Introduction
Renal cell carcinoma (RCC) arises mainly in the
renal parenchyma, and accounts for >90% of kidney carcinomas
worldwide (1). RCC has the highest
mortality rate among the types of genitourinary cancer with cancer
statistics for 2018 indicating that RCC constitutes 3% of adult
malignant tumors and accounts for 2% of all cancer mortalities
(2). A significant histological
subtype of RCC is clear cell (cc)RCC. Although the treatment method
of nephrectomy has improved survival times, advanced ccRCC remains
lethal due to tumor metastasis and recurrence (3). Furthermore, the pathogenesis of ccRCC
is not well understood and the underlying genetic defects are
widespread and complex. The molecular mechanism of ccRCC tumor
growth and metastasis is also unclear and its clarification will
accelerate the identification of novel targets for treatment and
intervention.
N-lysine methyltransferase KMT5A, a member of the
SET domain-containing methyltransferase family, specifically
catalyzes the monomethylation of histone H4 lysine 20 (4,5). KMT5A
helps maintain genome integrity and regulates cell cycle
progression and development (6–11).
Previous studies have reported that KMT5A is overexpressed in
several tumors, including hepatocellular carcinoma, breast cancer,
colorectal cancer, thyroid cancer, esophageal squamous cell
carcinoma, melanoma, prostate cancer and ovarian cancer, and
promotes tumor progression (12–22). The
crucial role of KMT5A in human cancer progression has triggered an
interest in investigating the potential association between KMT5A
and ccRCC. Whether KMT5A serves a role in ccRCC invasion and
metastasis remains unknown.
The present study aimed to investigate the function
role of KMT5A in ccRCC development progression. Firstly, the
present study analyzed the expression of KMT5A and its clinical
significance in the ccRCC tissue samples using transwell assays to
evaluate the function of KMT5A in RCC cell migration and
invasiveness in vitro. The relationship between KMT5A and
CDH1 were detected in the cell line and ccRCC samples, and the
association between the combination of KMT5A and CDH1 expression
and patient overall survival (OS) time were analyzed.
Materials and methods
Tissue samples
Data from 522 ccRCC tissue samples, including gene
expression and patient survival information, were obtained from The
Cancer Genome Atlas database (https://portal.gdc.cancer.gov/). Additionally, tumor
tissue specimens were collected from 20 patients with ccRCC who
underwent resection treatment at the First Hospital of Quanzhou
Affiliated to Fujian Medical University during 2016. These patients
assigned randomly by computer (including 10 females and 10 males,
with an age range of 50–70 years). The present study was approved
by the ethics committee of the First Hospital of Quanzhou
Affiliated to Fujian Medical University.
Plasmid constructs
Lentiviral vector pLKO.1-TRC (Addgene plamid 10879)
was used for generating recombinant lentiviruses. In order to
achieve RNA interference of KMT5A, DNA fragments encoding hairpin
precursors for shKMT5A#1 (5′-CGCAACAGAATCGCAAACTTA-3′) and
shKMT5A#2 (5′-GAATCGCAAACTTACGGATTT-3′) (GENEWIZ, Suzhou, China)
were inserted into pLKO.1-TRC. A scrambled siRNA precursor (SCR,
5′-TTCTCCGAACGTGTCACGT-3′) (GENEWIZ, Suzhou, China) was used as
negative control. For re-expression of KMT5A in the ccRCC and
papillary RCC cells with silenced KMT5A expression, the full-length
KMT5A cDNA was cloned into pWPI.1 (Addgene plamid 12254).
Synonymous mutations were introduced an shKMT5A#1
(5′-CGCAGCAAAACCGTAAGCTCA-3′, mutations underlined) (GENEWIZ,
Suzhou, China) target sequences in KMT5A coding sequences to create
the RNAi resistant shKMT5A-RES plasmid.
Cell lines, transfection and
lentiviruses
Human ccRCC 786-O cells, papillary RCC ACHN cells
and 293 cells were cultured in Dulbecco's modified Eagle medium
(DMEM, 10% fetal bovine serum, Gibco; Thermo Fisher Scientific,
Inc.) and maintained at 37°C with 5% CO2 condition.
pMD2.G (2.5 µg, Addgene plamid 12259) and pSPAX2 (7.5 µg, Addgene
plamid 12260) used as helper plasmids were together with
pLKO.1-based plasmids (10 µg, Addgene plamid 10879) to transfected
into 293 cells to assemble the recombinant lentiviruses. The
transfection efficiency of 293 cells is almost 100%, the
transfection efficiency was evaluated using GFP plasmid (Clontech,
Mountain View, CA, USA). Transfection was performed using
Lipofectamine 2000 Reagent (Themo Fisher Scientific, Inc., Waltham
MA, ISA) according to the manufacturers' protocols. At 48 h later,
supernatants from co-transfections were used directly for infection
of Human ccRCC 786-O cells.
RNA extraction and RT-qPCR assay
TRIzol (Invitrogen, Thermo Fisher Scientific, Inc.)
reagent was used to extract the total RNA from the ccRCC cells
(786-O-SCR and 786-O-shKMT5A), and PrimeScript RT reagent kit
(TaKaRa Biotechnology Co., Ltd., Dalian, China) was used to convert
the mRNA to cDNA at 37°C according to the manufacturer's protocol.
The corresponding primer sequences were as follows: KMT5A forward,
5′-TCACTCTGTTTCACGCC-3′; and reverse, 5′-CATTCCTCCATCTCATC-3′; CDH1
forward, 5′-GTGAGAGGAATCCAAAGCC-3′; and reverse,
5′-AATGGCAGGGAGTTGGGG-3′; and β-actin forward,
5′-CCCTGGAGAAGAGCTAC-3′; and reverse, 5′-TAGTTTCGTGGATGCCAC-3′. The
qPCR was performed with the following conditions: 95°C/30 sec, 40
cycles of 95°C/5 sec, 60°C/15 sec and 72°C/10 sec on MXP3000 cycler
(Stratagene, La Jolla, CA, USA) for detecting the relative level of
KMT5A expression using SYBR Premix Ex Taq (TaKaRa Biotechnology
CO., Ltd.) and quantified by the 2−ΔΔCq method using
β-actin as the control and expressed as 2^(−ΔΔCq)
(23).
Transwell assays
For transwell invasion assays, matrigel (BD
biosciences, Franklin Lakes, NJ, USA) was allowed to polymerize at
the base of the top chamber of a 24-well transwell plate (8 µm,
Corning Costar Corp, Corning, NY, USA) for 45 min at 37°C. 786-O
(SCR, shKMT5A and shKMT5A-RES) and ACHN (SCR, shKMT5A and
shKMT5A-RES) cells (1×105 cells/well) were starved in
serum- and growth factor-free medium for 24 h and added to the top
chambers. The bottom chambers were filled serum-containing medium
(10% fetal bovine serum, Gibco; Thermo Fisher Scientific, Inc.).
Cultures were maintained for 48 h, and was followed by the removal
of non-invading cells from the upper surface of the membrane with
cotton swabs. Cells that had invaded were fixed in 4%
paraformaldehyde and stained with 0.1% crystal violet for 15 min at
room temperature.
Western blotting
The 786-O (SCR, shKMT5A#1 and #2) cells were lysed
in RIPA buffer (Thermo Fisher Scientific, Inc.) supplemented with
protease inhibitors (Thermo Fisher Scientific, Inc.). The protein
concentration of each sample was measured by the BCA Protein sssay
kit (Thermo Fisher Scientific, Inc.). Then 10 µg of protein lysed
buffer were subjected to a 10% SDS-PAGE and transferred to a PVDF
membrane (Millipore, MA, USA). Following blocking with 5% non-fat
milk for 2 h at room temperature, membranes were incubated with
specific primary antibody at 4°C overnight (KMT5A: 1:2,000, Abcam,
#ab3798, Cambridge, MA, USA; CDH1: 1:2,000, #14472, Cell Signaling
Technology, Inc., Danvers, MA, USA; β-actin: 1:10,000, Thermo
Fisher Scientific, Inc., #MA5-15739-HRP). Following washing with
TBST three times, membranes were incubated with HRP-conjugated
secondary antibody (1:5,000, Cell Signaling Technology, #7076,
Inc., Danvers, MA, USA) for 1 h at room temperature. Then proteins
were photographed using ECL detection reagents (Thermo Fisher
Scientific, Inc.) and detected with a Bio-Rad GelDoc XR+ system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Luciferase reporter assay
Dual Luciferase Report Assay were performed
according to the manufacturer's protocol (Dual-Glo®
Luciferase assay system, #E2940, Promega Corporation, Madison, WI,
USA). In brief, 0.6 µg luciferase reporter and were cotransfected
along with 0.3 µg pCMV-lacZ (Promega Corporation) plasmid into the
786-O cells (SCR and shKMT5A). At 48 h following transfection,
these cells were harvested and lysed in 1× reporter lysis buffer
(Promega Corporation). Luciferase activities were determined with
luciferase assay system (Promega Corporation) according to the
manufacturer's protocol. The method of normalization was the
Firefly comparison with Renilla luciferase activity.
Chromatin immunoprecipitation (CHIP)
assay
Chromatin immunoprecipitation was performed
following a published protocol (24). Antibody against KMT5A (Abcam) was
used to immunoprecipitate sonicated chromatins prepared from 786-O
cells. Five percent of each post-sonication sample was saved as the
input control and pre-immune IgG was used for specificity control.
DNA extracted from precipitated chromatins were quantitated using
qrtPCR in triplicates with primers for CDH1 promoter (forward,
5′-GCAGGTCCCATAACCCACCT-3′; and reverse,
5′-CATAGACGCGGTGACCCTCT-3′). DNA extracted from saved input were
quantitated in parallel (Ct[Input]), and results from IP by
anti-KMT5A mAb or pre-immnue IgG (Ct[IP]) were then used to
calculate relative specific occupancy or non-specific background
using the equation: 2^(Ct[IP]-Ct[Input]) ×100%.
Statistical analysis
Statistical analysis was performed with SPSS version
22.0 (IBM Corp., Armonk, NY, USA). The Kaplan-Meier analysis with
log-rank test, Pearson's test, Student's t-test One-way ANOVA and
Bonferroni's post-hoc test were used for compare the differences
between the variables. P<0.05 was considered to indicate
statistically significant differences.
Results
High KMT5A expression is associated
with shorter OS time in ccRCC
Firstly, the KMT5A mRNA levels were detected in 20
ccRCC and paired adjacent renal tissue specimens from the First
Hospital of Quanzhou Affiliated to Fujian Medical University. The
KMT5A mRNA levels were significantly upregulated in the ccRCC
tissues compared with those in the adjacent normal tissues
(Fig. 1A). Subsequently, the KMT5A
expression levels in the postoperative tumor tissues obtained from
522 patients with ccRCC were analyzed. These samples were grouped
according to the KMT5A mRNA level (high KMT5A group: The expression
range from 500.9 to 1870.24, n=261 and low KMT5A group: The
expression range from 150.65 to 500.8, n=261) and the association
between that and the OS time of the patients was assessed using
Kaplan-Meier analysis. The OS time was significantly poorer in the
high-expression group compared with the low-expression group
(Fig. 1B). Together, these findings
suggest that KMT5A serves a role in ccRCC development.
KMT5A knockdown hinders RCC cell
migration and invasiveness
In order to evaluate the function of KMT5A in RCC
cell migration and invasiveness in vitro, ccRCC and
papillary RCC cells with silenced KMT5A expression (786-O-shKMT5A
and ACHN-shKMT5A) and their corresponding controls (786-O-SCR and
ACHN-SCR) were used. Transwell assays were performed to assess the
migration and invasion ability of the cells. The cells with
silenced KMT5A expression displayed sharp declines in cell
migration and invasiveness compared with the corresponding control
groups, which was prevented by re-expression of KMT5A (Fig. 2A-D). These data demonstrate that
KMT5A enhances RCC cell migration and invasiveness in
vitro.
 | Figure 2.KMT5A improves ccRCC cell migration
and invasion. Representative Transwell migration assay images and
quantification of migratory cells for renal call carcinoma (A)
786-O and (B) ACHN cells, which had been transfected with SCR
sequence, had the KMT5A expression silenced with shKMT5A, and had
the KMT5A expression rescued with shKMT5A-RES. Representative
Transwell invasion assay images and quantification of invasive
cells for renal call carcinoma (C) 786-O and (D) ACHN cells, which
had been transfected with SCR sequence, had the KMT5A expression
silenced with shKMT5A, and had the KMT5A expression rescued with
shKMT5A-RES. Significant differences were confirmed by one-way
analysis of variance. **P<0.01, shKMT5A group compared with the
SCR group. KMT5A, N-lysine methyltransferase KMT5A; SCR, scrambled
control; shKMT5A, short hairpin RNA targeting KMT5A,
shKMT5A-RES. |
KMT5A downregulates CDH1
expression
In order to explore whether KMT5A activates EMT in
order to promote ccRCC metastasis, the effect of KMT5A on CDH1, a
central participating factor in EMT, was investigated. Firstly, the
question of whether KMT5A decreases CDH1 protein levels in ccRCC
cells was addressed. Western blot analysis was used to determine
any effects on the CDH1 protein expression levels in the
KMT5A-knockout and control cells, and revealed that knockdown of
KMT5A expression markedly upregulated CDH1 protein levels in 786-O
cells (Fig. 3A). Next, the CDH1 mRNA
levels were measured, demonstrating that the knockdown of KMT5A
increased CDH1 transcription levels (P<0.01; Fig. 3B). Furthermore, the luciferase
reporter assay revealed that silencing KMT5A in 786-O cells led to
the activation of CDH1 promoter activity (P<0.01; Fig. 3C). Finally, the chromatin
immunoprecipitation (ChIP) assay demonstrated that
immunoprecipitation using an anti-KMT5A antibody on chromatin
fragments from 786-O cells specifically enriched CDH1 promoter
sequences (P<0.01; Fig. 3D),
indicating that endogenous KMT5A is associated with the CDH1
promoter in the ccRCC cells. These data collectively indicate that
KMT5A downregulates CDH1 transcription in the ccRCC cells, likely
via the CDH1 promoter.
Prognostic value of KMT5A and CDH1 for
patients with ccRCC
Following the finding of upregulation in the
expression level of CDH1 in the 786-O cells upon KMT5A knockdown,
the correlation between the expression levels of KMT5A and CDH1 in
the tumor tissues from the aforementioned patients with ccRCC was
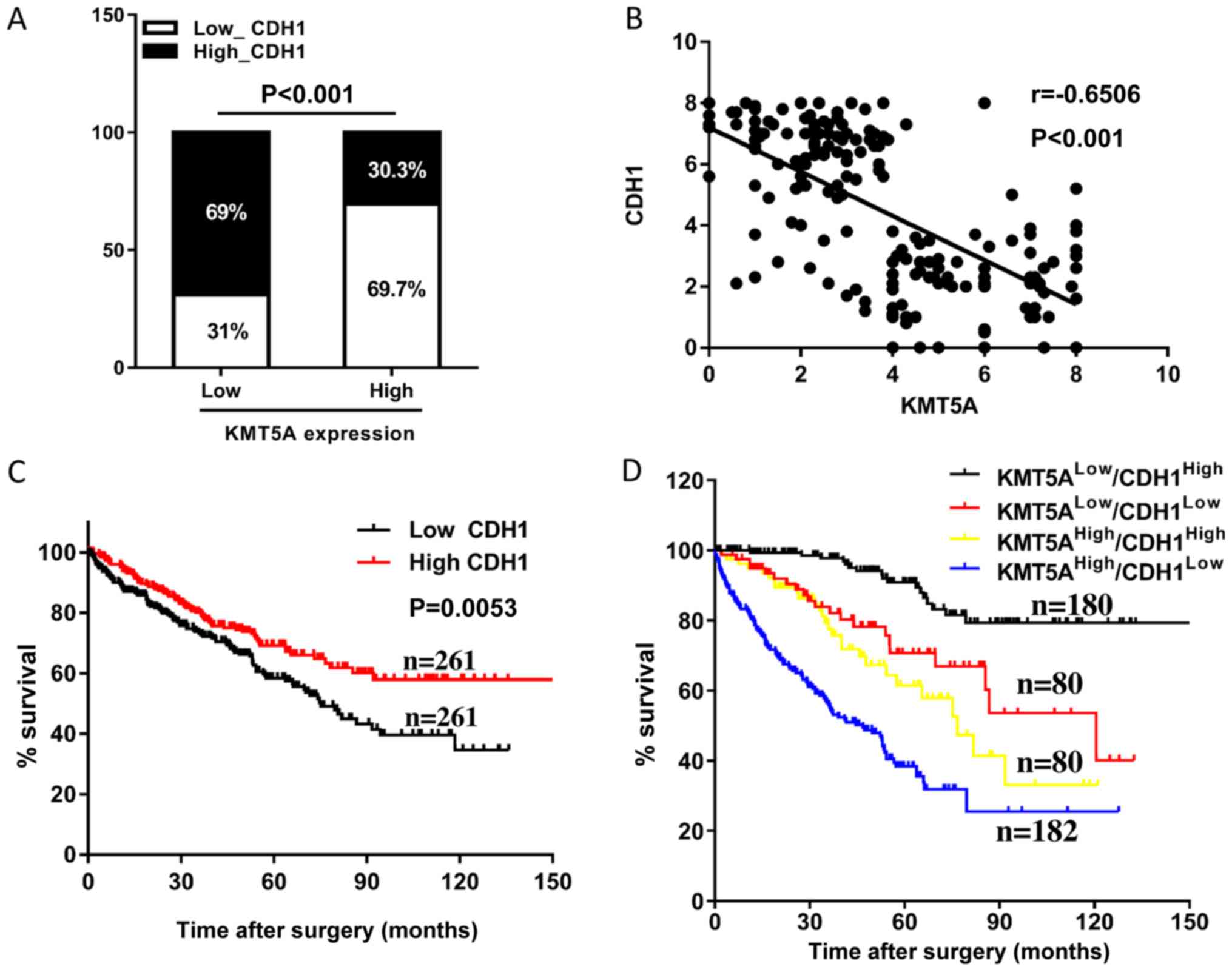
investigated. High levels of CDH1 (high CDH1 group: The expression
range from 1949.34 to 39322.42, n=261 and low CDH1 group: The
expression range from 3.37 to 1939.41, n=261) were observed in
30.3% (79/261 samples) of the high-KMT5A group in comparison with
69.0% (180/261 samples) in the low-KMT5A group (P<0.001;
Fig. 4A). Furthermore, a negative
correlation was revealed between the expression levels of KMT5A and
CDH1 in the ccRCC samples (r=−0.6506; P<0.001; Fig. 4B). The patients whose samples
expressed high CDH1 levels had longer OS times compared with those
whose samples expressed low levels (Fig.
4C). The patients with high KMT5A and low CDH1 expression
levels were associated with the shortest OS times, and those with
low KMT5A and high CDH1 expression levels were associated with the
longest (Fig. 4D). The combination
of KMT5A and CDH1 was demonstrated to be a promising independent
prognostic indicator for OS time (P<0.001), more accurate than
KMT5A or CDH1 alone.
Discussion
ccRCC is one of the most common types of kidney
cancer worldwide, and is associated with poor prognosis owing to
high rates of metastasis and recurrence (25–27). The
underlying molecular mechanism in ccRCC metastasis is not well
known. Numerous studies have reported that the EMT is an important
process in ccRCC cell metastasis (28–31). In
the present study, supporting evidence was collected for the
hypothesis that KMT5A enhances EMT through inhibiting CDH1
transcription levels, and consequently inducing ccRCC cells
metastasis. Consistent with these findings, ccRCC patients with
high KMT5A expression levels were associated with shorter OS time.
Overall, KMT5A is identified as an important factor in ccRCC cell
metastasis.
Previous studies demonstrated that KMT5A serves a
vital role in tumor development and progression (12–22). To
the best of our knowledge, the association between KMT5A and ccRCC
has not been reported to date. In the present study, it was
confirmed that the KMT5A expression level was significantly
upregulated in ccRCC tissues compared with the adjacent normal
tissues. Furthermore, ccRCC patients with KMT5A-high expression
levels in their tumor tissue were revealed to be associated with
shorter postoperative OS time. Together, these data suggest that
KMT5A may be a novel target for ccRCC treatment.
The epithelial-mesenchymal transition (EMT) is
considered to be the initial step in ccRCC metastasis (28) and CDH1 is a central factor in this
mechanism (29–31). At present, no studies have reported
on the association between KMT5A and CDH1 in ccRCC, to the best of
our knowledge. The results of the present study demonstrated that
the knockdown of KMT5A expression inhibits the migration and
invasiveness of ccRCC 786-O cells. In order to investigate the
molecular mechanism underlying the promotional effects of KMT5A on
ccRCC metastasis, the present investigated the relationship between
KMT5A and CDH1. The knockdown of KMT5A expression in the 786-O
cells upregulated CDH1 mRNA and protein levels. Furthermore,
downregulation of KMT5A increased CDH1 promoter luciferase activity
in 786-O cells. Finally, the ChIP assay revealed that endogenous
KMT5A was associated with the CDH1 promoter in the ccRCC cells.
Together, these findings indicate that KMT5A may be an important
factor in ccRCC cell migration and invasion by inhibiting CDH1
transcription.
ccRCC is associated with a markedly poor prognosis
due to high rates of metastasis and recurrence. At present, no
biomarkers are available for ccRCC diagnosis and prediction. The
present findings demonstrated that KMT5A expression level was
significantly upregulated in ccRCC tissues compared with adjacent
normal tissues. Patients with high KMT5A expression levels were
revealed to be associated with shorter postoperative OS time. In
agreement with the clinical results, in vitro experiments
demonstrated that KMT5A enhances ccRCC cell migration and invasion
by inhibiting CDH1 expression. Patients with ccRCC who exhibited
high KMT5A and low CDH1 levels had the poorest prognosis, with the
shortest OS time, and those with low KMT5A and high CDH1 expression
had the best prognosis. The combination of KMT5A and CDH1 was
confirmed to be an independent prognostic indicator for OS
(P<0.001), which was more accurate than monitoring KMT5A or CDH1
expression alone. Consequently, KMT5A serves an important role in
ccRCC development and progression, and it may be a novel target for
ccRCC treatment and prevention.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZL, DSM, YBC, JMZ, HHC and JJJ performed the
experiments and analyzed the data. ZZL and ZSZ designed the
experiments and wrote the manuscript.
Ethics approval and consent to
participate
The human sample collection was approved by the
ethical committee of the First Hospital of Quanzhou Affiliated to
Fujian Medical University. Patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martinez-Salamanca JI, Huang WC, Millan I,
Bertini R, Bianco FJ, Ciancio G, Hernández C, Herranz F, Haferkamp
A, et al: Prognostic impact of the 2009 UICC/AJCC TNM staging
system for renal cell carcinoma with venous extension. Eur Urol.
59:120–127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fang J, Feng Q, Ketel CS, Wang H, Cao R,
Xia L, Erdjument-Bromage H, Tempst P, Simon JA and Zhang Y:
Purification and functional characterization of SET8, a nucleosomal
histone H4-lysine 20-specific methyltransferase. Curr Biol.
12:1086–1099. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nishioka K, Rice JC, Sarma K,
Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P,
Tempst P, Steward R, et al: PR-Set7 is a nucleosome-specific
methyltransferase that modifies lysine 20 of histone H4 and is
associated with silent chromatin. Mol Cell. 9:1201–1213. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oda H, Okamoto I, Murphy N, Chu J, Price
SM, Shen MM, Torres Padilla ME, Heard E and Reinberg D:
Monomethylation of histone H4-lysine 20 is involved in chromosome
structure and stability and is essential for mouse development. Mol
Cell Biol. 29:2278–2295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Houston SI, McManus KJ, Adams MM, Sims JK,
Carpenter PB, Hendzel MJ and Rice JC: Catalytic function of the
PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for
mitotic entry and genomic stability. J Biol Chem. 283:19478–19488.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jorgensen S, Elvers I, Trelle MB, Menzel
T, Eskildsen M, Jensen ON, Helleday T, Helin K and Sorensen CS: The
histone methyltransferase SET8 is required for S-phase progression.
J Cell Biol. 179:1337–1345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abbas T, Shibata E, Park J, Jha S, Karnani
N and Dutta A: CRL4(Cdt2) regulates cell proliferation and histone
gene expression by targeting PR-Set7/Set8 for degradation. Mol
Cell. 40:9–21. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Centore RC, Havens CG, Manning AL, Li JM,
Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC and Zou L:
CRL4(Cdt2)-mediated destruction of the histone methyltransferase
Set8 prevents premature chromatin compaction in S phase. Mol Cell.
40:22–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu S, Wang W, Kong X, Congdon LM, Yokomori
K, Kirschner MW and Rice JC: Dynamic regulation of the PR-Set7
histone methyltransferase is required for normal cell cycle
progression. Genes Dev. 24:2531–2542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo Z, Wu C, Wang X, Wang C, Zhang R and
Shan B: A polymorphism at the miR-502 binding site in the
3′-untranslated region of the histone methyltransferase SET8 is
associated with hepatocellular carcinoma outcome. Int J Cancer.
131:1318–1322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang F, Sun L, Li Q, Han X, Lei L, Zhang H
and Shang Y: SET8 promotes epithelial-mesenchymal transition and
confers TWIST dual transcriptional activities. EMBO J. 31:110–123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song F, Zheng H, Liu B, Wei S, Dai H,
Zhang L, Calin GA, Hao X, Wei Q, Zhang W and Chen K: An
miR-502-binding site single-nucleotide polymorphism in the
3′-untranslated region of the SET8 gene is associated with early
age of breast cancer onset. Clin Cancer Res. 15:6292–6300. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu N, Huangyang P, Yang X, Han X, Yan R,
Jia H, Shang Y and Sun L: microRNA-7 suppresses the invasive
potential of breast cancer cells and sensitizes cells to DNA
damages by targeting histone methyltransferase SET8. J Biol Chem.
288:19633–19642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu B, Zhang X, Song F, Zheng H, Zhao Y,
Li H, Zhang L, Yang M, Zhang W and Chen K: MiR-502/SET8 regulatory
circuit in pathobiology of breast cancer. Cancer Lett. 376:259–267.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mosallayi M, Simonian M, Khosravi S,
Salehi AR, Khodadoostan M, Sebghatollahi V, Baradaran A and Salehi
R: Polymorphism (rs16917496) at the miR-502 binding site of the
lysine methyltransferase 5A (SET8) and Its correlation with
colorectal cancer in iranians. Adv Biomed Res. 6:772017.PubMed/NCBI
|
|
18
|
Liao T, Wang YJ, Hu JQ, Wang Y, Han LT, Ma
B, Shi RL, Qu N, Wei WJ, Guan Q, et al: Histone methyltransferase
KMT5A gene modulates oncogenesis and lipid metabolism of papillary
thyroid cancer in vitro. Oncol Rep. 39:2185–2192. 2018.PubMed/NCBI
|
|
19
|
Wang C, Wu J, Zhao Y and Guo Z: miR-502
medaited histone methyltransferase SET8 expression is associated
with outcome of esophageal squamous cell carcinoma. Sci Rep.
6:329212016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benamar M, Guessous F, Du K, Corbett P,
Obeid J, Gioeli D, Slingluff CL Jr and Abbas T: Inactivation of the
CRL4-CDT2-SET8/p21 ubiquitylation and degradation axis underlies
the therapeutic efficacy of pevonedistat in melanoma. EBioMedicine.
10:85–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yao L, Li Y, Du F, Han X, Li X, Niu Y, Ren
S and Sun Y: Histone H4 Lys 20 methyltransferase SET8 promotes
androgen receptor-mediated transcription activation in prostate
cancer. Biochem Biophys Res Commun. 450:692–696. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang C, Guo Z, Wu C, Li Y and Kang S: A
polymorphism at the miR-502 binding site in the 3′ untranslated
region of the SET8 gene is associated with the risk of epithelial
ovarian cancer. Cancer Genet. 205:373–376. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using realtime quantitative PCR and
the 2(Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee TI, Johnstone SE and Young RA:
Chromatin immunoprecipitation and microarray-based analysis of
protein location. Nat Protoc. 1:729–748. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Janzen NK, Kim HL, Figlin RA and
Belldegrun AS: Surveillance after radical or partial nephrectomy
for localized renal cell carcinoma and management of recurrent
disease. Urol Clin North Am. 30:843–852. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jemal A, Siegel R, Ward E, Murray T, Xu J,
Smigal C and Thun MJ: Cancer statistics, 2006. CA Cancer J Clin.
56:106–130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Myszczyszyn A, Czarnecka AM, Matak D,
Szymanski L, Lian F, Kornakiewicz A, Bartnik E, Kukwa W, Kieda C
and Szczylik C: The role of hypoxia and cancer stem cells in renal
cell carcinoma pathogenesis. Stem Cell Rev. 11:919–943. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krishnamachary B, Zagzag D, Nagasawa H,
Rainey K, Okuyama H, Baek JH and Semenza GL: Hypoxia-inducible
factor-1-dependent repression of E-cadherin in von Hippel- Lindau
tumor suppressor-null renal cell carcinoma mediated by TCF3,
ZFHX1A, and ZFHX1B. Cancer Res. 66:2725–2731. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Russell RC and Ohh M: The role of VHL in
the regulation of E-cadherin: A new connection in an old pathway.
Cell Cycle. 6:56–59. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Harten SK, Shukla D, Barod R, Hergovich A,
Balda MS, Matter K, Esteban MA and Maxwell PH: Regulation of renal
epithelial tight junctions by the von Hippel-Lindau tumor
suppressor gene involves occludin and claudin 1 and is independent
of E-cadherin. Mol Biol Cell. 20:1089–1101. 2009. View Article : Google Scholar : PubMed/NCBI
|