of FoxP4 significantly promoted the
migration and invasion of HCC cells by regulation of EMT. In
epithelial cells, E-cadherin is an important protein that regulates
cell-cell adhesion (31). The
expression of E-cadherin is markedly decreased during EMT, which
results in loss of cell-cell adhesion as well as gain of cell
motility. Previous studies have indicated that the reduction in
E-cadherin expression is mainly regulated by the transcriptional
repressors of E-cadherin gene (CDH1) (18,32). A
major transcriptional repressor of CDH1 is the zinc finger factor,
Slug (17). Notably, ectopic
expression of FoxP4 resulted in an increased expression of Slug.
These results indicated that Slug may be involved in FoxP4-mediated
EMT.
However, there are a number of limitations present
in the study. First, an immunohistochemistry assay is the optimal
method to verify the expression of FoxP4 in tissue samples. Second,
the roles of FoxP4 in vivo require further investigation.
Lastly, the ChIP-seq assay is necessary to extensively investigate
FoxP4-target genes in HCC. Further examination is required of the
underlying mechanism of FoxP4 in HCC.
In conclusion, our present study revealed that FoxP4
promoted EMT in HCC cells through transcriptional regulation of
Slug expression. Moreover, upregulation of FoxP4 also promoted
cellular proliferation, thus indicating that FOXP4 may have
potential as a novel therapeutic target for the treatment of
patients with HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
GZ and GYZ conceived and designed the present study.
GZ and GYZ constructed expression plasmids, prepared proteins and
performed experiments. GZ analyzed the data. GZ and GYZ wrote the
manuscript. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Rizhao Hospital of Traditional Chinese Medicine and each patient
was well informed and signed informed consent forms.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Waly Raphael S, Yangde Z and Yuxiang C:
Hepatocellular carcinoma: Focus on different aspects of management.
ISRN Oncol. 2012:4216732012.PubMed/NCBI
|
|
2
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bosch FX, Ribes J, Diaz M and Cleries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology. 127:S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Faivre S, Bouattour M and Raymond E: Novel
molecular therapies in hepatocellular carcinoma. Liver Int. 31
Suppl:S151–S160. 2011. View Article : Google Scholar
|
|
6
|
Pan FC and Wright C: Pancreas
organogenesis: From bud to plexus to gland. Dev Dyn. 240:530–565.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Sun J, Cui M, Zhao F, Ge C, Chen
T, Yao M and Li J: Downregulation of FOXP1 inhibits cell prolife
ration in hepatocellular carcinoma by inducing G1/S phase cell
cycle arrest. Int J Mol Sci. 17:E15012016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Datta J, Kutay H, Nasser MW, Nuovo GJ,
Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et
al: Methylation mediated silencing of MicroRNA-1 gene and its role
in hepatocellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan X, Zhou H and Zhang T, Xu P, Zhang S,
Huang W, Yang L, Gu X, Ni R and Zhang T: Downregulation of FOXP2
promoter human hepatocellular carcinoma cell invasion. Tumour Biol.
36:9611–9619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Z, Lin X, Tian M and Chang W:
microRNA196b promotes cell migration and invasion by targeting
FOXP2 in hepatocellular carcinoma. Oncol Rep. 39:731–738.
2018.PubMed/NCBI
|
|
11
|
Zhang Y, Zhang S, Wang X, Liu J, Yang L,
He S, Chen L and Huang J: Prognostic significance of FOXP1 as an
oncogene in hepatocellular carcinoma. J Clini Pathol. 65:528–533.
2012. View Article : Google Scholar
|
|
12
|
Shi JY, Ma LJ, Zhang JW, Duan M, Ding ZB,
Yang LX, Cao Y, Zhou J, Fan J, Zhang X, et al: FOXP3 Is a HCC
suppressor gene and Acts through regulating the TGF-beta/Smad2/3
signaling pathway. BMC Cancer. 17:6482017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Katoh M and Katoh M: Human FOX gene family
(Review). IntJ Oncol. 25:1495–1500. 2004.
|
|
14
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lam EW, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: Tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–495. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barasch J: Genes and proteins involved in
mesenchymal to epithelial transition. Curr Opin Nephrol Hypertens.
10:429–436. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nieto MA: The snail superfamily of
zinc-finger transcription factors. Nat Rev Mol Cell Biol.
3:155–166. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu SQ, Qiu Y, Dai WJ and Zhang XY: FOXR2
promotes the proliferation, invasion, and epithelial-mesenchymal
transition in human colorectal cancer cells. Oncol Res. 25:681–689.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H
and Cai J: MiR-186-5p upregulation inhibits proliferation,
metastasis and epithelial-to-mesenchymal transition of colorectal
cancer cell by targeting ZEB1. Arch Biochem Biophys. 640:53–60.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun C, Tao Y, Gao Y, Xia Y, Liu Y, Wang G
and Gu Y: F-box protein 11 promotes the growth and metastasis of
gastric cancer via PI3K/AKT pathway-mediated EMT. Biomed
Pharmacother. 98:416–423. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu L, Wu B, Cai H, Li D, Ma Y, Zhu X, Lv
Z, Fan Y and Zhang X: Tiam1 promotes thyroid carcinoma metastasis
by modulating EMT via Wnt/beta-catenin signaling. Exp Cell Res.
362:532–540. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nordenstedt H, White DL and El-Serag HB:
The changing pattern of epidemiology in hepatocellular carcinoma.
Dig Liver Dis. 42 Suppl:S206–S214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang JD, Harmsen WS, Slettedahl SW,
Chaiteerakij R, Enders FT, Therneau TM, Orsini L, Kim WR and
Roberts LR: Factors that affect risk for hepatocellular carcinoma
and effects of surveillance. Clin Gastroenterol Hepatol. 9:617–623.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng FD, Wei JC, Qu K, Wang ZX, Wu QF, Tai
MH, Liu HC, Zhang RY and Liu C: FoxM1 overexpression promotes
epithelial-mesenchymal transition and metastasis of hepatocellular
carcinoma. World J Gastroenterol. 21:196–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sanchez-Tillo E, Lazaro A, Torrent R,
Cuatrecasas M, Vaquero EC, Castells A, Engel P and Postigo A: ZEB1
represses E-cadherin and induces an EMT by recruiting the SWI/SNF
chromatin-remodeling protein BRG1. Oncogene. 29:3490–3500. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tam WL and Weinberg RA: The epigenetics of
epithelial-mesenchymal plasticity in cancer. Nature Med.
19:1438–1449. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|