Introduction
Quantitative structure-activity relationship (QSAR)
models are mathematical tools used to predict the physical,
chemical or biological characteristics of chemical substances from
their chemical structure, as expressed through a variety of
‘chemical descriptors’ (1). In the
famous statistical aphorism of George Box, ‘all models are wrong
but some are useful’ (2); QSAR
models might be imperfect, but they have proven useful in a
plethora of applications (3), from
drug design (being frequently used for virtual screening, as well
as lead optimization) (4) to
toxicological predictions (being used to predict toxicity for a
large number of substances for which wet lab experiments have not
yet been performed and may be unlikely to be performed in the near-
or mid-term future (5), or from
protein binding (6) to cytochrome
P450 interaction forecasts (7).
Melanoma is considered the most threatening form of
skin neoplasm, having fast progression and metastasizing, as well
as a high burden of death, particularly if detected late (8). Although an important number of
therapies have recently been approved for advanced stage melanoma,
the disease is far from being vanquished, resistance development
through mutations or alternative signaling pathways, cancer
heterogeneity and serious adverse events limiting the efficacy and
potential benefits of the newer treatments, at least in a
proportion of the patients (9,10).
Therefore, although therapeutic options are now better for patients
with advanced melanoma than they were a decade ago, there is still
a need for developing new drugs targeting melanoma, and a variety
of approaches are still explored, from evaluating new targets
(11) to exploring new delivery
systems for old compounds (12).
SK-MEL-5 is a human melanoma cell line derived from a metastatic
axillary node of a young female patient, and is characterized by a
high level of expression of the V600E mutation of B-Raf, of the
wild-type N-Ras (13), as well as by
relatively high levels of the ABCB1 transcript (14). This is unlike SK-MEL-2 melanoma cell
line, which has wild-type B-Raf, but normal N-Ras (11). It has been used in various studies to
explore new therapies against melanoma in various in vitro
experiments (15–17).
In the present study, we report on our attempts to
develop QSAR models, able to forecast the cytotoxic effects of
different chemical compounds on the SK-MEL-5 melanoma cell line,
using the data available on PubChem. Such data are derived from
different laboratories, have been generated at different times,
most likely with different reagents and laboratory equipment;
moreover, whereas most QSAR studies are focused on a well-defined
biological target, the cytotoxicity data are inherently more
heterogeneous, as different molecules may induce cytotoxicity
through a variety of biochemical pathways. Thus, it is to be
expected that QSAR modelling of such data is more challenging than
for compounds targeting specific proteins or other unambiguous cell
targets. Kalliokoski et al (18), based on a data set filtered using
certain validity criteria have shown that the standard deviation
for IC50 is only approximately 25% higher than that of
ki; we have used GI50, which is similar to
IC50, in our models, as ki data are not available for
cytotoxicity measurements on cultured cell lines (ki is applicable
to distinct protein targets). Because of these considerations, as
well as due to the relatively large structural diversity of the
dataset, we used a binary classification approach (not regression
models) (19) and have focused on 4
machine learning techniques extensively made use of in the area of
data prediction: Random forest (RF), gradient boosting (BST),
support vector machine (SVM) and k-nearest neighbor (KNN).
Materials and methods
Dataset
The dataset of cytotoxic and inactive compounds on
the SK-MEL-5 cell line was downloaded from the PubChem data base
(https://pubchem.ncbi.nlm.nih.gov) in
June 2017. We have retained the data for all chemical compounds for
which cytotoxicity results expressed by GI50 was
recorded. Other assessment criteria for the same cell line (e.g.,
LC50 or ED50) were not preferred and selected
because the number of records was much lower for these measures (35
observations for the former, 138 for the latter). We downloaded the
PubChem canonical SMILES and used ChemAxon Standardizer v. 18.8.0
(ChemAxon, Budapest, Hungary) for the standardization of the
molecules. Duplicates were removed in two steps: First, we detected
duplicates in R, based on the canonical SMILES, and replacing the
GI50 with the mean value of the duplicates. This
procedure identified most of the duplicates. In a second step we
used the ISIDA/Duplicates (http://infochim.u-strasbg.fr; University of
Strasbourg, France) software following the structure
standardization and this detected an additional duplicate.
Standardized SMILES were converted to 2D chemical structures using
Discovery Studio Visualizer v16.1.0.15350 (Dassault Systèmes
BIOVIA, San Diego, CA, USA). We defined a compound as ‘active’ if
the GI50 was less than 1 µM and ‘inactive’ if the
GI50 was higher than the 1 µM threshold. We started with
a number of 445 observations and, following removal of duplicates
ended up with 422 observations, of which 174 labelled as ‘active’
and 248 as ‘inactive’; the ratio of inactive:active compounds was
~1.42. Having a balanced data set is important for a good
performance of machine learning algorithms, especially when the
target class is underrepresented (20). We therefore also assessed the effect
of balancing the data through over-, under-, and a combination of
over- and under-sampling, but the benefit was in most cases rather
limited, if at all. We randomly divided the data set in a training
(learning) set (316 compounds) and a testing set (106 compounds),
using the rminer package of the R statistical tool (21).
Descriptors
Thirteen blocks of molecular descriptors were
computed with the Dragon 7 program (version 7.0, https://chm.kode-solutions.net; Kode SRL, Milano,
Italy): Constitutional descriptors (n=47), ring descriptors (n=32),
topological indices (n=75), walk and path counts (n=46),
information indices (n=50), 2D matrix-based descriptors (n=607),
2D-autocorrelations (n=213), Burden eigenvalues (n=96), P-VSA-like
descriptors (n=55), ETA indices (n=23), Edge adjacency indices
(n=324), and molecular properties (n=20). We have also used the
whole set of 1D and 2D descriptors (264 descriptors after the
removal of constant, quasi-constant and highly correlated
variables), in order to assess whether models based on a larger
pool of descriptors have better performance with the chosen
classifiers than models based on a narrow and well-defined family
of descriptors. Thus, the total number of descriptor blocks used
for building classification models was 13. Because the models based
on the molecular properties had poor performance we did not include
the results of those models here.
Pre-processing and feature
selection
We generated distinct QSAR models with each of the
15 blocks of descriptors and pre-processed the data using R, v.
3.4.4 (22), and ‘mlr’ package, v.
2.12.1 (23). For this purpose,
within each block of descriptors we removed variables with constant
or near constant values (using a threshold value of 0.1%, i.e.,
features for which less than 0.1% differed from their mode value
were removed). Features containing missing values were also
removed, because it is likely that for virtual screening purposes
models built with such features will not be applicable for a part
of the new compounds. Features highly correlated were also removed,
using a threshold value of the coefficient correlation of 0.80. For
each subset, after such pre-processing we selected maximum 7
features using two methods: i) RF importance (‘random forest’ R
package) (24); and ii) symmetrical
uncertainty (‘FSelector’ R package) (25).
Classifiers
We made use of four machine learning algorithms to
build classification models able to predict with reasonable
accuracy the effect of substances against the SK-MEL-5 melanoma
cell line: RF, BST, SVM, and KNN.
RFs, first proposed by Ho in 1995 (26) and improved by Breiman in 2001
(27) use a large number of decision
trees (hence the name, ‘forests’), which are aggregated through
bootstrap (bagging), and prediction for unseen samples are made
through averaging or a majority vote. It has been described as
‘among the most accurate methods’ in the field of QSAR (28). It is implemented in the R package
‘random forest’ (24).
Gradient boosting machines (GBMs) represent an
algorithm able to combine weak learners in a strong one, building,
in an iterative manner, additional base-learners that have a
maximal correlation with the negative slope of a cost function, a
variety of such functions being available (29). In QSAR models GBMs have shown good
results with respect to performance of prediction, speed and
robustness (30). The algorithm was
run under ‘mlr’ R package based on the implementation carried out
in ‘bst’ (31) and ‘rpart’ (32) R packages.
Support vector machines (SVMs), proposed for the
first time and developed by Vladmir Vapnik, makes use of a
hyperplane separating the data from the variable space into
classes. Variables are first mapped in a high-dimensional space
through a variety of kernel functions, then the algorithm
identifies in this high-dimensional space the maximal margin
hyperplane, thus separating the compounds in classes (33). Its chief advantage consists in the
fact that it makes use of the structure risk minimization (SRM)
principle, which is more efficient than the conventional empirical
risk minimization (ERM) (34). We
used the implementation of the algorithm available in the ‘e1071’ R
package (35).
KNN is a classification method, in which the
separation of variables in classes is performed using the nearest
training observations from the variable space (36), more precisely, a test instance is
classified with the help of majority decision using the data of its
KNN, as computed from the learning set (37). The algorithm was run under ‘mlr’ R
package based on the implementation carried out in the ‘rknn’ R
package (38).
Performance measures and model
validation
A nested (double) cross validation method was used
to tune the hyper-parameters for each algorithm and to assess the
performance and robustness of the model thus developed (guiding the
decision by the best performance in terms of Cohen's kappa). This
is considered the most appropriate procedure for cross-validation,
the data being partitioned into a learning subset and a test
subset, the learning subset being used in the internal loop, for
the model building and selection, whereas the test subset is being
used for the assessment of the performance of the model picked in
the inner loop. The inner loop used a 5-fold cross-validation,
whereas the outer loop used a 10-fold cross-validation. The nested
cross-validation method was performed on the 316 compounds
constituting the initial training set (which was thus, successively
divided in training and test subsets). To externally assess the
reliability of the model performance on data unseen by the model,
we used the 106 compounds of the (initial) test set.
The purpose of developing the models was to identify
compounds with a high likelihood of being active; in other words,
we were not equally interested in classifying both positive and
negative observations correctly, but rather in avoiding false
positives. Therefore, the most relevant performance measure was the
selectivity (true negative rate, tnr), indicating the proportion of
observations rightly classified in the negative category, and we
are interested in maximizing it; its complementary value (1-tnr)
gives the false positive rate, our interest being in its
minimization. Sensitivity (true positive rate, tpr), defined as the
proportion of observations in the positive class properly
classified, is also relevant, although for our purposes it is
preferably to have a higher selectivity and lower sensitivity than
the other way round. The positive predictive value (PPV,
precision), calculated as tp/(tp+fp), where tp is the sum of all
true positive values correctly classified and fp the false
positives (misclassified observations from the positive class), is
a composite measure reflecting both selectivity and sensitivity.
Although not the most important for our purposes, for a better
understanding of performance we also looked at the balanced
accuracy (defined as the mean of tpr and tnr) and mean
misclassification error (MMCE), defined as the proportion of cases
where the response (classification result for a particular
observation) is different from the truth (the real class of a
particular observation). All these measures are implemented in the
mlr package (23).
Besides 10-fold nested cross-validation and external
testing, Y-scrambling was applied to assess the robustness of the
models, ruling out to a reasonable extent the possibility that the
models were the result of chance associations. The IC50
value was randomly scrambled using 500 permutations (R package
‘gtools’) (39) and then several
different models were re-built from zero (i.e., repeating the
process of feature selection, so as to correspond to the new
(scrambled) activity values) and the performance measures were
computed for the new models thus re-built.
We assessed the applicability domain (AD) of the
models developed employing the KNN approach developed by Sahigara
et al (2013) (40) and the
method proposed by Roy et al (2015) (37), which assumes normal distribution of
the descriptor values, using code written by us in R. We have also
explored the local density methods implemented in the R package
‘ldbod’ (41), using arbitrary
thresholds of 5 and 10% for the ranked values of the local
density-based outlier scores computed against the reference values
of the train set. The same techniques were used to investigate and
detect outliers among the train set values.
Results
Assessment of the dataset chemical
diversity
To ensure a reasonable predictive accuracy of QSAR
models it is important to have a data set sufficiently diverse
(42) and in the literature various
ways of the chemical diversity assessment have been used. We have
computed a dissimilarity matrix based on the Gower distance, which
is an appropriate measure for data sets containing combinations of
numerical and categorical or binary variables and returns a
distance that is already scaled, i.e., is always a number between 0
(identical values, no dissimilarity) and 1 (very distinct values,
maximal dissimilarity) (43). For
the dissimilarity matrix we used all 1D and 2D descriptors computed
by Dragon Program, v. 7.0 after minimal processing for the removal
of constant and near constant features (1,920 remaining
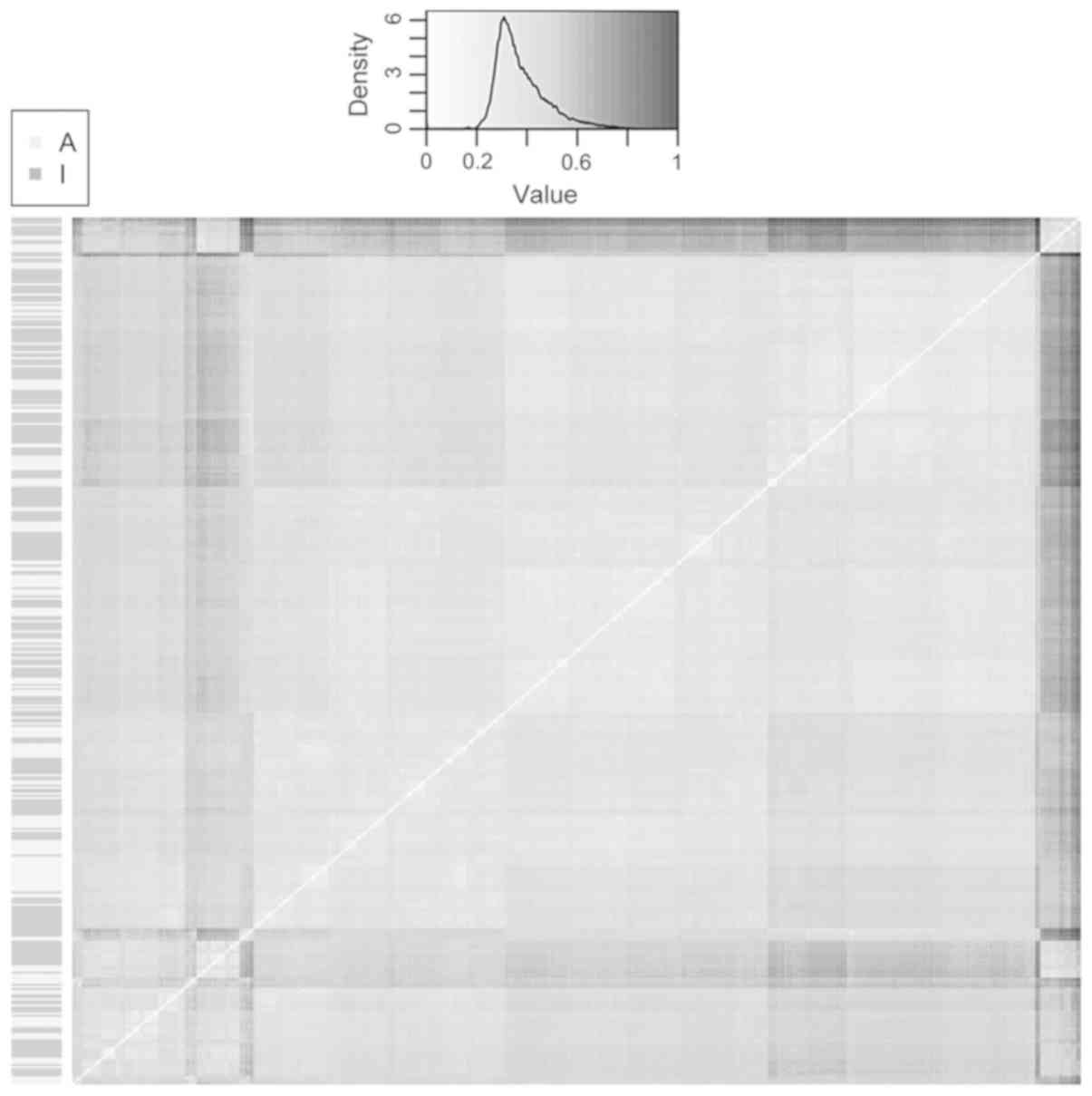
descriptors). To get a quick understanding of the differences, a
heat map of the dissimilarity matrix was drawn and examined
(Fig. 1). As indicated by the
(smaller) density plot, most of the observations have a
dissimilarity coefficient of 0.2–0.6, i.e., there is a moderate
chemical diversity in the whole dataset.
We also used the technique of Xu et al
(42), who used a scatter plot of
the molecular weight and AlogP for the substances from the learning
and test subsets to assess whether the latter were distributed in
the same chemical space as the former compounds. The graph showed
that most test points were close to one or more several train
points, but there were also a few outliers which seemed to be out
of the AD of the models (Fig.
2).
The exploration of the AD for the seven best
performing models with the first two methods (based on the KNN and
local probability density) has shown that for most only a small
proportion (3.77–12.3% for the different sets of features and
depending on the method used for the assessment) of the test set
observations were outside the AD; moreover, in most cases despite
the fact that those cases were outside the AD, most of them were
predicted correctly (for instance all of the nine values identified
by the KNN-based method as outside AD were predicted correctly by
the RF model based on the first set of topological descriptors and
oversampling, and 11 out of 13 values identified by the Roy method
(37) as outside AD were also
correctly classified for this method; in the case of
2D-autocorrelations, for the KNN method out of four values outside
AD, three were correctly classified, all five values identified by
probability density methods at the 5% threshold were correctly
predicted and four out of five identified by the Roy method were
correctly labeled by this model.
In the case of informational indices, the number of
test observations outside AD identified by the KNN method was
surprisingly high (29.25%, almost one in every three observations),
and slightly more than half of those cases (51.61%) were wrongly
classified. The Roy method identified only five outliers and two of
them were wrongly classified. The probability density methods
suggested that slightly more than half of the values outside AD for
this model were wrongly classified (3 out of 5 and 6 out of 10 most
extreme values based on the outlier scores were wrongly
predicted).
Performance of nested cross
validation
We attempted to use the connectivity indexes but all
descriptors of this subset had some values not available and
therefore we preferred to discard this subset and not to build
classification models based on these descriptors.
Using 4 classifiers, 13 different sets of
descriptors, as well as ‘synthetic’ samples obtained by
over-sampling or a combination of over- and under-sampling
(‘smote’) different models were build, the performance of which was
assessed through nested cross validation. Because we used 2
different algorithms for feature selection, which in most cases
identified two partially different subsets of features (in rarer
cases a single set of features), the total number of models
evaluated was 186 (not counting those built with molecular
properties, whose performance was poor). We report here only those
models (n=28) with an acceptable performance [positive predictive
value (PPV) higher than 75% in both the nested cross-validation and
on the previously unseen dataset] (Tables I and II). The performance of each model in the
nested cross-validation and on the independent data set is shown in
the Tables SI and SII.
 | Table I.Performance of selected
classification models with PPV higher than 75% for the 10-fold
nested cross-validation. |
Table I.
Performance of selected
classification models with PPV higher than 75% for the 10-fold
nested cross-validation.
| Models | Specificity | Sensitivity | PPV | Balanced
accuracy | MMCE |
|---|
| Topological
descriptors-RF (1) | 0.9374 | 0.3583 | 0.8424 | 0.6479 | 0.3022 |
| Topological
descriptors-RF (2) | 0.9298 | 0.3628 | 0.7964 | 0.6463 | 0.3105 |
| Topological
descriptors-RF (1), over | 0.9148 | 0.5752 | 0.8749 | 0.745 | 0.2548 |
| Topological
descriptors-RF (1), smote | 0.8946 | 0.499 | 0.8158 | 0.6968 | 0.3086 |
| Walk and path-RF
(1) | 0.9465 | 0.285 | 0.7587 | 0.6158 | 0.3231 |
| Information
indices-RF (1) | 0.9486 | 0.3434 | 0.8368 | 0.646 | 0.3003 |
| Information
indices-RF (2) | 0.9685 | 0.3448 | 0.8848 | 0.6566 | 0.2878 |
| Information
indices-RF (1), over | 0.9022 | 0.634 | 0.8715 | 0.7681 | 0.2319 |
| Information
indices-RF (1), smote | 0.9023 | 0.5438 | 0.851 | 0.723 | 0.2776 |
| Information
indices-BST (1), smote | 0.78 | 0.7536 | 0.7803 | 0.7668 | 0.2344 |
|
2D-autocorrelation-RF (1) | 0.927 | 0.3414 | 0.776 | 0.6342 | 0.3063 |
|
2D-autocorrelation-RF (2) | 0.9687 | 0.3005 | 0.8707 | 0.6346 | 0.3063 |
|
2D-autocorrelation-RF (2), over | 0.9453 | 0.611 | 0.9201 | 0.7782 | 0.2289 |
|
2D-autocorrelation-RF (2), smote | 0.9174 | 0.4858 | 0.8583 | 0.7016 | 0.2993 |
| Burden
eigenvalues-RF (2) | 0.941 | 0.3373 | 0.7943 | 0.6391 | 0.3063 |
| Burden
eigenvalues-RF (2), over | 0.8803 | 0.6373 | 0.8417 | 0.7588 | 0.2427 |
| Burden
eigenvalues-RF (2), smote | 0.8445 | 0.6265 | 0.8057 | 0.7355 | 0.2641 |
| P-VSA-like-RF
(1) | 0.9327 | 0.3528 | 0.7825 | 0.6428 | 0.3058 |
| P-VSA-like-RF
(2) | 0.9332 | 0.3716 | 0.7996 | 0.6524 | 0.2967 |
| P-VSA-like-RF
(2), over | 0.9149 | 0.6159 | 0.8891 | 0.7654 | 0.2369 |
| P-VSA-like-RF
(2), smote | 0.8919 | 0.5541 | 0.8273 | 0.723 | 0.283 |
| Eta indices-RF
(2) | 0.9384 | 0.3807 | 0.8394 | 0.6596 | 0.2872 |
| Edge adjacency-RF
(1) | 0.9412 | 0.3453 | 0.8242 | 0.6432 | 0.307 |
| Edge adjacency-RF
(2) | 0.9301 | 0.3652 | 0.8006 | 0.6477 | 0.3038 |
| Edge adjacency-RF
(1), over | 0.9031 | 0.6477 | 0.8635 | 0.7754 | 0.2239 |
| Edge adjacency-SVM
(1), over | 0.7663 | 0.7113 | 0.7519 | 0.7388 | 0.2696 |
| Global-BST
(1), over | 0.793 | 0.8137 | 0.7899 | 0.8034 | 0.1994 |
| Global-BST
(1), smote | 0.7974 | 0.7957 | 0.7927 | 0.7966 | 0.202 |
 | Table II.Performance of selected
classification models with PPV higher than 75% on the independent
data set. |
Table II.
Performance of selected
classification models with PPV higher than 75% on the independent
data set.
| Models | Specificity | Sensitivity | PPV | Balanced
accuracy | MMCE |
|---|
| Topological
descriptors-RF (1) | 0.9194 | 0.5 | 0.8148 | 0.7097 | 0.2547 |
| Topological
descriptors-RF (2) | 0.9194 | 0.5227 | 0.8214 | 0.721 | 0.2453 |
| Topological
descriptors-RF (1), over | 0.9355 | 0.5682 | 0.8621 | 0.7518 | 0.217 |
| Topological
descriptors-RF (1), smote | 0.9516 | 0.5909 | 0.8966 | 0.7713 | 0.1981 |
| Walk and path-RF
(1) | 0.9516 | 0.2727 | 0.8 | 0.6122 | 0.3302 |
| Information
indices-RF (1) | 1 | 0.5 | 1 | 0.75 | 0.2075 |
| Information
indices-RF (2) | 0.9839 | 0.5227 | 0.9583 | 0.7533 | 0.2076 |
| Information
indices-RF (1), over | 1 | 0.5227 | 1 | 0.7614 | 0.1981 |
| Information
indices-RF (1), smote | 1 | 0.5682 | 1 | 0.7841 | 0.1792 |
| Information
indices-BST (1), smote | 0.9355 | 0.75 | 0.8919 | 0.8427 | 0.1415 |
|
2D-autocorrelation-RF (1) | 0.9355 | 0.3864 | 0.8095 | 0.6609 | 0.2924 |
|
2D-autocorrelation-RF (2) | 0.9677 | 0.4091 | 0.9 | 0.6884 | 0.2642 |
|
2D-autocorrelation-RF (2), over | 0.9032 | 0.5 | 0.7857 | 0.7016 | 0.2642 |
|
2D-autocorrelation-RF (2), smote | 0.9194 | 0.4773 | 0.8077 | 0.6983 | 0.2642 |
| Burden
eigenvalues-RF (2) | 0.9516 | 0.4773 | 0.875 | 0.7144 | 0.2453 |
| Burden
eigenvalues-RF (2), over | 0.9516 | 0.5909 | 0.8966 | 0.7713 | 0.1981 |
| Burden
eigenvalues-RF (2), smote | 0.9355 | 0.5682 | 0.8621 | 0.7518 | 0.217 |
| P-VSA-like-RF
(1) | 0.9783 | 0.6562 | 0.9545 | 0.8173 | 0.1538 |
| P-VSA-like-RF
(2) | 0.9783 | 0.6875 | 0.9565 | 0.8329 | 0.141 |
| P-VSA-like-RF
(2), over | 0.9783 | 0.7812 | 0.9615 | 0.8798 | 0.1026 |
| P-VSA-like-RF
(2), smote | 0.9783 | 0.9062 | 0.9667 | 0.9423 | 0.0513 |
| Eta indices-RF
(2) | 0.9032 | 0.4318 | 0.76 | 0.6675 | 0.2924 |
| Edge adjacency-RF
(1) | 0.9839 | 0.4545 | 0.9524 | 0.7192 | 0.2358 |
| Edge adjacency-RF
(2) | 0.9839 | 0.3864 | 0.9444 | 0.6851 | 0.2642 |
| Edge adjacency-RF
(1), over | 0.9516 | 0.4545 | 0.8696 | 0.7031 | 0.2547 |
| Edge adjacency-SVM
(1), over | 0.9023 | 0.6364 | 0.8235 | 0.7698 | 0.2076 |
| Global-BST
(1), over | 0.8871 | 0.9318 | 0.8542 | 0.9095 | 0.0943 |
| Global-BST
(1), smote | 0.9032 | 0.9318 | 0.8723 | 0.9175 | 0.0849 |
Among the 186 models reported in the Tables SI and SII, none had a PPV higher than 0.90 in
both nested cross-validation and on the external dataset, but seven
models had a PPV higher than 0.85 in both evaluations, all seven
using the RF algorithm as a classifier and topological descriptors,
information indices, 2D-autocorrelation descriptors, P-VSA-like
descriptors, and edge-adjacency descriptors as sets of features
used for classification. For 16 models PPV was higher than 80% with
the two assessment methods (cross-validation and external
evaluation). Using the pool of all descriptors and two feature
selection algorithms did not lead to better results than using
smaller blocks of descriptors: None of the 16 models developed with
the pool of all 1D and 2D descriptors had a PPV higher than 80% in
both cross-validation and external testing and only two of those 16
models had a PPV higher than 75% in both evaluations. We have not
explored a larger range of feature selection options for this large
pool of descriptors, but with the two also applied on the smaller
blocks there was no clear advantage in using the larger number of
descriptors as a start. Thus, on the subject of descriptor
efficiency more is not necessarily better, in our case less was
rather more.
The nitrogen percentage, oxygen atom numbers and
oxygen percentage, number of multiple bonds, of heavy atoms, and of
terminal atoms, as well as the average molecular weight, were the
most important constitutional descriptors. The sense of the
interactions between nitrogen percentage and average molecular
weight, and between nitrogen percentage and number of terminal
atoms in the RF model based on the unbalanced data is shown for
exemplification in Figs. S1 and
S2. Among the ring descriptors, the
first two most important were the molecular cyclized degree and
aromatic ratio, both being easy to compute and easy to interpret; a
sense of their interaction in an RF model is shown in Fig. S3.
The y-scrambling test was associated with
considerably worse performance of the models re-built through the
same steps as the initial models, with respect to all performance
measures employed (e.g., PPV not higher than 0.50 and sensitivity
lower than 5%), thus strongly suggesting that the good performance
of the models was not the result of chance, but rather of a real
association between the cytotoxic effect on the melanoma cell line
SK-MEL-5 and the descriptor blocks used in those models.
Discussion
A small number of ‘local’ QSAR models have been
published (44–47), focused on the cytotoxicity of a
limited number of similar substances against one or several cancer
cell lines, but such models have a narrow range of chemical
structures and a narrow domain of applicability (48). Our study is one of the few where
cytotoxicity assessed on a cancer cell line (SK-MEL-5) is explored
through ‘global’ QSAR modelling. Such an approach is more
challenging, because even for a single therapeutic target (a
protein) median efficacy values (such as IC50) are more
heterogeneous and likely to be affected by multiple sources of
errors and to differ from one laboratory to another and from one
experiment to another, depending on the experimental conditions. It
is of notoriety that assays based on MTT and analogues rarely give
consistent IC50 values. In the case of cisplatin effect
on the SKOV-3 cell lines, the IC50 values reported in 17
published study sources varied between 2 and 40 µM, and although at
the beginning it was thought that those inconsistencies were
related to the reagents and their way of using them in various
laboratories, it was later discovered that IC50 remained
inconsistent even when the assay was carried out by the same
researcher in the same laboratory (49). Moreover, as it has been stated in the
literature with respect to the methodology used in computing such
efficacy values, ‘just because a value is obtained does not mean it
is accurate’ (50). For these
reasons, QSAR modeling of IC50 is more challenging and
this was the reason why we preferred the use of classification
techniques instead of modeling directly the IC50 values
through methods for continuous variables and our results show that
developing QSAR models with reasonable performance in these
conditions is feasible.
All seven best performing models used RF algorithm
as a classifier, as were all 16 models with PPV higher than 80% in
both nested 10-fold cross-validation and external testing. Two BST
models and one using SVM had PPV higher than 75%, but for the
latter algorithms the performance tended to be lower than that of
RFs. These classifiers were more prone to overfit, having good
performance with the artificially balanced data set (oversampling
and smote technique), but rather poor performance in the external
evaluation. In an independent study RFs also were reported to have
better performance than BST (51),
and in a comparative study it was reported that BST was more
sensitive to noise than other machine learning algorithms (52). Balancing the data, irrespective of
the classifier used tended to increase the sensitivity with a
slight cost in specificity.
Of the thirteen descriptor blocks assessed by us to
build the QSAR models, the best performing models (PPV higher than
80% in both cross-validation and external testing) used five of
these blocks: Topological descriptors, information indices,
2D-autocorrelation descriptors, P-VSA-like descriptors and edge
adjacency indices.
Of the topological descriptors, the Balaban centric
index (BAC) had the largest importance. It has been described as
reflecting the molecular shape, but as little importance in other
models published up to now (53).
Other important topological descriptors were: Path/walk-2-randic
shape index (PW2), which has been described as important in
describing the antiviral activity of azolo-adamantanes (54); lopping centric index (LOC), which has
been used previously in QSAR models for cytotoxic compounds on
cancer cell lines (55,56); and Narumi harmonic topological index,
which also has been shown useful in developing predictive
cytotoxicity models (57).
Information indices best associated with the
cytotoxic activity on the SK-MEL-5 were the mean information
content on the vertex degree equality (IVDE), which has been
previously shown to be important in predicting the COX-2 (58) and p56lck protein tyrosine kinase
(59) inhibitory activities, Balaban
U index (relevant in previous models for describing sweetness
(60). Structural information
content index (neighborhood symmetry of 0-order, SIC0), also used
earlier for COX-2 inhibition prediction (61), as well as in toxicity models
(62) turned out to be important in
our models. Other information indices pertinent for the prediction
of the anti-melanoma cell activity were the Balaban V index (shown
to be relevant for the inhibitory effect on MATE1 transporter)
(63), mean information content on
the distance equality (IDE) used beforehand in models for HDM2
inhibitors (64), the Balaban Y
index, Kier symmetry index, and the relative number of symmetry
classes (rGES; not identified as important in other published QSAR
models).
Among the 2D-autocorrelations, the most important
descriptors were geary autocorrelation of lag 1 weighted by
polarizability, used earlier to model cyclooxygenase-2 inhibitors
(GATS1p) (65); moran
autocorrelation of lag 3 weighted by Sanderson electronegativity
(MATS3e), used previously to describe the antimalarial activity
(66); geary autocorrelation of lag
3 weighted by Sanderson electronegativity (GATS3e), reported as
significant in describing the antitubercular activity of
1,4-dihydropyridine-3,5-dicarboxamides (67), moran autocorrelation of lag 3 and 2,
respectively, weighted by ionization potential (MATS3i and MATS2i),
geary autocorrelation of lag 2 weighted by mass (GATS2m), and moran
autocorrelation of lag 6 weighted by polarizability (MATS6p), not
identified in previous publications as important for other QSAR
models.
P-VSA-like descriptors have been scarcely used in
QSAR models, as shown by the scarce studies including them. Among
this group of descriptors, the most important used by us in
building models with a reasonably good performance were: P_VSA-like
on LogP, bin 5, P_VSA-like on mass, bin 4 (P_VSA_m_4), P_VSA-like
on potential pharmacophore points, aromatic atoms, P_VSA-like on
LogP, bin 1, P_VSA-like on potential pharmacophore points, L -
lipophilic, P_VSA-like on Molar refractivity, bin 1, and P_VSA-like
on Molar refractivity, bin 2. Of this group, only the P_VSA-like on
mass, bin 4 (P_VSA_m_4) was reported in models on olfactory
properties (68), whereas the
remainder have not been reported in other QSAR models as being
significant features. The same is true for the relevant
edge-adjacency descriptors used in building our models: Although a
number of other studies reported the use of different
edge-adjacency descriptors, none of those found by the feature
selection algorithms applied by us were reported in published
models: SpMAD_AEA(ed)-spectral mean absolute deviation from
augmented edge adjacency matrix weighted by edge degree;
SpMAD_EA(bo)-normalized leading eigenvalue from augmented edge
adjacency matrix weighted by bond order; Eig02_AEA(bo)-eigenvalue
n. 2 from augmented edge adjacency matrix weighted by bond order;
SpDiam_EA(bo)-spectral diameter from edge adjacency matrix weighted
by bond order; SpMAD_AEA(dm)-spectral mean absolute deviation from
augmented edge adjacency matrix weighted by dipole moment;
SpDiam_EA(dm)-spectral diameter from edge adjacency matrix weighted
by dipole moment; SpMaxA_EA(dm)-normalized leading eigenvalue from
edge adjacency matrix weighted by dipole moment.
Simpler, more easily interpretable descriptors, such
as constitutional ones, ring descriptors or molecular properties
led to models with lower performance (but models with PPV higher
than 70% could be built with the constitutional and ring
descriptors).
Exploring a variety of descriptor blocks to produce
QSAR models able to anticipate the cytotoxicity of chemical
compounds on the cancer cell line SK-MEL-5, we were able to build
models with good performance in terms of selectivity and PPV, but
with relatively low sensitivity. In other words, the models built
have good performance in having a low rate of false positives, but
this is done at the cost of labelling about half of the active
compounds as ‘inactive’. Of the four classification algorithms
applied, RF was the most effective, all models with PPV higher than
85% in both (nested) cross-validation and external evaluation being
built with this classifier. The descriptors most appropriate to
describe the effect on the cancer cell line SK-MEL-5 were
topological, information indices, 2D-autocorrelation descriptors,
P-VSA-like descriptors and edge adjacency indices. All these groups
are rather hard to interpret in a simple manner, but simpler
descriptors (e.g., constitutional descriptors, ring descriptors,
molecular properties) led to less successful models.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was partially supported by a grant of
Romanian Ministry of Research and Innovation (CCCDI-UEFISCDI)
(project no. 61PCCDI⁄2018 PN-III-P1-1.2-PCCDI-2017-0341; Bucharest,
Romania) within PNCDI–III.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RA was responsible for the conception and design of
the study, checked the primary data and performed the modelling. IN
collected and analysed the primary data. MD, IN, FGL, DB
contributed to the design and interpretation of the data and
writing the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
RA has received consultancy and speakers' fees from
various pharmaceutical companies. MD, IN, FGL and DB declare they
have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
BST
|
gradient boosting
|
|
ERM
|
empirical risk minimization
|
|
KNN
|
k-nearest neighbors
|
|
PPV
|
positive predictive value
|
|
QSAR
|
quantitative structure-activity
relationship
|
|
RF
|
random forests
|
|
SRM
|
structure risk minimization
|
|
SVM
|
support vector machines
|
References
|
1
|
European Chemical Agency (ECHA): Practical
guide, . How to use and report (Q)SARs. Version 3.1. ECHA;
Helsinki: 2016, https://echa.europa.eu/documents/10162/13655/pg_report_qsars_en.pdfJuly.
2016
|
|
2
|
Launer RL and Wilkinson GN: Robustness in
the strategy of scientific model building. Robustness in Statistics
(1st). Elsevier. 201–236. 1979.
|
|
3
|
Aouidate A, Ghaleb A, Ghamali M, Chtita S,
Ousaa A, Choukrad M, Sbai A, Bouachrine M and Lakhlifi T: QSAR
study and rustic ligand-based virtual screening in a search for
aminooxadiazole derivatives as PIM1 inhibitors. Chem Cent J.
12:322018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lima MNN, Melo-Filho CC, Cassiano GC,
Neves BJ, Alves VM, Braga RC, Cravo PVL, Muratov EN, Calit J,
Bargieri DY, et al: QSAR-driven design and discovery of novel
compounds with antiplasmodial and transmission blocking activities.
Front Pharmacol. 9:1462018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qin L, Zhang X, Chen Y, Mo L, Zeng H and
Liang Y: Predictive QSAR models for the toxicity of disinfection
byproducts. Molecules. 22:16712017. View Article : Google Scholar
|
|
6
|
Sun L, Yang H, Li J, Wang T, Li W, Liu G
and Tang Y: In silico pediction of compounds binding to human
plasma proteins by QSAR models. ChemMedChem. 13:572–581. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nembri S, Grisoni F, Consonni V and
Todeschini R: In silico prediction of cytochrome P450-drug
interaction: QSARs for CYP3A4 and CYP2C9. Int J Mol Sci.
17:9142016. View Article : Google Scholar
|
|
8
|
Garmpis N, Damaskos C, Garmpi A,
Dimitroulis D, Spartalis E, Margonis GA, Schizas D, Deskou I, Doula
C, Magkouti E, et al: Targeting histone deacetylases in malignant
melanoma: A future therapeutic agent or just great expectations?
Anticancer Res. 37:5355–5362. 2017.PubMed/NCBI
|
|
9
|
Stueven NA, Schlaeger NM, Monte AP, Hwang
SL and Huang CC: A novel stilbene-like compound that inhibits
melanoma growth by regulating melanocyte differentiation and
proliferation. Toxicol Appl Pharmacol. 337:30–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marra A, Ferrone CR, Fusciello C,
Scognamiglio G, Ferrone S, Pepe S, Perri F and Sabbatino F:
Translational research in cutaneous melanoma: New therapeutic
perspectives. Anticancer Agents Med Chem. 18:166–181. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Theodosakis N, Micevic G, Langdon CG,
Ventura A, Means R, Stern DF and Bosenberg MW: p90RSK blockade
inhibits dual BRAF and MEK inhibitor-resistant melanoma by
targeting protein synthesis. J Invest Dermatol. 137:2187–2196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mioc M, Pavel IZ, Ghiulai R, Coricovac DE,
Farcaş C, Mihali CV, Oprean C, Serafim V, Popovici RA, Dehelean CA,
et al: The cytotoxic effects of betulin-conjugated gold
nanoparticles as stable formulations in normal and melanoma cells.
Front Pharmacol. 9:4292018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Memorial Sloan Kettering and Cancer
Center: SK-MEL-5: Human Melanoma Cell Line (ATCC HTB 70).
https://www.mskcc.org/research-advantage/support/technology/tangible-material/human-melanoma-cell-line-sk-mel-5August
30–2018
|
|
14
|
Al-Qathama A, Gibbons S and Prieto JM:
Differential modulation of Bax/Bcl-2 ratio and onset of caspase-3/7
activation induced by derivatives of Justicidin B in human melanoma
cells A375. Oncotarget. 8:95999–96012. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carbone C, Martins-Gomes C, Pepe V, Silva
AM, Musumeci T, Puglisi G, Furneri PM and Souto EB: Repurposing
itraconazole to the benefit of skin cancer treatment: A combined
azole-DDAB nanoencapsulation strategy. Colloids Surf B
Biointerfaces. 167:337–344. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Al-Sanea MM, Ali Khan MS, Abdelazem AZ,
Lee SH, Mok PL, Gamal M, Shaker ME, Afzal M, Youssif BG and Omar
NN: Synthesis and in vitro antiproliferative activity of new
1-phenyl-3-(4-(pyridin-3-yl)phenyl)urea scaffold-based compounds.
Molecules. 23:2972018. View Article : Google Scholar
|
|
17
|
Plitzko B, Kaweesa EN and Loesgen S: The
natural product mensacarcin induces mitochondrial toxicity and
apoptosis in melanoma cells. J Biol Chem. 292:21102–21116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kalliokoski T, Kramer C, Vulpetti A and
Gedeck P: Comparability of mixed IC50 data: A
statistical analysis. PLoS One. 8:e610072013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Niu AQ, Xie LJ, Wang H, Zhu B and Wang SQ:
Prediction of selective estrogen receptor beta agonist using open
data and machine learning approach. Drug Des Devel Ther.
10:2323–2331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Datta S and Das S: Near-bayesian support
vector machines for imbalanced data classification with equal or
unequal misclassification costs. Neural Netw. 70:39–52. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cortez P: Package ‘rminer’: Data Mining
Classification and Regression Methods. Version 1.4.2. https://cran.r-project.org/web/packages/rminer/rminer.pdfSeptember
2–2016
|
|
22
|
R Core Team R, . A Language and
Environment for Statistical Computing. R Foundation for Statistical
Computing; Vienna: 2018
|
|
23
|
Bischl B, Lang M, Kotthoff L, Schiffner J,
Richter J, Studerus E, Casalicchio G and Jones ZM: mlr: Machine
learning in R. J Mach Learn Res. 17:1–5. 2016.
|
|
24
|
Liaw A and Wiener M: Classification and
regression by randomForest. R News. 2:18–22. 2002.
|
|
25
|
Romanski P and Kotthoff L: FSelector:
Selecting Attributes. R package. version 0.31. https://cran.r-project.org/web/packages/FSelector/index.htmlNovember
19–2018
|
|
26
|
Ho TK: Random decision forests. 1. IEEE
Computer Society Press; Washington, DC: pp. 278–282. 1995
|
|
27
|
Breiman L: Random forests. Mach Learn.
45:5–32. 2001. View Article : Google Scholar
|
|
28
|
Svetnik V, Liaw A, Tong C, Culberson JC,
Sheridan RP and Feuston BP: Random forest: A classification and
regression tool for compound classification and QSAR modeling. J
Chem Inf Comput Sci. 43:1947–1958. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Natekin A and Knoll A: Gradient boosting
machines, a tutorial. Front Neurorobot. 7:212013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He T, Heidemeyer M, Ban F, Cherkasov A and
Ester M: SimBoost: a read-across approach for predicting
drug-target binding affinities using gradient boosting machines. J
Cheminformatics. 9:242017. View Article : Google Scholar
|
|
31
|
Wang Z: Package ‘bst’: Gradient Boosting.
Version 0.3–15. https://cran.r-project.org/web/packages/bst/bst.pdfJuly
23–2018
|
|
32
|
Therneau T and Atkinson B: Package
‘rpart’: Recursive Partitioning and Regression Trees. Version
4.1–13. https://cran.r-project.org/web/packages/rpart/rpart.pdfFebruary
23–2018August 30–2018
|
|
33
|
Luo M, Wang XS, Roth BL, Golbraikh A and
Tropsha A: Application of quantitative structure-activity
relationship models of 5-HT1A receptor binding to
virtual screening identifies novel and potent 5-HT1A
ligands. J Chem Inf Model. 54:634–647. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pourbasheer E, Vahdani S, Malekzadeh D,
Aalizadeh R and Ebadi A: QSAR Study of 17β-HSD3 inhibitors by
genetic algorithm-support vector machine as a target receptor for
the treatment of prostate cancer. Iran J Pharm Res. 16:966–980.
2017.PubMed/NCBI
|
|
35
|
Meyer D, Dimitriadou E, Hornik K,
Weingessel A and Leisch F; e1071: Misc Functions of the Department
of Statistics, . Probability Theory Group (Formerly: E1071), TU
Wien. Version 1.6–8. https://rdrr.io/rforge/e1071/May 31–2017
|
|
36
|
Cai C, Fang J, Guo P, Wang Q, Hong H,
Moslehi J and Cheng F: In silico pharmacoepidemiologic evaluation
of drug-induced cardiovascular complications using combined
classifiers. J Chem Inf Model. 58:943–956. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roy K, Kar S and Ambure P: On a simple
approach for determining applicability domain of QSAR models.
Chemometr Intell Lab Syst. 145:22–29. 2015. View Article : Google Scholar
|
|
38
|
Li S: Package ‘rknn’: Random KNN
Classification and Regression. https://cran.r-project.org/web/packages/rknn/rknn.pdfJune
7–2015
|
|
39
|
Warnes GR, Bolker B and Lumley T: gtools:
various R programming tools. R Foundation for Statistical
Computing; Vienna: 2015
|
|
40
|
Sahigara F, Ballabio D, Todeschini R and
Consonni V: Defining a novel k-nearest neighbours approach to
assess the applicability domain of a QSAR model for reliable
predictions. J Cheminform. 5:272013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Williams K: Package ‘ldbod’: Local
Density-Based Outlier Detection. Version 0.1.2. https://cran.r-project.org/web/packages/ldbod/ldbod.pdfMay
26–2017
|
|
42
|
Xu C, Cheng F, Chen L, Du Z, Li W, Liu G,
Lee PW and Tang Y: In silico prediction of chemical Ames
mutagenicity. J Chem Inf Model. 52:2840–2847. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gower JC: A general coefficient of
similarity and some of its properties. Biometrics. 27:8571971.
View Article : Google Scholar
|
|
44
|
Miladiyah I, Jumina J, Haryana SM and
Mustofa M: Biological activity, quantitative structure-activity
relationship analysis, and molecular docking of xanthone
derivatives as anticancer drugs. Drug Des Devel Ther. 12:149–158.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yadav DK, Kumar S, Saloni S, Singh H, Kim
MH, Sharma P, Misra S and Khan F: Molecular docking, QSAR and ADMET
studies of withanolide analogs against breast cancer. Drug Des
Devel Ther. 11:1859–1870. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gaikwad R, Ghorai S, Amin SA, Adhikari N,
Patel T, Das K, Jha T and Gayen S: Monte Carlo based modelling
approach for designing and predicting cytotoxicity of
2-phenylindole derivatives against breast cancer cell line MCF7.
Toxicol In Vitro. 52:23–32. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Abdelhaleem EF, Abdelhameid MK, Kassab AE
and Kandeel MM: Design and synthesis of thienopyrimidine urea
derivatives with potential cytotoxic and pro-apoptotic activity
against breast cancer cell line MCF-7. Eur J Med Chem.
143:1807–1825. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Feher M and Ewing T: Global or local QSAR:
Is there a way out? QSAR Comb Sci. 28:850–855. 2009. View Article : Google Scholar
|
|
49
|
He Y, Zhu Q, Chen M, Huang Q, Wang W, Li
Q, Huang Y and Di W: The changing 50% inhibitory concentration
(IC50) of cisplatin: A pilot study on the artifacts of the MTT
assay and the precise measurement of density-dependent
chemoresistance in ovarian cancer. Oncotarget. 7:70803–70821. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sebaugh JL: Guidelines for accurate
EC50/IC50 estimation. Pharm Stat. 10:128–134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kryshchyshyn A, Devinyak O, Kaminskyy D,
Grellier P and Lesyk R: Development of predictive QSAR models of
4-thiazolidinones antitrypanosomal activity using modern machine
learning algorithms. Mol Inform. 37:e17000782018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cortes-Ciriano I, Bender A and Malliavin
TE: Comparing the influence of simulated experimental errors on 12
machine learning algorithms in bioactivity modeling using 12
diverse data sets. J Chem Inf Model. 55:1413–1425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dearden JC: The use of topological indices
in QSAR and QSPR modeling. Advances in QSAR modeling. Roy K: 24.
Springer International Publishing; Cham: pp. 57–88. 2017,
View Article : Google Scholar
|
|
54
|
Karbakhsh R and Sabet R: Application of
different chemometric tools in QSAR study of azolo-adamantanes
against influenza A virus. Res Pharm Sci. 6:23–33. 2011.PubMed/NCBI
|
|
55
|
Prachayasittikul V, Pingaew R,
Anuwongcharoen N, Worachartcheewan A, Nantasenamat C,
Prachayasittikul S, Ruchirawat S and Prachayasittikul V: Discovery
of novel 1,2,3-triazole derivatives as anticancer agents using QSAR
and in silico structural modification. Springerplus. 4:5712015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fereidoonnezhad M, Faghih Z, Mojaddami A,
Rezaei Z and Sakhteman A: A comparative QSAR analysis, molecular
docking and PLIF studies of some N-arylphenyl-2,
2-dichloroacetamide analogues as anticancer agents. Iran J Pharm
Res. 16:981–998. 2017.PubMed/NCBI
|
|
57
|
Edraki N, Das U, Hemateenejad B, Dimmock
JR and Miri R: Comparative QSAR analysis of 3,5-bis
(arylidene)-4-piperidone derivatives: The development of predictive
cytotoxicity models. Iran J Pharm Res. 15:425–437. 2016.PubMed/NCBI
|
|
58
|
Akbari S, Zebardast T, Zarghi A and
Hajimahdi Z: QSAR modeling of COX-2 inhibitory activity of some
dihydropyridine and hydroquinoline derivatives using multiple
linear regression (MLR) method. Iran J Pharm Res. 16:525–532.
2017.PubMed/NCBI
|
|
59
|
Fassihi A and Sabet R: QSAR study of
p56(lck) protein tyrosine kinase inhibitory activity of flavonoid
derivatives using MLR and GA-PLS. Int J Mol Sci. 9:1876–1892. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rojas C, Todeschini R, Ballabio D, Mauri
A, Consonni V, Tripaldi P and Grisoni F: A QSTR-based expert system
to predict sweetness of molecules. Front Chem. 5:532017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Mohanapriya A and Achuthan D: Comparative
QSAR analysis of cyclo-oxygenase2 inhibiting drugs. Bioinformation.
8:353–358. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chavan S, Nicholls IA, Karlsson BC,
Rosengren AM, Ballabio D, Consonni V and Todeschini R: Towards
global QSAR model building for acute toxicity: Munro database case
study. Int J Mol Sci. 15:18162–18174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wittwer MB, Zur AA, Khuri N, Kido Y,
Kosaka A, Zhang X, Morrissey KM, Sali A, Huang Y and Giacomini KM:
Discovery of potent, selective multidrug and toxin extrusion
transporter 1 (MATE1, SLC47A1) inhibitors through prescription drug
profiling and computational modeling. J Med Chem. 56:781–795. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Dai Y, Chen N, Wang Q, Zheng H, Zhang X,
Jia S, Dong L and Feng D: Docking analysis and multidimensional
hybrid QSAR model of 1,4-benzodiazepine-2,5-diones as HDM2
antagonists. Iran J Pharm Res. 11:807–830. 2012.PubMed/NCBI
|
|
65
|
Sharma BK, Singh P, Pilania P, Shekhawat M
and Prabhakar YS: QSAR of 2-(4-methylsulphonylphenyl) pyrimidine
derivatives as cyclooxygenase-2 inhibitors: Simple structural
fragments as potential modulators of activity. J Enzyme Inhib Med
Chem. 27:249–260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sharma BK, Verma S and Prabhakar YS:
Topological and physicochemical characteristics of
1,2,3,4-Tetra-hydroacridin-9(10H)-ones and their antimalarial
profiles: A composite insight to the structure-activity relation.
Curr Comput Aided Drug Des. 9:317–335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Rasouli Y and Davood A: Hybrid Docking -
QSAR studies of 1,4-dihydropyridine-3, 5-dicarboxamides as
potential antitubercular agents. Curr Comput Aided Drug Des.
14:35–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li H, Panwar B, Omenn GS and Guan Y:
Accurate prediction of personalized olfactory perception from
large-scale chemoinformatic features. Gigascience. 7:72018.
View Article : Google Scholar
|