Introduction
Gastric cancer (GC) is a malignancy that originates
from the glandular epithelial cells of gastric mucosa. It is the
second most common cause of cancer-associated mortality in China
(1). According to the International
Agency for Research on Cancer (https://www.iarc.fr/), GC was the fifth most common
type of cancer worldwide in 2012 with 951,000 new cases identified,
and represented the third most common causes of cancer-associated
mortality, with 723,000 cases. In addition, >70% of new cases of
GC occur in developing countries, and ~50% originate from Eastern
Asia, mainly China (2). Treatment of
GC essentially comprises surgery, chemotherapy and radiotherapy.
However, the high rate of GC spreading to local lymph nodes, liver
and peritoneal cavity contributes to a poor prognosis and a low
5-year survival rate (<30%) (3,4).
Numerous studies suggest that the biological evolution of GC is
complex. In China, GC is typically diagnosed at an advanced stage.
It is therefore crucial to improve early diagnosis of GC and
determine its underlying molecular mechanism in order to increase
the 5-year survival rate and improve the quality of life of
patients with GC (5,6).
Long non-coding RNA (lncRNA) is an RNA molecule of
>200 nucleotides, which is not translated into protein (7). LncRNA was originally considered to be
‘noise’ in genome transcription. However, an increasing number of
lncRNAs have been identified to serve a pivotal role in cancer
development (8). A previous study
(9) reported that lncRNA acts as a
potential therapeutic target, which provides a novel direction for
treatment development. Other studies (10,11) have
revealed that lncRNAs are associated with stomach cancer
development. A previous study identified that GACAT1 expression is
correlated with tumor stage, metastasis, differentiation and
invasion depth of GC (12). Yang
et al (13) reported that H19
is overexpressed in GC and correlated with cancer cell
proliferation and apoptosis. In addition, H19 interacts with p53 to
suppress its activity, which leads to downregulation of the
downstream Bax gene, and therefore results in increased cell
proliferation and decreased cell apoptosis (13). Colon cancer-associated transcript 1
(CCAT1), which is overexpressed in GC, is associated with tumor
size and lymphatic or distant metastasis (13,14).
In vitro studies indicated that CCAT1 is associated with
stomach carcinoma cell migration, which may result from c-Myc and
E-box interaction (15). However,
the expression profile and underlying mechanism of lncRNA in GC
remain unclear. It is therefore crucial to investigate further the
functions and underlying molecular mechanism of lncRNAs in GC.
High-mobility group AT-hook 2 (HMGA2) is a crucial
protein, which is highly expressed during embryonic development and
responsible for cell differentiation (16). The gene coding for HMGA2 is located
on loci 13–15 of the long arm of chromosome 12 (17). Previous studies have reported that
HMGA2 is overexpressed in various types of cancer, including
thyroid (17), colon (18), non-small cell lung (19), squamous cell (20) and gastric (21) cancer, and is associated with invasion
and metastasis of certain types of malignancy. Furthermore, HMGA2
is an independent factor that can predict the prognosis of certain
malignant tumors (20). As a
structural transcriptional regulator, HMGA2 is involved in numerous
biological processes. For example, HMGA2 mediates glioblastoma
cancer stem cell proliferation via forkhead box M1 and plasminogen
activator urokinase regulation (22). In addition, HMGA2 serves a role in
DNA damage repair (23,24). In addition, previous studies
identified that HMGA2 promotes the epithelial-mesenchymal
transition (EMT) by targeting Twist family basic helix-loop-helix
transcription factor 1 (Twist1), Snail family transcriptional
repressor 2 and Wnt/β signaling pathways in tumors (25–28). In
summary, HMGA2 serves important roles in the biological evolution
of cancer. HIT000218960 is an lncRNA which has not been extensively
investigated. A recent study identified that HMGA2 regulates
HIT000218960 expression in thyroid carcinoma tissues (29); however, the association between HMGA2
and HIT000218960 in GC remains unclear.
To the best of our knowledge, the present study is
the first to investigate the role of HIT000218960 and HMGA2 in GC.
The present study clarified the regulatory effects of HIT000218960
and HMGA2 on cell proliferation and migration in gastric tumor,
which may provide a novel strategy and potential target for the
treatment of GC.
Materials and methods
Tissue samples and clinical
characteristics of patients with GC
In total, 60 tumor samples were obtained from
patients with GC who were admitted to The First Affiliated Hospital
of Zhejiang Chinese Medical University (Hangzhou, China) between
March 2016 and March 2017. A total of 25 normal tissues adjacent to
cancer were selected as controls. All fresh tissues were quickly
frozen in liquid nitrogen. The clinical characteristics of patients
with GC were collected, including age, sex, tumor size,
Tumor-Node-Metastasis (TNM) staging and lymph node metastasis. The
present study was approved by the Ethics Committee of The First
Affiliated Hospital of Zhejiang Chinese Medical University. All
subjects provided written informed consent.
Cell culture and transfection
The HGC27 and NCI-N87 cell lines (purchased from
Shanghai Huiying Biotechnology Co., Ltd., Shanghai, China), which
are commonly used as GC cell lines, were selected for the present
study (30,31). HGC27 and NCI-N87 cells were cultured
in RPMI-1640 (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
containing 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and placed at 37°C in a
humidified incubator containing 5% CO2. Culture medium
was changed after 2 days and replaced according to the cell culture
conditions. Cell lines were transfected with small interfering
(si)-negative control (NC) (5′-UCCGGCGACCGGGUAACCUTT-3′),
si-HIT000218960 (5′-CCUAACCCAGAGCUCUGAUUATT-3′) and
si-HIT000218960+pcDNA-HMGA2, which were designed and synthesized by
GenePharma Biotech Corp., Taipei, Taiwan. A total of 5 µl si-NC,
si-HIT000218960 and pcDNA-HMGA2 were dissolved in 250 µl
transfection dilution solution, containing LipoHigh transfection
reagent (catalog no. E607403; Sangon Biotech Co., Ltd., Shangai
China). The mixture was incubated at room temperature for 20 min.
The cells were washed three times with PBS prior to transfection
and the PBS was then replaced with fresh serum-free culture medium.
The cells were incubated with the transfection mixture at 37°C in a
5% CO2 incubator and the medium was changed 6 h
post-transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues samples and
cancer cells using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.). RNA purity and concentration were detected using
a Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific,
Inc.). The reverse transcription was operated using a PrimeScript™
RT kit (Takara Bio, Inc., Otsu, Japan) according to the
manufacturer's protocol. Following reverse transcription, cDNA was
diluted with diethy; pyrocarbonate at 1:2 and premixed.
Subsequently, 1 µl cDNA, primers, premixed liquid and
double-distilled water were mixed into a 20 µl volume in PCR tubes.
Following centrifugation at 800 × g for 5 min at 4°C, PCR
amplification was performed under the recommended parameters: 40
cycles of 50°C for 5 min, 95°C for 5 min, 95°C for 5 sec, 60°C for
15 sec and 72°C for 15 sec, followed by 94°C for 15 sec. The
fluorescence signals were collected and analyzed. The primer
sequences were as follows: HIT000218960 forward,
5-CCACCTACCCATCTGACTTTG-3′ and reverse, 5-CCACTATTTCCCACTGCCTT-3;
HMGA2 forward, 5-AGAATCTGGTGCAGGAATGG-3 and reverse,
5-TCGTATTTAGTGTCTCCAGCC-3; and β-actin forward,
5′-CCTCGCCTTTGCCGATCC-3′ and reverse,
5′-GGATCTTCATGAGGTAGTCAGTC-3′. The relative RNA expression levels
were analyzed using the 2−ΔΔCq method (32).
Western blotting
Cells were washed with PBS, and lysed using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China) for 10 min on ice. The BCA method was
used for protein quantification. Following centrifugation at 14,000
× g for 15 min at 4°C, the supernatant was collected. Proteins (30
µg) were separated by SDS-PAGE (12% gel), and transferred onto a
polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA,
USA). Membranes were washed with Tris-buffered saline containing
Tween-20 (TBST), and blocked with 5% skimmed milk with TBST at 25°C
for 1 h. Membranes were then incubated with monoclonal primary
antibody at 4°C overnight. The following primary antibodies were
used: Anti-HMGA2 (1:500; catalog no. ab52039; Abcam, Cambridge, UK)
and anti-GAPDH (1:500; catalog no. ab9485; Abcam). Membranes were
washed with TBST for 30 min and incubated at 25°C for 1 h with goat
anti-rabbit IgG H&L secondary antibody (1:1,000; catalog no.
ab6940; Abcam). Enhanced chemiluminescence reagent (Thermo Fisher
Scientific, Inc.) was used to detect the signal on the
membrane.
Cell Counting Kit-8 (CCK-8) assay
Cells (2,000 cells/well) were seeded in 96-well
plates. CCK-8 reagent (10 µl; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) was added to each well at 4, 24, 48, 72, 96 and
120 h after seeding for 1.5 h. Each group comprised 6 wells and
contained a blank control. The absorbance value of each well was
determined using a microplate reader at a wavelength of 490 nm.
Migration assay
A Transwell assay was performed to determine the
cell migratory ability. HCG2 and NCI-N87 cells (25×104)
were digested, suspended in serum-free RPMI-1640 medium and seeded
onto the upper non-Matrigel-coated chamber with a membrane pore
size of 8.0-µm (Corning, Inc., Corning, NY, USA). The lower chamber
was filled with 500 µl RPMI-1640 medium containing 10% FBS. After 2
h, cells that had invaded the lower chamber were fixed for 20 min
with 95% ethanol at room temperature, stained for 20 min with 0.5%
crystal violet at room temperature and washed with water.
Subsequently, images were captured with a light microscope
(magnification, ×400) and cells were counted.
Statistical analysis
SPSS software (version 22.0; IBM Corp., Armonk, NY,
USA) was used to analyze data. Images were edited with GraphPad
Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
Clinical characteristics were evaluated using a χ2 test.
Student's t-test was applied for comparing data between two groups.
Comparison between multiple groups was performed using one-way
analysis of variance followed a least significant difference
multiple-range post hoc test. Pearson correlation analysis was used
to determine the correlation between one dependent variable and
single independent variable. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression levels of HIT000218960 and
HMGA2 in GC tissue samples
A total of 60 GC tissue samples and 25 normal
control samples were collected. Expression levels of HIT000218960
and HMGA2 in the two groups were detected using RT-qPCR. The
results indicated that HIT000218960 and HMGA2 expression levels
were significantly higher in GC samples compared with in control
samples (P<0.001; Fig. 1A and B,
respectively). In addition, western blotting revealed that HMGA2
protein expression was higher in GC samples compared with in
control samples (Fig. 1C). Further
analysis indicated a positive correlation between HIT000218960 and
HMGA2 expression levels in GC tissues (P<0.001;
R2=0.6020; Fig. 1D).
However, there was no difference in HIT000218960 and HMGA2
expression levels between HGC27 and NCI-N87 cell lines (Fig. 1E).
Association between HIT000218960 and
clinical characteristics of patients with GC
To clarify the function of HIT000218960 in GC
tissue, the association between the clinical characteristics and
HIT000218960 expression levels of the 60 patients with GC was
analyzed. Patients were divided into lower-expression and
higher-expression groups according to the median level of
HIT000218960. Patients with an expression level ≥2.1 were placed in
the higher-expression group and patients with an expression level
<2.1 were placed in the lower-expression group, and each group
consisted of 30 patients (Table I).
Analysis using a χ2 test indicated that the tumor size
of patients in the higher-expression group was significantly larger
compared with those in the lower-expression group (P=0.0019). In
addition, compared with the lower-expression group, the rate of
advanced TNM staging (P=0.0008) and the incidence of lymph node
metastasis (P=0.0195) were higher in the higher-expression group
(P=0.0008 and P=0.0195, respectively). These data suggested that
the level of HIT000218960 expression was associated with tumor
size, TNM stage and lymph node metastasis in patients with GC.
 | Table I.Association between HIT000218960
expression level and clinical characteristics of patients with
gastric cancer (n=60). |
Table I.
Association between HIT000218960
expression level and clinical characteristics of patients with
gastric cancer (n=60).
|
|
| HIT000218960
expression level |
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristic | n | Low (n=30) | High (n=30) | P-value |
|---|
| Age, years |
|
|
|
|
|
≤60 | 31 | 14 | 17 | 0.4383 |
|
>60 | 29 | 16 | 13 |
|
| Sex |
|
|
|
|
|
Male | 28 | 12 | 16 | 0.3006 |
|
Female | 32 | 18 | 14 |
|
| Tumor size, cm |
|
|
|
|
| ≤3 | 28 | 20 | 8 | 0.0019a |
|
>3 | 32 | 10 | 22 |
|
| TNM stage |
|
|
|
|
|
I–II | 31 | 22 | 9 | 0.0019a |
|
III–IV | 29 | 8 | 21 |
|
| Lymph node
metastasis |
|
|
|
|
|
Negative | 27 | 18 | 9 | 0.0195a |
|
Positive | 33 | 12 | 21 |
|
HIT000218960 silencing inhibits HGC27
and NCI-N87 cell proliferation and migration
RT-qPCR analysis indicated that si-HIT000218960
transfection significantly decreased HIT000218960 expression levels
in HGC27 and NCI-N87 cell lines (P<0.001; Fig. 2A). CCK-8 assay demonstrated that
HGC27 and NCI-N87 cell proliferation was significantly decreased
following si-HIT000218960 transfection compared with si-NC
(P<0.001; Fig. 2B). The results
of the Transwell assay indicated that HIT000218960 silencing
decreased HGC27 and NCI-N87 cell migratory ability (Fig. 2C). These results suggested that
decreasing HIT000218960 expression decreased GC cell proliferative
and migratory ability.
HMGA2 mediates the regulatory effect
of HIT000218960 on GC cell proliferation and migration
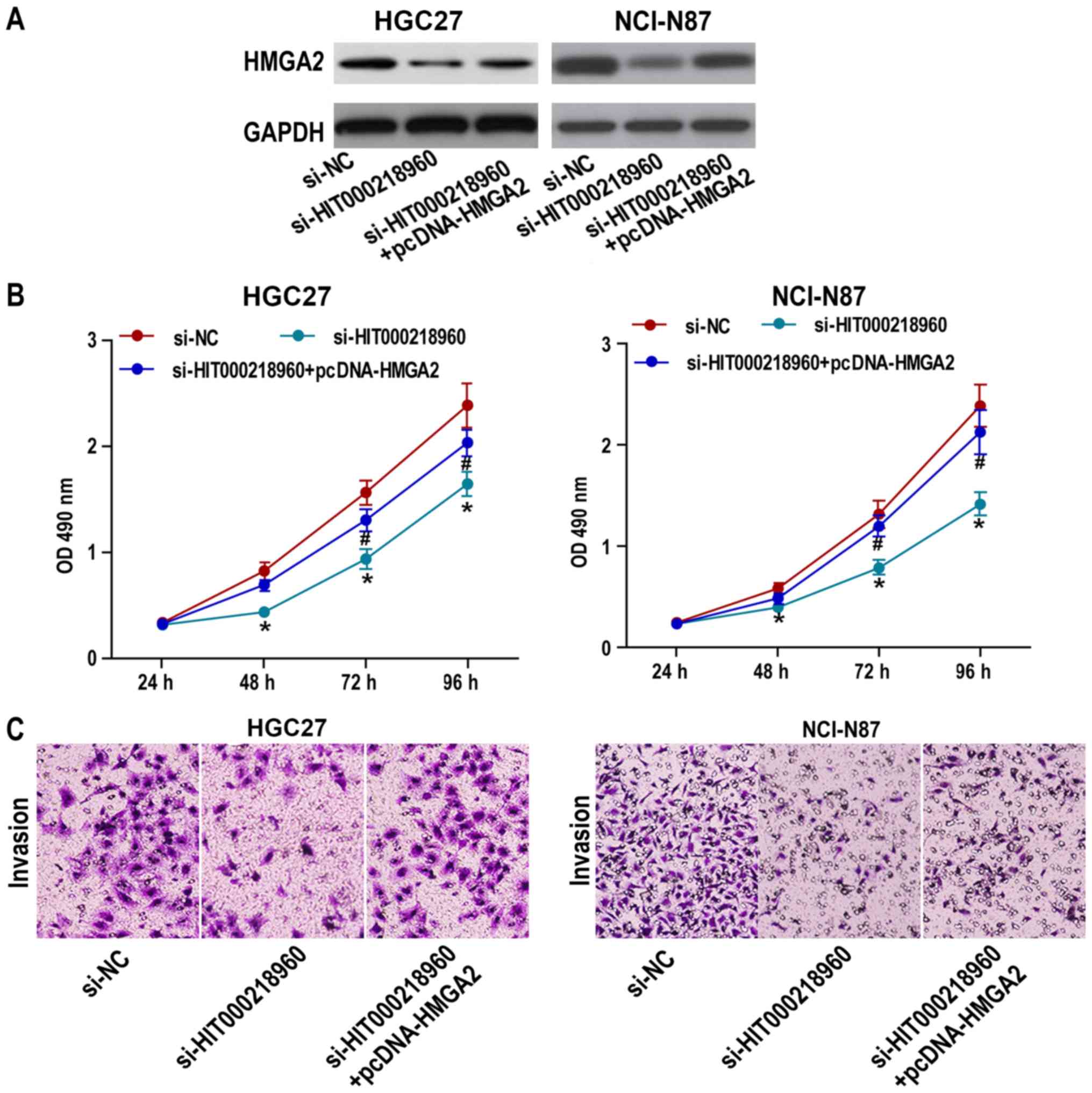
Western blotting indicated that HIT000218960
knockdown induced HMGA2 downregulation in HGC27 and NCI-N87 cells,
whereas pcDNA-HMGA2 reversed this phenomenon (Fig. 3A). In order to further clarify the
association between HIT000218960 and HMGA2, HGC27 and NCI-N87 cells
were transfected with si-NC, si-HIT000218960 or
si-HIT000218960+pcDNA-HMGA2. A CCK-8 assay revealed that HMGA2
overexpression reversed the si-HIT000218960-induced decrease in
cell proliferation (Fig. 3B) and
migration (Fig. 3C) of HGC27 and
NCI-N87 cell lines. These results suggested that HIT000218960 may
promote the proliferative and migratory ability of HGC27 and
NCI-N87 cells through HMGA2 upregulation.
Discussion
GC is a common gastrointestinal cancer in China,
which leads to high mortality and low 5-year survival rates. The
treatment of GC is essentially based on conventional surgery;
however, there is currently a lack of research on the mechanisms of
occurrence and development of GC, compared with other
gastrointestinal malignancies. It has been reported that lncRNAs
are involved in numerous biological processes, and that
dysregulation of RNA expression is associated with various types of
disease, including cancer. Subsequently, studies have identified
the underlying mechanisms of lncRNA in cancer. For example, the
antisense non-coding RNA in the INK4 locus and HOX transcript
antisense RNA can determine chromatin localization by recruiting
chromatin-modifying complexes and can regulate histone
modifications (33,34). To the best of our knowledge, only a
few studies on HIT000218960 have been performed. A recent study
reported that HIT000218960 is highly expressed in thyroid carcinoma
tissues, and that HIT000218960 inhibition induces a decrease in the
papillary thyroid carcinoma cell proliferative, migratory ability
and invasive capacity (29). They
also reported for the first time that HMGA2 regulates HIT00021896
expression. However, the effect of HIT000218960 in GC has not been
yet reported. Further investigation on the mechanism of action of
HIT000218960 in GC is therefore required. The aim of the present
study was to investigate the effect of HIT000218960 in GC. The
results demonstrated for the first time that HIT000218960
expression level was significantly higher in GC tissues compared
with normal tissues, and that HIT000218960 knockdown inhibited
HGC27 and NCI-N87 cell proliferative and migratory ability.
A previous study reported that HMGA2 overexpression
is closely associated with the occurrence and development of GC.
Motoyama et al (33) observed
that HMGA2 expression level in cancerous tissue was high in 110
patients with gastric cancer and was positively correlated with
infiltration depth and lymph node metastasis. In addition,
follow-up analysis revealed that patients with a high HMGA2
expression level had a shorter survival time compared with that of
healthy subjects. This was consistent with the results from the
present study, where HMGA2 expression was higher in GC tissues
compared with in normal tissues. In addition, HIT000218960
expression level was positively correlated with HMGA2 expression
level in GC tissues. HMGA2, which is a non-histone protein present
in the nucleus, can change chromosome structure and regulate the
expression of various target genes (15). HMGA2 is now considered to be a
oncogene (27). However, the
underlying molecular mechanism of HMGA2 in tumors is not fully
understood and may be associated with activation or translocation
of certain oncogenes. Motoyama et al (33) reported that in the transforming
growth factor β signaling pathway, Smad upregulates its downstream
target genes, including Snail, Twist1 and Snail family
transcriptional repressor 2 through HMGA2 and functions as an
epithelial (E-)cadherin transcription inhibitor. This results in
downregulation of E-cadherin expression and loss of intercellular
polarity, which induces EMT. Currently, the association between
HMGA2 and the clinical characteristics of patients is being
investigated to determine whether HMGA2 can be used to distinguish
benign from malignant tumors. For example, a high HMGA2 expression
level is associated with N staging, 2-year metastasis and poor
prognosis in patients with nasopharyngeal carcinoma (35,36). In
the present study, the expression levels of HIT000218960 and HMGA2
were associated with the clinical characteristics of patients with
GC. These results suggested that HIT000218960 may serve a
tumorigenic role by upregulating HMGA2 expression.
In conclusion, the results of the present study
indicated that HIT000218960 and HMGA2 were significantly
overexpressed in GC tissues and HGC27 an NCI-N87 cell lines.
HIT000218960 and HMGA2 expression levels were positively correlated
in GC tissues, and the HIT000218960 expression level in patients
with GC was associated with tumor size, TNM stage and lymph node
metastasis. In addition, HIT000218960 promoted the HGC27 and
NCI-N87 cell proliferative and migratory capacity by upregulating
HMGA2 expression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LS and SR designed the study and performed the
experiments. JY, PW and MS collected the data. JY and PW analyzed
the data. LS and SR prepared the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
The First Affiliated Hospital of Zhejiang Chinese Medical
University (Hangzhou, China). All subjects and/or guardians
provided written informed consent.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ilson DH: Adjuvant treatment for gastric
cancer: Too much is not enough. Lancet Oncol. 15:788–789. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bagcchi S: Radiotherapy for testicular
cancer increases gastric cancer risk. Lancet Oncol. 15:e5932014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Konishi H, Ichikawa D, Komatsu S, Shiozaki
A, Tsujiura M, Takeshita H, Morimura R, Nagata H, Arita T,
Kawaguchi T, et al: Detection of gastric cancer-associated
microRNAs on microRNA microarray comparing pre- and post-operative
plasma. Br J Cancer. 106:740–747. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zirtiloglu A, Cil I, Mehmet VF and Tural
D: What are the optimal treatment modalities according to age group
in gastric cancer patients? J Buon. 22:312–319. 2017.PubMed/NCBI
|
|
7
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xia Y, Yan Z, Wan Y, Wei S, Bi Y, Zhao J,
Liu J, Liao DJ and Huang H: Knockdown of long noncoding RNA GHET1
inhibits cellcycle progression and invasion of gastric cancer
cells. Mol Med Rep. 18:3375–3381. 2018.PubMed/NCBI
|
|
10
|
Lv Z, Zhang Y, Yu X, Lin Y and Ge Y: The
function of long non-coding RNA MT1JP in the development and
progression of gastric cancer. Pathol Res Pract. 214:1218–1223.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu C, Ma J, Li Y, Zhang Y and Da M: Low
expression of long noncoding RNA MT1JP is associated with poor
overall survival in gastric cancer patients: Protocol for
meta-analysis. Medicine (Baltimore). 97:e103942018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi X, Wang X and Hua Y: LncRNA GACAT1
promotes gastric cancer cell growth, invasion and migration by
regulating miR-149-mediated of ZBTB2 and SP1. J Cancer.
9:3715–3722. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. Febs J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu G, Xiang T, Wu QF and Wang WX:
Curcumin suppresses the proliferation of gastric cancer cells by
downregulating H19. Oncol Lett. 12:5156–5162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y
and Fang G: Long noncoding RNA CCAT1, which could be activated by
c-Myc, promotes the progression of gastric carcinoma. J Cancer Res
Clin Oncol. 139:437–445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Henriksen J, Stabell M, Meza-Zepeda LA,
Lauvrak SA, Kassem M and Myklebost O: Identification of target
genes for wild type and truncated HMGA2 in mesenchymal stem-like
cells. BMC Cancer. 10:3292010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun T, Zhao N, Zhao XL, Gu Q, Zhang SW,
Che N, Wang XH, Du J, Liu YX and Sun BC: Expression and functional
significance of Twist1 in hepatocellular carcinoma: Its role in
vasculogenic mimicry. Hepatology. 51:545–556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wagenblast E, Soto M, Gutierrez-Angel S,
Hartl CA, Gable AL, Maceli AR, Erard N, Williams AM, Kim SY,
Dickopf S, et al: A model of breast cancer heterogeneity reveals
vascular mimicry as a driver of metastasis. Nature. 520:358–362.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Williamson SC, Metcalf RL, Trapani F,
Mohan S, Antonello J, Abbott B, Leong HS, Chester CP, Simms N,
Polanski R, et al: Vasculogenic mimicry in small cell lung cancer.
Nat Commun. 7:133222016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang JY, Sun T, Zhao XL, Zhang SW, Zhang
DF, Gu Q, Wang XH, Zhao N, Qie S and Sun BC: Functional
significance of VEGF-a in human ovarian carcinoma: Role in
vasculogenic mimicry. Cancer Biol Ther. 7:758–766. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Baeten CI, Hillen F, Pauwels P, de Bruine
AP and Baeten CG: Prognostic role of vasculogenic mimicry in
colorectal cancer. Dis Colon Rectum. 52:2028–2035. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhong X, Liu X, Li Y, Cheng M, Wang W,
Tian K, Mu L, Zeng T, Liu Y, Jiang X, et al: HMGA2 sustains
self-renewal and invasiveness of glioma-initiating cells.
Oncotarget. 7:44365–44380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fujikane R, Komori K, Sekiguchi M and
Hidaka M: Function of high-mobility group A proteins in the DNA
damage signaling for the induction of apoptosis. Sci Rep.
6:317142016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu H, Lim HH, Tjokro NO, Sathiyanathan P,
Natarajan S, Chew TW, Klonisch T, Goodman SD, Surana U and Droge P:
Chaperoning HMGA2 protein protects stalled replication forks in
stem and cancer cells. Cell Rep. 6:684–697. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tan EJ, Thuault S, Caja L, Carletti T,
Heldin CH and Moustakas A: Regulation of transcription factor twist
expression by the DNA architectural protein high mobility group A2
during epithelial-to-mesenchymal transition. J Biol Chem.
287:7134–7145. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tan EJ, Kahata K, Idas O, Thuault S,
Heldin CH and Moustakas A: The high mobility group A2 protein
epigenetically silences the Cdh1 gene during
epithelial-to-mesenchymal transition. Nucleic Acids Res.
43:162–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Zhao Z, Xu C, Zhou Z, Zhu Z and You
T: HMGA2 induces transcription factor Slug expression to promote
epithelial-to-mesenchymal transition and contributes to colon
cancer progression. Cancer Lett. 355:130–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Singh I, Mehta A, Contreras A, Boettger T,
Carraro G, Wheeler M, Cabrera-Fuentes HA, Bellusci S, Seeger W,
Braun T and Barreto G: Hmga2 is required for canonical WNT
signaling during lung development. BMC Biol. 12:212014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li T, Yang XD, Ye CX, Shen ZL, Yang Y,
Wang B, Guo P, Gao ZD, Ye YJ, Jiang KW and Wang S: Long noncoding
RNA HIT000218960 promotes papillary thyroid cancer oncogenesis and
tumor progression by upregulating the expression of high mobility
group AT-hook 2 (HMGA2) gene. Cell Cycle. 16:224–231. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu C, Ren L, Wang Y, Liu Y and Xiao J:
The interaction between RACK1 and WEE1 regulates the growth of
gastric cancer cell line HGC27. Oncol Lett. 14:4784–4792. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kawai S, Kato S, Imai H, Okada Y and
Ishioka C: Suppression of FUT1 attenuates cell proliferation in the
HER2-overexpressing cancer cell line NCI-N87. Oncol Rep. 29:13–20.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Motoyama K, Inoue H, Nakamura Y, Uetake H,
Sugihara K and Mori M: Clinical significance of high mobility group
A2 in human gastric cancer and its relationship to let-7 microRNA
family. Clin Cancer Res. 14:2334–2340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xia YY, Yin L, Tian H, Guo WJ, Jiang N,
Jiang XS, Wu J, Chen M, Wu JZ and He X: HMGA2 is associated with
epithelial-mesenchymal transition and can predict poor prognosis in
nasopharyngeal carcinoma. Onco Targets Ther. 8:169–176. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xia YY, Yin L, Jiang N, Guo WJ, Tian H,
Jiang XS, Wu J, Chen M, Wu JZ and He X: Downregulating HMGA2
attenuates epithelial-mesenchymal transition-induced invasion and
migration in nasopharyngeal cancer cells. Biochem Biophys Res
Commun. 463:357–363. 2015. View Article : Google Scholar : PubMed/NCBI
|