Introduction
Liver cancer is acknowledged as a health burden
worldwide, of which hepatocellular carcinoma (HCC) is the most
frequently occuring type and the leading cause of death associated
with cancer (1,2). Additionally, it has been documented
that liver cancer is one of the highest mortality tumors in China
and its morbidity increases annually (3). As reported, the number of new cases
augmented by 75% from 1990 to 2015. Besides, there are 854,000
newly diagnosed cases and 810,000 deaths reported in 2015, which
makes the treatment of liver cancer still a priority in medical
field across the world (2).
According to a published report on cancer statistics in China, the
upward trend concerning liver cancer may be attributed to the
growth and aging of population (4).
If patients with HCC cannot be diagnosed at an early stage, they
may suffer from liver failure and cancer symptoms. Worse still, HCC
at an advanced stage cannot be treated and patients commonly
succumb to HCC after 3–6 months (5).
Hereby, it is of significance to allocate resources for prevention,
diagnosis, palliative care, screening and treatment to make sure
proper interventions are effectively performed on patients with
liver cancer (6).
Gene therapy is a promising treatment strategy for
cancer by means of immunosuppressors, tumor suppressor genes,
tumor- or tissue-specific promoter-driven suicide genes or
antiangiogenic genes (7). However,
gene therapy commonly induces apoptosis of normal cells along with
the tumor cells, indicating that the tumor suppressive effects are
not specific. Melanoma differentiation associated gene-7, also
called interleukin 24 (IL-24) (MDA-7), is a tumor suppressor gene,
which is firstly identified from melanoma cells (8). MDA-7 has been proved to exert selective
tumor-suppressive effects without any normal cells affected
(9). Besides, it has been
demonstrated that MDA-7 can inhibit liver cancer cell proliferation
(10), yet the mechanism has not
been revealed. In addition, B-cell lymphoma 2 (Bcl-2), a recognized
anti-apoptotic factor, can manipulate the intrinsic pathway in
regulating the mitochondrial membrane permeability (11). Noteworthy, Bcl-2 has been pointed out
to be effective in cancer therapies either alone or in combination
with other therapies to facilitate the development of novel cancer
therapies (12). Therefore, in the
present study, we attempted to develop a liver cancer-specific gene
therapy system by constructing recombinant eukaryotic expression
vector pcDNA3-MDA-7 containing human MDA-7 to explore the
underlying mechanism of the selective killing of liver cancer cells
of MDA-7 with the involvement of Bcl-2 in an attempt to provide a
theoretical basis for gene therapy of liver cancer.
The study was approved by the Ethics Committee of
The Third Affiliated Hospital of Qiqihar Medical University
(Qiqihar, China).
Materials and methods
Construction of eukaryotic expression
vector (pcDNA3-MDA-7)
The full-length polymerase chain reaction (PCR)
product of MDA-7 (F: 5′-CTCTAGAGGGGCTGTGAAAGA
CACTAT-3′; and R: 5′-CCTCGAGGGCATCCAGGTCAGA AGAA-3′;
length: 630 bp) and the eukaryotic expression vector pcDNA3 were
digested by the restriction endonucleases XbaI and
XhoI (cat. nos. 1093A and 1094A), both from Takara Bio,
Inc., (Otsu, Japan) followed by purification, recovery and
ligation. The ligation product was transformed into DH5α competent
cells of Escherichia coli (preserved in our laboratory) by
heat shock in water bath at 42°C for 90 sec. Next, culture in the
plate containing ampicillin sodium (A7490; Solarbio Science &
Technology Co., Ltd., Beijing, China) for 12–16 h, single colony
was selected for plasmid extraction and restriction enzyme
digestion (XbaI and XhoI). The colony supplemented
with clone of MDA-7 was screened out. The plasmid identified by
restriction enzyme digestion was allowed to undergo sequencing and
the correctly identified plasmid was used for subsequent
experiments.
Cell line transfection using
MDA-7
The pcDNA3-MDA-7 and empty vector pcDNA3 were
transfected into HepG2 and L02 cell lines (cat. nos. HB8065 and
CRL-12461; ATCC, Manassas, VA, USA) (both preserved in our
laboratory) according to the instructions of Lipofectamine™ 3000
(cat. no. L3000001; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), respectively. Then, the cells were cultured in medium
containing 15% calf serum (MB5175; Melone Pharmaceutical Co., Ltd.,
Dalian, China) at 37°C with 5% CO2 for 48 h. The medium
was then replaced with selective culture medium containing G418
until the resistant clones were observed. The monoclone was
selected for amplification culture. The transfected cell line with
stable passage was established and stored at −80°C for further
use.
MDA-7 determination by reverse
transcription quantitative PCR (RT-qPCR)
The total RNA was extracted according to the
instructions of the TRIzol RNA extraction kit (WLA088a, Wanlei
Biotechnology Co., Ltd., Shenyang, China) for reverse transcription
reaction. The reaction conditions are: 37°C, for 15 min; 98°C for 5
min and 4°C for 5 min. The above reverse transcription product was
used as a template for RT-qPCR cycle, and the β-actin was the
internal reference. The RT-qPCR kit (RR037A, Takara Bio, Inc.) was
used for reverse transcription and amplification. The required
primers synthesized by Sangon Biotech were shown in Table I. The amplification conditions are
95°C for 5 sec, 55°C for 10 sec and 72°C for 15 sec. The RT-qPCR
products were treated with 1% agarose gel electrophoresis for
observation. All expression levels were calculated using the
2−ΔΔCq method (13).
 | Table I.Primer sequences for amplification of
Mda7/IL-24. |
Table I.
Primer sequences for amplification of
Mda7/IL-24.
| Items | Forward primers | Reverse primers |
|---|
| Mda7/IL-24 (90
bp) |
5′-GACTCGAGATGAATTTTCAACAGA-3′ |
5′-GTCCCTCTGGTCCTGTAAG-3′ |
| β-actin (219 bp) |
5′-CCTTCCTGGGCAATGGAGTCCT-3′ |
5′-GGAACAATGATCTTGATCTT-3′ |
MDA-7 determination by western blot
analysis
The liver cancer cell lines of HepG2-pcDNA3-MDA-7,
HepG2-pcDNA3 and HepG2 along with the normal liver cell lines of
L02-pcDNA3-MDA-7, L02-pcDNA3 and L02 were cultured until they
reached the logarithmic growth phase. The culture medium was
removed and the cells were washed with 1 ml phosphate buffer saline
(PBS) 3 times. The cells were lysed with 200 µl protein lysis
buffer (protease inhibitor was added at a ratio of 1:100) and
incubated at 37°C for 2 h. The cell-free supernatant was collected,
followed by centrifugation (12,000 × g, at 4°C for 5 min). The
supernatant was the protein extract, which was quantified using the
bicinchonininc acid (BCA) protein assay kit. The total protein in
cells of each group was adjusted to 10 µg. A total of 15 ml of
sample was added. Following 10% sodium dodecyl
sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), the protein
was transferred to the polyvinylidene fluoride (PVDF) membrane in a
wet manner. The membrane was then blocked in 1X PBS containing 5%
skimmed milk for 4 h at room temperature and incubated with primary
antibody, MDA-7 monoclonal antibody (cat. no. AF1965-SP; 1:1,000;
R&D Systems, Inc., Minneapolis, MN, USA) at 4°C overnight.
After being washed with Tris-buffered saline with Tween-20 (TBST) 3
times (5 min per wash), the membrane was further incubated with the
secondary antibody, rabbit anti-sheep IgG (cat. no. 6912-100;
1:2,000; BioVision, Inc., Milpitas, CA, USA) at room temperature
for 2 h, followed by washing with TBST for 3 times (5 min per
wash). The membrane was developed using enhanced chemiluminescence
(ECL, GE Healthcare Life Sciences, Logan, UT, USA).
Cell proliferation detected by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
The liver cancer cell lines of HepG2-pcDNA3-MDA-7,
HepG2-pcDNA3 and HepG2 along with the normal liver cell lines of
L02-pcDNA3-MDA-7, L02-pcDNA3 and L02 were cultured (3 wells per
group) in serum-free medium until they reached the logarithmic
growth phase. The period of growth in each group was controlled to
be the same. The cells were then added into a 96-well plate at a
density of 104 cells/well (100 µl/well), cultured for 24
h and incubated with 20 µl MTT (5 mg/ml, 40201ES72, Shanghai Yeasen
Biotechnology Co., Ltd., Shanghai, China) at 37°C for 4 h. The
culture medium was removed and 150 µl dimethyl sulphoxide (DMSO)
was added, followed by 15 min incubation at 37°C. The absorbance
value (A) was measured by a microplate reader (ELX-800; BioTek
Instruments, Inc., Winooski, VT, USA) at the wavelength of 540 nm.
The average value and the number of cells were calculated.
Cell apoptosis detected by flow
cytometry
The liver cancer cell lines of HepG2-pcDNA3-MDA-7,
HepG2-pcDNA3 and HepG2 along with the normal liver cell lines of
L02-pcDNA3-MDA-7, L02-pcDNA3 and L02 were cultured in the 24-well
plate and then treated with trypsin. The density of harvested cells
was adjusted to 106 cells/ml and the binding buffer was
added. Then, 100 µl of the sample was added with 5 µl Annexin V and
10 µl propidium iodide (PI) dye (MA0220, Dalian Meilun
Biotechnology Co., Ltd.). The sample was allowed to react for 15–20
min under conditions void of light and analyzed by flow cytometry
(BD Accuri C6; BD Biosciences, San Jose, CA, USA). Cell Quest
software (version 0.9.13 alpha; BD Biosciences) was used to acquire
data, and FlowJo v10 software (FlowJo, LLC; Ashland, OR 97520, USA)
was used to analyse.
Mitochondrial protein extraction
The liver cancer cell lines HepG2-pcDNA3-MDA-7,
HepG2-pcDNA3 and HepG2 were cultured in the 24-well plate until
they reached the logarithmic growth phase. The cells were collected
and centrifuged (600 × g at 4°C for 8 min) with the supernatant was
removed. After re-suspension in 10 ml pre-cooled PBS (4°C),
centrifugation was conducted again (600 × g at 4°C for 8 min) and
the supernatant was discarded. The cells were re-suspended in 1 ml
cytoplasm buffer. After standing on ice for 10 min, the cell
suspension was ground in a pre-cooled grinder for 10–20 min. Then,
the well-ground fluid was transferred to a centrifuge tube for
centrifugation (600 × g at 4°C for 8 min) with the precipitate
abandoned. The supernatant was subjected to centrifugation again
(10,000 × g at 4°C for 35 min). The obtained supernatant was
considered as cytoplasmic protein. The obtained precipitate was
dissolved in 100 µl mitochondrion buffer and triturated with a
pipette. The well-treated precipitate was considered as
mitochondrial protein solution.
Mitochondrial protein determination by
western blot analysis
The concentration of the obtained protein was
determined by the protein analysis system (Bio-Rad Laboratories
Inc., Hercules, CA, USA). The total protein in each group was
adjusted to 10 µg. Following 10% SDS-PAGE, the protein was
transferred onto the PVDF membrane in a wet manner. The membrane
was then blocked in 1X PBS containing 5% skimmed milk for 4 h at
room temperature and incubated with the corresponding rabbit
anti-human antibodies at 4°C overnight, including Bcl-2 (cat. no.
LS-B6548; 1:1,000), Bcl-2 associated X protein (Bax, cat. no.
LS-B1333; 1:1,000), caspase 9 (cat. no. LS-C148247; 1:1,000),
cytochrome c (cat. no. LS-C208738; 1:1,000) and actin
(cat.no. LS-B11095; 1:1,000) all purchased from LifeSpan
BioSciences, Inc. (Seattle WA, USA). After washing with TBST 3
times (5 min per wash), the membrane was further incubated with the
secondary antibody, mouse anti-rabbit IgG (cat. no. LS-C60914;
1:3,000, GE Healthcare Life Sciences, Little Chalfont, UK) at room
temperature for 2 h, and then washed with TBST 3 times (5 min per
wash) and developed by ECL (Amersham, Little Chalfont,
Buckinghamshire, UK). All antibodies used in the procedure were
purchased from LifeSpan BioSciences Inc., and diluted by the
blocking fluid.
Statistical analysis
All data were calculated by SPSS Statistics 23.0
software (IBM Corp., Armonk, NY, USA). Test of significance was
analyzed by independent sample t-test and analysis of variance.
P-value <0.05 was considered to indicate a statistically
significant difference.
Results
Successfully constructed pcDNA3-MDA-7
vector
After the construction of pcDNA3-MDA-7 vector, the
extracted plasmid was digested by XbaI and XhoI and
then confirmed by electrophoresis (Fig.
1). The correct bands (630 bp length) representing the fragment
of MDA-7 of the three plasmids were identified. The three plasmids
were then subjected to sequencing. Blast analysis was conducted to
align the sequences and the match rate with the target sequence was
99.99%. The results indicated that recombinant plasmid pcDNA3-MDA-7
was successfully constructed.
MDA-7 gene is expressed in HepG2 and
L02 cells transfected with pcDNA3-MDA-7
The mRNA expression of MDA-7 in liver cells was
identified by RT-qPCR (Fig. 2). The
corresponding bands of MDA-7 were detected in HepG2 and L02 cells
transfected with pcDNA3-MDA-7. However, no band was identified in
cells transfected with pcDNA3 or the untransfected HepG2 and L02
cells. The findings indicated that MDA-7 gene could be expressed in
HepG2 and L02 cells after transfection of pcDNA3-MDA-7.
MDA-7 protein is expressed in HepG2
and L02 cells transfected with pcDNA3-MDA-7
The protein expression of MDA-7 in HepG2 and L02
cells was measured by western blot analysis (Fig. 3). The corresponding protein bands
were detected in the HepG2 and L02 cells that were transfected with
pcDNA3-MDA-7. While in the cells transfected with the pcDNA3 and
the untransfected HepG2 and L02 cells, there was no band in the
corresponding region, indicating no MDA-7 protein expression.
Therefore, it was concluded that MDA-7 protein can be expressed in
HepG2 and L02 cells transfected with pcDNA3-MDA-7.
MDA-7 inhibits liver cancer cell
proliferation
The effect of MDA-7 on the cell proliferation was
evaluated through MTT assay (Fig.
4). The cell growth curves in each group were drawn. It was
shown that pcDNA3-MDA-7 transfection could significantly inhibit
the growth of the liver cancer cell line HepG2 (P<0.05) while no
inhibitory effect was found in the normal liver cell line L02
(P>0.05). Thus, MDA-7 exerted inhibitory effect on the
proliferation of liver cancer cells.
MDA-7 promotes liver cancer cell
apoptosis
Cell apoptosis affected by MDA-7 was evaluated by
flow cytometry. As depicted in Fig.
5, compared with the untransfected HepG2 and the pcDNA3
transfected HepG2 cells, the HepG2 cells transfected with
pcDNA3-MDA-7 exhibited evidently higher apoptosis rate (P<0.05),
which demonstrated that MDA-7 promoted HepG2 cell apoptosis. As for
the normal liver cells, the apoptosis rates were all lower in L02
cells without transfection, with transfection of empty vector
pcDNA3 or pcDNA3-MDA-7. Hence, the apoptosis of the liver cancer
cells could be facilitated by MDA-7.
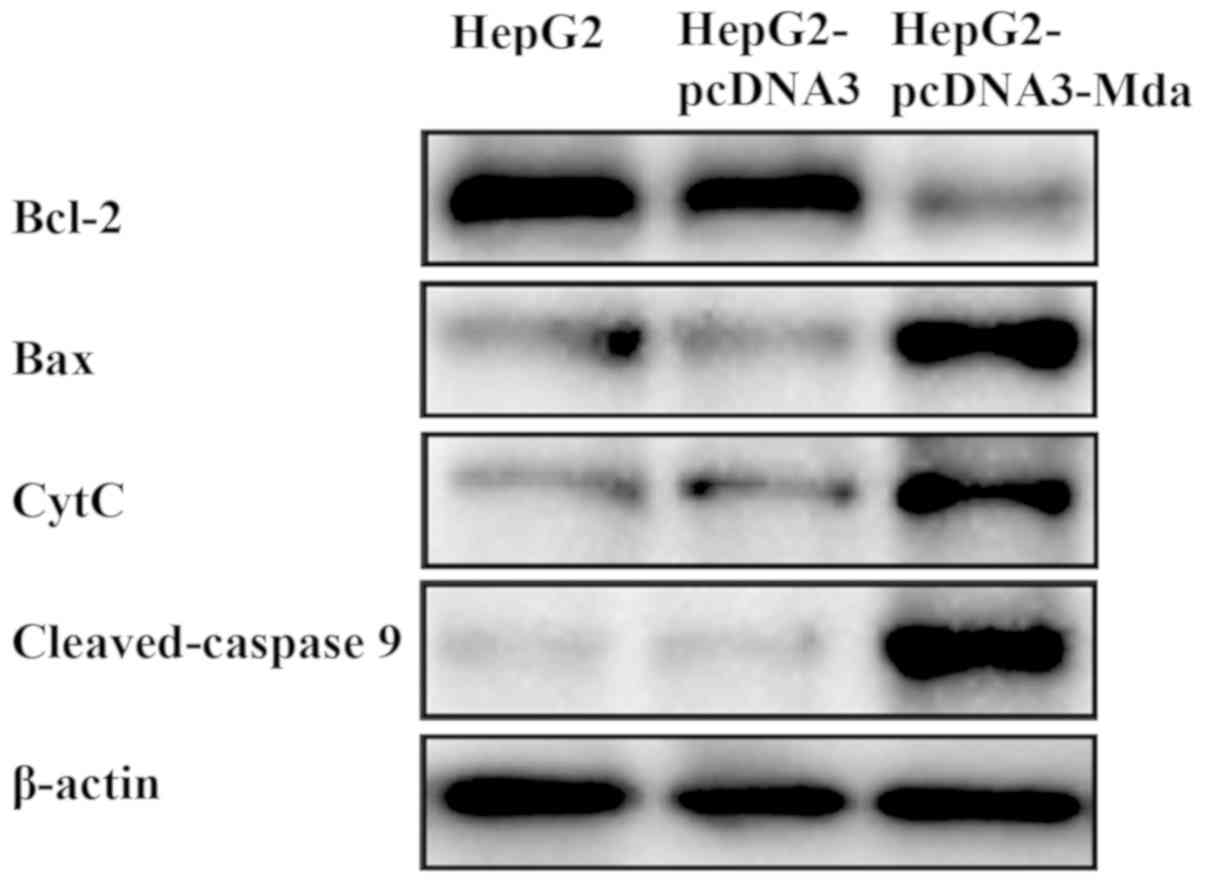
MDA-7 regulates the levels of
mitochondrial apoptosis pathway-related proteins
The mechanism of MDA-7 on the suppression of liver
cancer was subsequently investigated (Fig. 6). Compared with untransfected HepG2
cells and the empty vector-transfected HepG2 cells, the expression
level of Bcl-2 (anti-apoptotic protein) decreased while that of Bax
(pro-apoptotic protein), cytochrome c and caspase 9 (marker
proteins in the cell apoptosis signaling pathway) increased in
HepG2 cells transfected with pcDNA3-MDA-7. Therefore, it was noted
that MDA-7 suppresses the development of liver cancer by regulating
the levels of the mitochondrial apoptosis pathway-related
proteins.
Discussion
Although the therapeutic level of liver cancer has
exhibited continuous improvement in recent years, the overall
survival rate of patients suffering from liver cancer still remains
unfavorable. There is no doubt that the development of gene therapy
and molecular oncology has brought great hope to cancer patients,
including those with liver cancer. Multiple studies have provided
evidence proving that MDA-7 can promote cell apoptosis in various
types of tumor cells (14–17), highlighting its potential role as a
targeting gene for tumor therapy. Therefore, the present study
emphasized the effects of MDA-7 on liver cancer cells with a
possible mechanism investigated, laying a theoretical foundation
for MDA-7 as a candidate gene for the treatment of liver
cancer.
Initially, the eukaryotic expression vector
pcDNA3-MDA-7 was successfully constructed in the present study. The
obtained results indicate that pcDNA3-MDA-7 could mediate the
expression of MDA-7 in the liver cancer cell line HepG2 and the
normal liver cell line L02. MDA-7 expression promoted apoptosis of
liver cancer cell, but it had no obvious effect on the normal liver
cells. Furthermore, the proliferation of the liver cancer cells was
suppressed by upregulated MDA-7 while the normal liver cells were
unaffected, indicating the specific functional role of MDA-7 on
liver cancer cells and strongly supporting the therapeutic
potential of MDA-7 for liver cancer. In addition, the changes of
expression levels of the extracted mitochondrial proteins
determined by western blot analysis suggested that Bcl-2 expression
was diminished significantly in HepG2 cells where Bax expression
was obviously enhanced. Besides, apoptosis of the liver cancer
cells was induced by stimulating the release of cytochrome c
from mitochondria and augmenting the expression of caspase 9. All
above-mentioned findings elucidated that MDA-7 could exert
pro-apoptotic effects on HepG2 liver cancer cells through
activation of the mitochondrial apoptotic pathway by downregulating
Bcl-2 expression.
Wang et al have delivered Ad.VGFP/IL-24 to
human liver cancer cell line SMMC-772l and applied intratumoral
injection in nude mice with liver cancer; as a result, the in
vitro experiments revealed that Ad. VGFP/IL-24 could shorten
the S phase of SMMC-772l cells and block G2/M phase while the
inhibitory effect on liver cancer cell growth was confirmed through
in vivo experiments (18).
Likewise, MDA-7 was found to promote liver cancer cell apoptosis
according to results of MTT assay and flow cytometry in our study.
It has been documented that MDA-7 can induce liver cancer cell
apoptosis via the death receptor pathway and the endoplasmic
reticulum stress signaling pathway (19). Consistently, our study indicated that
MDA-7 induces apoptosis of the liver cancer cells via the
mitochondrial apoptotic pathway in which cytochrome c and
caspase 9 were both upregulated, further leading to an in-depth
understanding of tumor cell apoptosis induced by MDA-7. However,
limitations are inevitable in the current study, for instance, the
specific mechanism of liver cancer cell cycle entry that can be
suppressed by MDA-7 is unclear and needs more exploration.
The anti-apoptotic protein Bcl-2, distributed on the
cytoplasmic surface, endoplasmic reticulum and mitochondrial outer
membrane of the nuclear membrane, has the ability to stabilize the
mitochondrial membrane and block the release of caspase and
cytochrome c (20). On the
contrary, Bax can promote the release of pro-apoptotic proteins
including cytochrome c by forming a mitochondrial
transmembrane channel through combination with the mitochondrial
membrane (21–23). The results of our study revealed that
MDA-7 promoted apoptosis of liver cancer cells by decreasing the
Bcl-2 expression and increasing the expression of Bax and
cytochrome c, which was in line with the aforementioned
studies.
The current study leads to further understanding of
the molecular mechanism of MDA-7-induced apoptosis in liver cancer
cells, and provides a theoretical basis for the future clinical
application of MDA-7 as a target therapy gene for liver cancer as
well as other tumors.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QM and LL wrote the manuscript and performed PCR,
western blot analysis and MTT assay. GB and TL were responsible for
the construction of the eukaryotic expression vector. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Third Affiliated Hospital of Qiqihar Medical University
(Qiqihar, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen Y, Liu R, Chu Z, Le B, Zeng H, Zhang
X, Wu Q, Zhu G, Chen Y, Liu Y, et al: High glucose stimulates
proliferative capacity of liver cancer cells possibly via
O-GlcNAcylation-dependent transcriptional regulation of GJC1. J
Cell Physiol. 234:606–618. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rodríguez-Hernández MA, González R, de la
Rosa AJ, Gallego P, Ordóñez R, Navarro-Villarán E, Contreras L,
Rodríguez-Arribas M, González-Gallego J, Álamo-Martínez JM, et al:
Molecular characterization of autophagic and apoptotic signaling
induced by sorafenib in liver cancer cells. J Cell Physiol.
234:692–708. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wei KR, Yu X, Zheng RS, Peng XB, Zhang SW,
Ji MF, Liang ZH, Ou ZX and Chen WQ: Incidence and mortality of
liver cancer in China, 2010. Chin J Cancer. 33:388–394.
2014.PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bruix J, Reig M and Sherman M:
Evidence-based diagnosis, staging, and treatment of patients with
hepatocellular carcinoma. Gastroenterology. 150:835–853. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Global Burden of Disease Cancer
Collaboration, ; Fitzmaurice C, Dicker D, Pain A, Hamavid H,
Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe
C, et al: The Global Burden of Cancer 2013. JAMA Oncol. 1:505–27.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li W, Li DM, Chen K, Chen Z, Zong Y, Yin
H, Xu ZK, Zhu Y, Gong FR and Tao M: Development of a gene therapy
strategy to target hepatocellular carcinoma based inhibition of
protein phosphatase 2A using the α-fetoprotein promoter enhancer
and pgk promoter: An in vitro and in vivo study. BMC Cancer.
12:5472012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang H, Lin JJ, Su ZZ, Goldstein NI and
Fisher PB: Subtraction hybridization identifies a novel melanoma
differentiation associated gene, mda-7, modulated during human
melanoma differentiation, growth and progression. Oncogene.
11:2477–2486. 1995.PubMed/NCBI
|
|
9
|
Menezes ME, Bhoopathi P, Pradhan AK, Emdad
L, Das SK, Guo C, Wang XY, Sarkar D and Fisher PB: Role of
MDA-7/IL-24 a multifunction protein in human diseases. Adv Cancer
Res. 138:143–182. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang CJ, Xue XB, Yi JL, Chen K, Zheng JW,
Wang J, Zeng JP and Xu RH: Melanoma differentiation-associated
gene-7, MDA-7/IL-24, selectively induces growth suppression,
apoptosis in human hepatocellular carcinoma cell line HepG2 by
replication-incompetent adenovirus vector. World J Gastroenterol.
12:1774–1779. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hassan M, Watari H, AbuAlmaaty A, Ohba Y
and Sakuragi N: Apoptosis and molecular targeting therapy in
cancer. BioMed Res Int. 2014:1508452014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thomas S, Quinn BA, Das SK, Dash R, Emdad
L, Dasgupta S, Wang XY, Dent P, Reed JC, Pellecchia M, et al:
Targeting the Bcl-2 family for cancer therapy. Expert Opin Ther
Targets. 17:61–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bhoopathi P, Lee N, Pradhan AK, Shen XN,
Das SK, Sarkar D, Emdad L and Fisher PB: mda-7/IL-24 induces cell
death in neuroblastoma through a novel mechanism involving AIF and
ATM. Cancer Res. 76:3572–3582. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin C, Liu H, Li L, Zhu Q, Liu H, Ji Z,
Liao J, Lang J, Wu J and Fan J: MDA-7/IL-24 inhibits cell survival
by inducing apoptosis in nasopharyngeal carcinoma. Int J Clin Exp
Med. 7:4082–4090. 2014.PubMed/NCBI
|
|
16
|
Menezes ME, Shen XN, Das SK, Emdad L, Guo
C, Yuan F, Li YJ, Archer MC, Zacksenhaus E, Windle JJ, et al:
MDA-7/IL-24 functions as a tumor suppressor gene in vivo in
transgenic mouse models of breast cancer. Oncotarget.
6:36928–36942. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park MA, Hamed HA, Mitchell C,
Cruickshanks N, Dash R, Allegood J, Dmitriev IP, Tye G, Ogretmen B,
Spiegel S, et al: A serotype 5/3 adenovirus expressing MDA-7/IL-24
infects renal carcinoma cells and promotes toxicity of agents that
increase ROS and ceramide levels. Mol Pharmacol. 79:368–380. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang X, Ye Z, Zhong J, Xiang J and Yang J:
Adenovirus-mediated IL-24 expression suppresses hepatocellular
carcinoma growth via induction of cell apoptosis and cycling arrest
and reduction of angiogenesis. Cancer Biother Radiopharm. 22:56–63.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gupta P, Su ZZ, Lebedeva IV, Sarkar D,
Sauane M, Emdad L, Bachelor MA, Grant S, Curiel DT and Dent P:
mda-7/IL-24: Multifunctional cancer-specific apoptosis-inducing
cytokine. Pharmacol Ther. 111:596–628. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai G, Zheng D, Guo W, Yang J and Cheng
AY: Cinobufagin induces apoptosis in osteosarcoma cells via the
mitochondria-mediated apoptotic pathway. Cell Physiol Biochem.
46:1134–1147. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ferrer PE, Frederick P, Gulbis JM, Dewson
G and Kluck RM: Translocation of a Bak C-terminus mutant from
cytosol to mitochondria to mediate cytochrome C release:
Implications for Bak and Bax apoptotic function. PLoS One.
7:e315102012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mondal S, Bhattacharya K, Mallick A,
Sangwan R and Mandal C: Bak compensated for Bax in p53-null cells
to release cytochrome c for the initiation of mitochondrial
signaling during Withanolide D-induced apoptosis. PLoS One.
7:e342772012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Renault TT, Floros KV and Chipuk JE:
BAK/BAX activation and cytochrome c release assays using isolated
mitochondria. Methods. 61:146–155. 2013. View Article : Google Scholar : PubMed/NCBI
|