Introduction
Breast cancer is the most significant health problem
in women worldwide, which accounted for an estimated 1.7 million
new cases and 521,900 cases of cancer-associated mortality globally
in 2012 (1). Histologically, breast
cancer can be classified into ductal carcinoma in situ,
ductal and lobular carcinoma, and invasive breast cancer, and this
classification is useful in selecting tumor lesions for surgical
resection; however, there is no or limited value for the selection
of targeted therapy. In addition, breast cancer can be molecularly
subtyped according to the expression of estrogen receptor (ER),
progesterone receptor (PR) and human epidermal growth factor 2
receptor (HER2/neu) (2), or even
according to gene signature (3). The
three receptor-positive types of breast cancer can be effectively
controlled by hormonal and targeted therapy, including tamoxifen or
trastuzumab (4,5). However, triple negative breast cancer
(TNBC), which does not express ER, PR or Her2/neu, is more
difficult to treat (2). TNBC
accounts for 15–20% of all breast cancer cases and has a high risk
of early recurrence (6) due to poor
response to conventional chemo- or radiotherapy; however, systemic
chemotherapy is the only strategy currently available for recurrent
or metastatic TNBC (7). The median
survival of patients with metastatic TNBC following conventional
chemotherapy is only 9–12 months (8). Therefore, the identification of novel
and effective strategies to control TNBC is urgently required.
Cell division cycle-associated protein (CDCA) 4,
also known as SEI-3/hematopoietic progenitor protein (HEPP), is a
target gene of the transcription factor E2F that can repress
E2F-dependent transcriptional activation and cell proliferation
(9). CDCA4 was initially identified
by Abdullah et al and termed HEPP due to its preferential
expression in fetal and adult hematopoietic progenitor cells and
mature blood cells (10). CDCA4
contains four highly conserved characteristic sequences: A cyclin A
binding domain, C-terminal motif, SERTA domain and plant
homeodomain (PHD)-bromine binding domain, which are closely
associated with the functions of the SEI family (11–13).
Therefore, CDCA4 is also referred to as SEI-3 or TRIP-Br3. Previous
studies have demonstrated that SEI-1 and SEI-2 are involved in
E2F-mediated cell cycle progression and tumorigenesis (14), while DNA damage induces the binding
of E2F-1 and p53 to the CDCA4 cyclin A binding domain to promote
apoptosis (15). In addition, the
SEI family proteins, including CDCA4, can regulate p53-dependent
transcriptional activity, and overexpression of the SEI family
proteins inhibits proliferation of HeLa and U2OS cell lines
(9) and suppresses c-JUN expression
(16), while the association of
CDCA4 with the formation and distribution of the spindle in early
and mid-mitotic stages may serve as a main transcription factor in
chromosome segregation and cytoplasmic division (17). Therefore, further studies concerning
this family of proteins, including CDCA4, could provide an improved
understanding of their role in tumorigenesis and may provide a
novel target for the clinical control of TNBC.
The present study investigated the effects of CDCA4
knockdown, using CDCA4 short hairpin (sh)RNA (shCDCA4), on the
regulation of TNBC cell proliferation in vitro and in
vivo. This provided novel insights into the role of CDCA4 in
TNBC MDA-MB-231 cells.
Materials and methods
Gene information
The online resource Metabolic gEne Rapid Visualizer
(MERAV: http://merav.wi.mit.edu/, accessed by
January 20, 2018) was used to generate boxplots of the expression
levels of CDCA4 in normal breast tissue and primary breast tumors
tissue (18).
Cell lines and culture
Human breast cancer (MDA-MB-231, MDA-MB-468 and
T-47D) cell lines and the normal human mammary gland Hs578BST cell
line were obtained from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China) and cultured in Dulbecco's modified
Eagle's medium (DMEM; GE Healthcare Life Sciences, Little Chalfont,
UK) supplemented with 10% fetal bovine serum (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) and 1% antimycotic
(Penicillin-Streptomycin-Amphotericin B solution; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified
chamber with 5% CO2.
RNA interference
shCDCA4 constructs targeting the CDCA4 cDNA sequence
(5′-CCTAGACCTAAGAGTAAATTA-3′) were synthesized and cloned into the
GV115 lentiviral vector (Shanghai GeneChem Co., Ltd., Shanghai,
China). Subsequently, 293T cells were co-infected with lentiviral
vector carrying the shCDCA4 or negative control shRNA (shCtrl;
5′-TTCTCCGAACGTGTCACGT-3′; Shanghai GeneChem Co., Ltd.) and
packaging plasmids. The lentiviruses were then harvested and the
virus titer was determined. Additionally, the lentiviral vectors
carried firefly luciferase and green fluorescent protein (GFP)
genes to label tumor cells. The TNBC cell lines were transfected
with lentivius (MOI=10) in a 24-well plate (5×104
cells/well). Following transfection for 24 h, the fresh complete
medium was replaced and the cells were cultured for an additional
72 h at 37°C. These lentiviruses were used to stably knockdown
CDCA4 expression in MDA-MB-231 and MDA-MB-468 breast cancer cell
lines, and shRNA-infected cells were selected in puromycin (5
ug/ml)-containing DMEM (Clontech Laboratories, Inc., Mountainview,
CA, USA). The images of infected cells were observed under a
phase-contrast and fluorescence microscope (Olympus Corporation,
Tokyo, Japan).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The concentration
of RNA samples was determined using a NanoDrop spectrophotometer
(NanoDrop Technologies, Thermo Fisher Scientific, Inc., Wilmington,
DE, USA) and reverse transcribed into cDNA using oligo (dT) primers
and a reverse transcriptase from Moloney murine leukemia virus
(Promega Corporation, Madison, WI, USA). The temperature protocol
for RT was as follows: 42°C for 1 h, 70°C for 5 min followed by
storage at 4°C. The resulting cDNA samples were subjected to qPCR
amplification using the SYBR Premix Ex Taq kit (Takara Bio, Inc.,
Otsu, Japan) and the ABI 7500 apparatus (Applied Biosystems; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. The optimized parameters for qPCR were set
to an initial cycle of 95°C for 10 min, followed by 40 cycles of
95°C for 5 sec, 60°C for 30 sec and 72°C for 5 sec, and then cooled
to and maintained at 4°C. The primer sequences were: Human CDCA4,
forward 5′-ATTTGAAACGCTGGAGACT-3′, reverse
5′-CCCATCATGCCTGTCAGTA-3′; and GAPDH, forward
5′-TGACTTCAACAGCGACACCCA-3′, and reverse
5′-CACCCTGTTGCTGTAGCCAAA-3′. GAPDH was used as the reference gene.
The 2−ΔΔCq method was used to calculate the relative
mRNA expression levels of CDCA4 as previously described (19).
MTT cell viability assay
MDA-MB-231 cells, infected with lentivirus carrying
shCDCA4 or shCtrl, were seeded into 96-well plates at a density of
2,000 cells/well and grown for up to 5 days. Subsequently, 20 µl
MTT (5 mg/ml; GenView, Tallahassee, FL, USA) was added to the cell
culture, and the cells were cultured for an additional 4 h at 37°C.
Subsequently, 100 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) replaced the cell culture medium to dissolve
the formazan crystals for 15 min. Optical density values were
measured using a microplate reader (Synergy H1; BioTek China,
Beijing, China) at 490 nm. The experiments were performed in
triplicate and repeated at least three times independently.
Cell counting assay
Logarithmic growth phase MDA-MB-231 cells, infected
with lentivirus carrying shCDCA4 or shCtrl, were seeded into
96-well plates at a density of 1,500 cells/well and incubated at
37°C with 5% CO2 for up to 5 days. Subsequently, the
cells were counted using the Celigo imaging cytometer (Nexcelom
Bioscience, LLC, Lawrence, MA, USA). The experiments were performed
in triplicate and repeated at least three times independently.
Flow cytometric cell cycle and Annexin
V-allophycocyanin (APC) apoptosis assays
MDA-MB-231 cell cycle distribution and the levels of
apoptosis were assessed using propidium iodide (PI; Sigma-Aldrich;
Merck KGaA) and the Annexin V Apoptosis Detection kit APC (Thermo
Fisher Scientific, Inc., Waltham, MA, USA), respectively, according
to the manufacturers' protocols. Briefly, following infection with
lentivirus carrying shCDCA4 or shCtrl, MDA-MB-231 cells were seeded
into a 6-well plate at a density of 1×105 cells/well and
grown for 5 days.
For the cell cycle assay, cells were harvested using
trypsin, washed twice in D-Hanks buffer (pH 7.2–7.4), fixed with
70% ethanol for 30 min at 20°C, and stored at 4°C overnight.
Subsequently, the cells were stained with 50 µg/ml PI solution
containing 100 µg/ml RNase A (Sigma-Aldrich; Merck KGaA) and
incubated for 1 h at room temperature in the dark. The cell cycle
distribution was analyzed using a fluorescence-activated cell
sorting (FACS) analyzer (EMD Millipore, Billerica, MA, USA).
For the apoptosis assay, cells were harvested using
trypsin, washed twice in D-Hanks buffer (pH 7.2–7.4), and
resuspended in the binding buffer from the kit. The cell suspension
(990 µl) was then supplemented with 10 µl Annexin V-APC solution
and incubated for 15 min at room temperature in the dark. The rate
of cell apoptosis was analyzed using the FACS analyzer. Analysis of
flow cytometery data was performed with the GuavaSoft foftware
package for Guava easyCyte HT systems (version 2.5; EMD
Millipore).
Animal experiments
To assess the effects of CDCA4 knockdown on the
regulation of TNBC cell xenograft formation and growth in
vivo, nude mouse MDA-MB-231 cell xenografts were established. A
total of 20 female BALB/C nude mice (specific-pathogen free; age, 4
weeks; weight, 17–24 g) were purchased from the Shanghai Animal
Laboratory Center (Shanghai, China) and randomized into two groups
(n=10) receiving either shCDCA4- or shCtrl-infected MDA-MB-231
cells. The knockdown group of nude mice was subcutaneously injected
with stable shCDCA4-infected MDA-MB-231 cells (1×107
cells in 200 µl) in the right axilla, while the negative control
group of mice was subcutaneously injected with the same number and
volume of MDA-MB-231 cells stably infected with shCtrl. The nude
mice were housed in laminar flow cabinets under a specific
pathogen-free environment with access to food and water ad
libitum (temperature, 25±1°C; relative humidity, 40–60%; 12 h
light/12 h dark cycle). Tumor xenograft formation and size were
recorded every 3 days using a Vernier caliper. In addition, nude
mice were anesthetized with pentobarbital (0.7%, 50 mg/kg;
Sigma-Aldrich; Merck KGaA) and were treated with D-luciferin (10
µl/g; Shanghai Qcbio Science & Technologies Co., Ltd.,
Shanghai, China) to measure tumor cell fluorescence; the total
tumor xenograft fluorescence radiant efficiency was measured on
days 22, 29 and 36 using the IVIS Lumina LT (PerkinElmer, Inc.,
Waltham, MA, USA). After 2 months, the nude mice were sacrificed
and tumor cell xenografts were isolated and weighed. All protocols
were approved by the Ethics Review Committee of The First
Affiliated Hospital of Guangxi Medical University (Nanning,
China).
Statistical analysis
All data are expressed as the means ± standard
deviation and were analyzed with SPSS v22.0 software (IBM Corp.,
Armonk, NY, USA). A Student's t-test was performed for two-group
comparisons, and one-way analysis of variance and least significant
difference post hoc test were performed for multiple-group
comparisons. All the experiments were repeated in triplicate.
P<0.05 was considered to indicate a statistically significant
difference.
Results
High expression of CDCA4 mRNA in
breast cancer tissues and cell lines
In the present study, CDCA4 expression data were
obtained from the online MERAV database (http://merav.wi.mit.edu/; accessed January 20, 2018)
to identify CDCA4 expression in normal breast and breast tumor
tissues (18). The boxplots of CDCA4
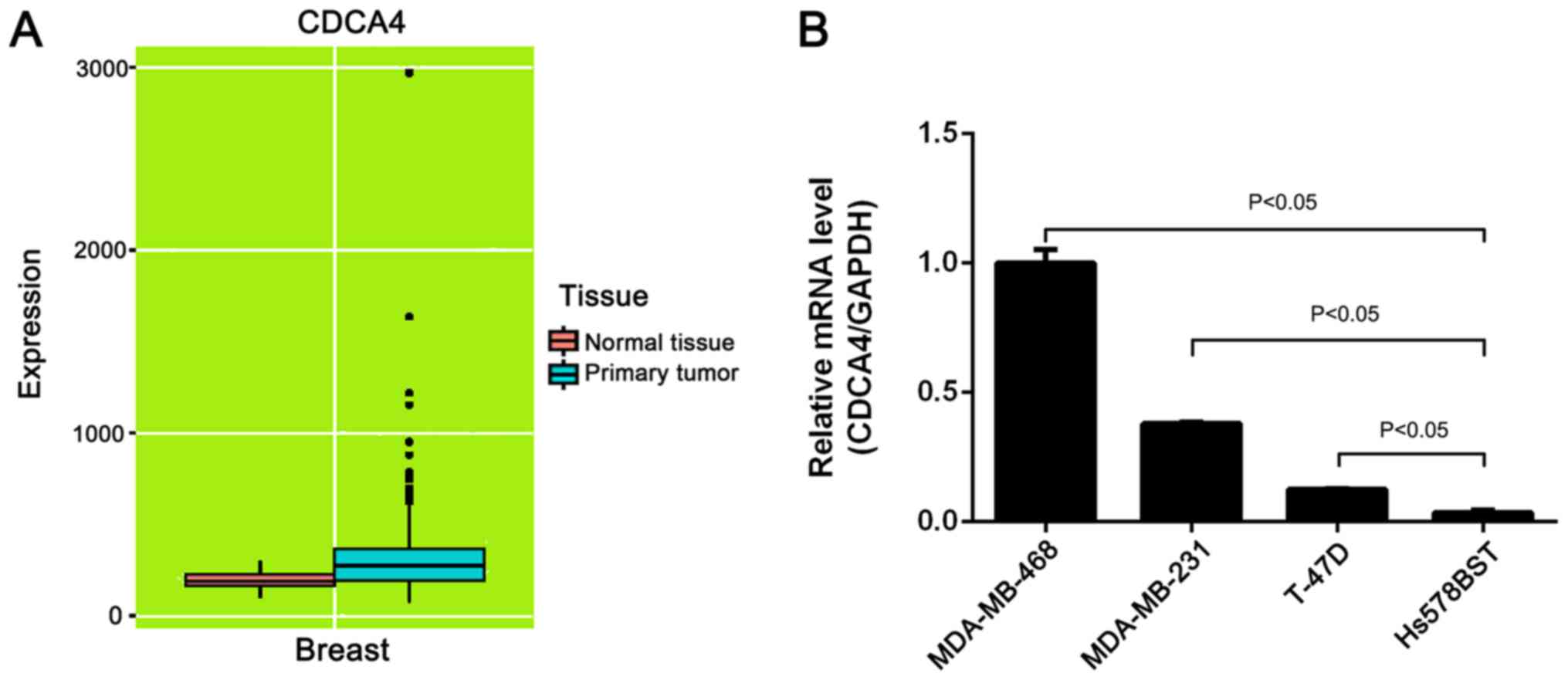
expression revealed that CDCA4 expression was higher in breast
cancer tissues than in normal tissues (Fig. 1A). Additionally, the mRNA expression
levels of CDCA4 in three breast cancer cell lines were higher than
in a normal mammary gland cell line (Fig. 1B).
Knockdown of CDCA4 expression in
breast cancer cell lines using lentivirus carrying shCDCA4 or
shCtrl
To investigate the effect of CDCA4 on breast cancer
cells, lentiviruses carrying shCDCA4 or shCtrl were prepared and
MDA-MB-231 and MDA-MB-468 breast cancer cells were infected. The
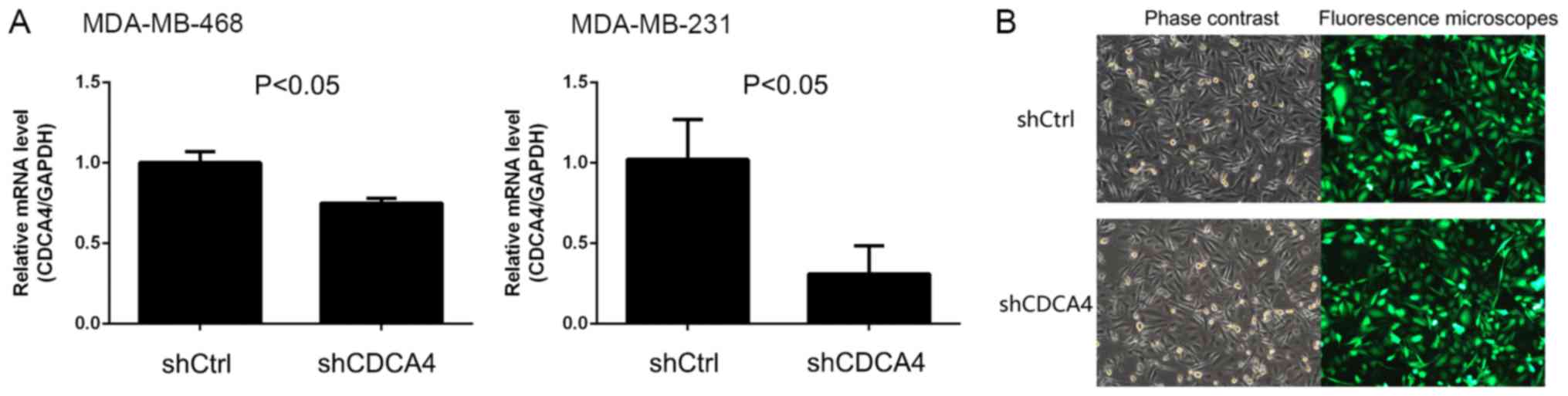
present study demonstrated that shCDCA4 was able to effectively
knockdown the mRNA expression levels of CDCA4 in TNBC MDA-MB-231
cells compared with the shCtrl; however, the knockdown efficiency
in MDA-MB-468 cells was <50% and not suitable for subsequent
experiments (Fig. 2A). Subsequently,
MDA-MB-231 cells were screened with puromycin and subjected to
fluorescence microscopy, which demonstrated that infection and GFP
expression rates were >80% (Fig.
2B). Therefore, the human TNBC MDA-MB-231 cell line was
selected as a model cell line to assess the effect of shCDCA4 on
breast cancer cells in vitro and in vivo.
shCDCA4 reduces MDA-MB-231 cell
proliferation in vitro
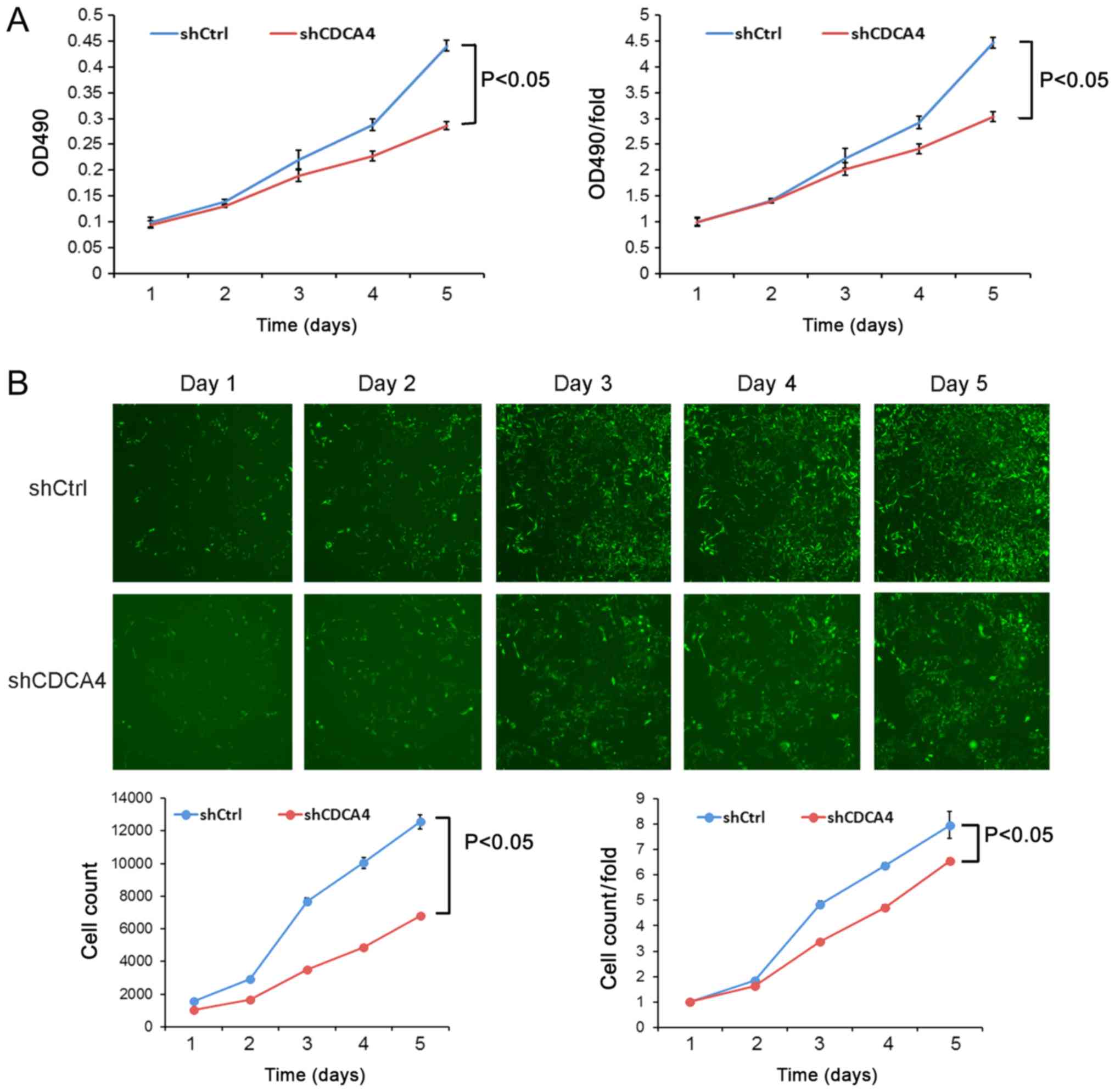
Following the knockdown of CDCA4 expression in TNBC
MDA-MB-231 cells, cell viability and cell counting assays were
performed. The cell viability following shCDCA4 infection was
significantly reduced compared with in the shCtrl group (Fig. 3A). Similarly, cell-counting Celigo
images revealed that the cell proliferation rate of the shCDCA4
group was significantly reduced (Fig.
3B). These data suggested that CDCA4 may enhance the
proliferation of MDA-MB-231 cells.
shCDCA4 induces MDA-MB-231 cell
apoptosis in vitro
The effect of CDCA4 knockdown on the regulation of
tumor cell apoptosis was assessed using FACS analysis. The data
demonstrated that, after 5 days of lentiviral infection with
shCtrl, the apoptosis rate of MDA-MB-231 cells was 3.72±0.09%,
whereas the apoptosis rate of the shCDCA4 group was 9.56±0.53%
(P<0.05, Fig. 4). These data
suggested that CDCA4 may negatively regulate apoptosis of
MDA-MB-231 cells.
shCDCA4 induces regulation of the cell
cycle
The effect of CDCA4 knockdown on regulation of the
tumor cell cycle was assessed using FACS analysis. For the
shCDCA4-infected cells, 53.05±3.51% of cells were in
G0/G1 phase, while 37.67±1.10% were in S
phase and 9.29±2.48% were in G2/M phase of the cell
cycle, which was significantly different from the percentages of
the shCtrl group (P<0.05, Fig.
5). Knockdown of CDCA4 led to increased accumulation of cells
in the S phase of the cell cycle. These data indicated that CDCA4
altered the cell cycle progression of MDA-MB-231 cells. The
inhibition of cell growth and proliferation following knockdown of
CDCA4 may be achieved by preventing the transition between S and
G2 phase.
shCDCA4 reduces the growth of
MDA-MB-231 cell xenografts in nude mice
Additionally, the effect of CDCA4 knockdown on the
regulation of breast cancer xenograft growth in vivo was
assessed by injecting MDA-MB-231 cells into nude mice following
stable infection with shCDCA4 or negative control shRNA. Tumor
volume and weight were significantly smaller in the knockdown group
compared with in the negative control group (Fig. 6A and B). In vivo small animal
imaging data also demonstrated smaller mean values for the
knockdown group, with the difference on days 29 being statistically
significant (Fig. 6C). Tumors
isolated from the nude mice were markedly smaller in the knockdown
group (Fig. 6D). The results of the
present study demonstrated that the knockdown of CDCA4 expression
suppressed the growth of MDA-MB-231 cell xenografts in
vivo.
Discussion
Due to a lack of treatment options, TNBC is a highly
invasive and metastatic malignancy (4,20). The
present study investigated the effects of CDCA4 knockdown on the
regulation of TNBC cell growth, apoptosis and xenograft growth
in vitro and in vivo. CDCA4 is a protein of the SEI
family, which contains common protein features, including the
cyclin A binding domain, C-terminal motif, SERTA domain and
PHD-bromine binding domain. MERAV database (18) analysis revealed that CDCA4 was highly
expressed in breast cancer tissue, indicating that CDCA4 may be
closely associated with breast cancer development and progression.
Our previous study demonstrated that the downregulation of CDCA4
expression significantly inhibited the proliferation of human
breast cancer doxorubicin-resistant MCF-7/ADM cells in vitro
(21). The present study revealed
that knockdown of CDCA4 expression significantly reduced the growth
of MDA-MB-231 cells and promoted their apoptosis in vitro.
Additionally, knockdown of CDCA4 expression suppressed nude mouse
MDA-MB-231 cell xenograft growth in vivo. In conclusion, the
results of the present study supported the hypothesis that CDCA4
overexpression in breast cancer tissues and cells contributes to
TNBC progression, and that targeting CDCA4 expression may be a
novel strategy in the future control of TNBC.
Notably, shCDCA4 lentivirus was infected into two
TNBC cell lines, MDA-MB-231 and MDA-MB-468. However, the silencing
efficiency of shCDCA4 in MDA-MB-468 cells was unsatisfactory
(knockdown ratio, 25.0% compared with shCtrl). Therefore, only
MDA-MB-231 cells (knockdown ratio, 68.9%) were utilized in the
present study. A possible reason for this difference may be the
poor infection efficiency of the RNA interference sequence selected
for the MDA-MB-468 cell line (20,22). In
the present study, a straightforward study design was followed by
assessing alterations in cell viability, proliferation and
apoptosis in vitro, and tumor cell xenograft growth in a
nude mouse model in vivo. The data indicated that the
knockdown of CDCA4 expression inhibited the proliferation of TNBC
cells in vitro and in vivo. These data were
consistent with those of previous studies (23), including our previous study (21).
A previous study reported that CDCA4 is an E2F
transcription factor-induced nuclear factor that regulates
E2F-dependent transcription as an E2F-downstream gene (24). CDCA4 protein is expressed in
different human cancer cell lines and induces progression of the
G1/S phase of the cell cycle (24). MicroRNA-15a-induced inhibition of
growth and invasiveness of malignant melanoma occurs by directly
targeting CDCA4 expression (23).
Furthermore, another previous study reported that the mRNA
expression levels of CDCA2, CDCA3, CDCA4, CDCA5, CDCA7 and CDCA8
are significantly higher in clinical tumor samples and cancer cell
lines compared with the control samples. Among them, the
overexpression of CDCA3, CDCA5 and CDCA8 genes is negatively
associated with the survival of patients with breast cancer
(25). Although this previous study
did not confirm the role of CDCA4 in breast cancer survival, there
are a number of factors contributing to survival of patients with
cancer. Therefore, joint survival analysis evaluating the
co-expression of multiple genes for patients with breast cancer
should be performed in future studies. Our recent study
demonstrated that CDCA4 is a downstream gene of the nuclear factor
erythroid 2 like 2 signaling pathway and that it upregulates the
proliferation of breast cancer MCF7/ADM cells (21). The present study did not explore the
underlying molecular events of CDCA4 action in TNBC cells due to
limited funding and time.
In conclusion, the present study demonstrated that
the downregulation of CDCA4 expression was able to inhibit the
proliferation and promote the apoptosis of MDA-MB-231 cells in
vitro, and the in vivo data supported the in
vitro data, demonstrating that knockdown of CDCA4 expression
suppressed the growth of MDA-MB-231 cell xenografts in vivo.
Combined with our recent study, it has been demonstrated that CDCA4
expression was not only associated with breast cancer drug
resistance but also promoted the growth of TNBC cells. Therefore, a
future study will investigate whether targeting CDCA4 expression
using shCDCA4 could be a novel strategy for treating TNBC.
Acknowledgements
Not applicable.
Funding
This study was supported in part by a grant from the
National Natural Science Foundation of China (grant no.
81260341).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW contributed to the study design and data
analysis, and the review and revision of the manuscript. SP
contributed to research design, data collection and analysis, and
drafting and revision of the manuscript. YX and JC assisted with
the statistical analysis and data interpretation. GL and JH
participated in the revision of the manuscript and statistical
analysis. All authors approved the final manuscript.
Ethics approval and consent to
participate
All protocols were approved by the Ethics Review
Committee of The First Affiliated Hospital of Guangxi Medical
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CDCA4
|
cell division cycle-associated protein
4
|
|
Ctrl
|
control
|
|
shRNA
|
short hairpin RNA
|
|
TNBC
|
triple negative breast cancer
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
de Ruijter TC, Veeck J, de Hoon JP, van
Engeland M and Tjan-Heijnen VC: Characteristics of triple-negative
breast cancer. J Cancer Res Clin Oncol. 137:183–192. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sotiriou C and Pusztai L: Gene-expression
signatures in breast cancer. N Engl J Med. 360:790–800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Romond EH, Perez EA, Bryant J, Suman VJ,
Geyer CE Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman
PA, et al: Trastuzumab plus adjuvant chemotherapy for operable
HER2-positive breast cancer. N Engl J Med. 353:1673–1684. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carlson RW, Allred DC, Anderson BO,
Burstein HJ, Carter WB, Edge SB, Erban JK, Farrar WB, Goldstein LJ,
Gradishar WJ, et al: Breast cancer. Clinical practice guidelines in
oncology. J Natl Compr Canc Netw. 7:122–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rastelli F, Biancanelli S, Falzetta A,
Martignetti A, Casi C, Bascioni R, Giustini L and Crispino S:
Triple-negative breast cancer: Current state of the art. Tumori.
96:875–888. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee A and Djamgoz MBA: Triple negative
breast cancer: Emerging therapeutic modalities and novel
combination therapies. Cancer Treat Rev. 62:110–122. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kassam F, Enright K, Dent R, Dranitsaris
G, Myers J, Flynn C, Fralick M, Kumar R and Clemons M: Survival
outcomes for patients with metastatic triple-negative breast
cancer: Implications for clinical practice and trial design. Clin
Breast Cancer. 9:29–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watanabe-Fukunaga R, Iida S, Shimizu Y,
Nagata S and Fukunaga R: SEI family of nuclear factors regulates
p53-dependent transcriptional activation. Genes Cells. 10:851–860.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abdullah JM, Jing X, Spassov DS, Nachtman
RG and Jurecic R: Cloning and characterization of Hepp, a novel
gene expressed preferentially in hematopoietic progenitors and
mature blood cells. Blood Cells Mol Dis. 27:667–676. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsu SI, Yang CM, Sim KG, Hentschel DM,
O'Leary E and Bonventre JV: TRIP-Br: A novel family of PHD zinc
finger- and bromodomain-interacting proteins that regulate the
transcriptional activity of E2F-1/DP-1. EMBO J. 20:2273–2285. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sugimoto M, Nakamura T, Ohtani N, Hampson
L, Hampson IN, Shimamoto A, Furuichi Y, Okumura K, Niwa S, Taya Y
and Hara E: Regulation of CDK4 activity by a novel CDK4-binding
protein, p34(SEI-1). Genes Dev. 13:3027–3033. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calgaro S, Boube M, Cribbs DL and Bourbon
HM: The Drosophila gene taranis encodes a novel trithorax group
member potentially linked to the cell cycle regulatory apparatus.
Genetics. 160:547–560. 2002.PubMed/NCBI
|
|
14
|
Cheong JK, Gunaratnam L, Zang ZJ, Yang CM,
Sun X, Nasr SL, Sim KG, Peh BK, Rashid SB, Bonventre JV, et al:
TRIP-Br2 promotes oncogenesis in nude mice and is frequently
overexpressed in multiple human tumors. J Transl Med. 7:82009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hsieh JK, Yap D, O'Connor DJ, Fogal V,
Fallis L, Chan F, Zhong S and Lu X: Novel function of the cyclin A
binding site of E2F in regulating p53-induced apoptosis in response
to DNA damage. Mol Cell Biol. 22:78–93. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tategu M, Nakagawa H, Hayashi R and
Yoshida K: Transcriptional co-factor CDCA4 participates in the
regulation of JUN oncogene expression. Biochimie. 90:1515–1522.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Zhu G, Yang D, Li Q, Li Y, Xu X,
He D and Zeng C: The spindle function of CDCA4. Cell Motil
Cytoskeleton. 65:581–593. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shaul YD, Yuan B, Thiru P, Nutter-Upham A,
McCallum S, Lanzkron C, Bell GW and Sabatini DM: MERAV: A tool for
comparing gene expression across human tissues and cell types.
Nucleic Acids Res. 44:D560–D566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lenz G: The RNA interference revolution.
Braz J Med Biol Res. 38:1749–1757. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu Y, Wu X, Li F, Huang D and Zhu W:
CDCA4, a downstream gene of the Nrf2 signaling pathway, regulates
cell proliferation and apoptosis in the MCF7/ADM human breast
cancer cell line. Mol Med Rep. 17:1507–1512. 2018.PubMed/NCBI
|
|
22
|
Kojima S and Borisy GG: An image-based,
dual fluorescence reporter assay to evaluate the efficacy of shRNA
for gene silencing at the single-cell level. F1000Res. 3:602014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alderman C, Sehlaoui A, Xiao Z and Yang Y:
MicroRNA-15a inhibits the growth and invasiveness of malignant
melanoma and directly targets on CDCA4 gene. Tumour Biol.
37:13941–13950. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hayashi R, Goto Y, Ikeda R, Yokoyama KK
and Yoshida K: CDCA4 is an E2F transcription factor family-induced
nuclear factor that regulates E2F-dependent transcriptional
activation and cell proliferation. J Biol Chem. 281:35633–35648.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Phan NN, Wang CY, Li KL, Chen CF, Chiao
CC, Yu HG, Huang PL and Lin YC: Distinct expression of CDCA3,
CDCA5, and CDCA8 leads to shorter relapse free survival in breast
cancer patient. Oncotarget. 9:6977–6992. 2018. View Article : Google Scholar : PubMed/NCBI
|