Introduction
Upper digestive tract cancers (UDTC) mainly include
esophageal cancer (EC) and gastric cancer (GC). GC can be defined
according to the tumor location as proximal or distal gastric
adenocarcinoma (1). EC is the
eleventh most common cancer and the sixth deadliest cancer
worldwide, and GC is ranked fifth for cancer incidence and third
for cancer-associated mortalities worldwide (2). Gastric cardia cancer (GCC), or
esophagogastric junction cancer, has also become a public health
concern (3). To date, several major
risk factors have been reported to be associated with UDTC,
including heavy smoking and alcohol consumption (4,5). It is
widely accepted that the development of UDTC is a result of complex
interactions between environmental triggers and genetic factors
(6–8). However, these interactions and the
exact mechanism of carcinogenesis are still not fully
understood.
Metabolites of tobacco and alcohol are first
metabolically activated by Phase I enzymes, including cytochrome
P4501A1 (CYP1A1) and cytochrome P4502E1 (CYP2E1),
into their final forms and then combine with DNA, forming
aromatic-DNA adducts that are considered to be an early stage in
carcinogenesis (9). These activated
forms are subsequently detoxified by Phase II enzymes, particularly
GSTM1, a member of the glutathione S-transferases (GSTs)
family (10). The CYP1A1
rs4646903 T>C polymorphism (MspI), also known as the m1 allele,
is a substitution of T to C in the non-coding 3′-flanking region
which appears to be associated with increased enzymatic activity
(11). The CYP2E1 rs2031920
C>T polymorphism (RsaI) also known as the c2 allele, involves a
C to T transition in the 5′-flanking region of the CYP2E1
gene, which appears to be associated with decreased enzymatic
activity (12). Individuals who
presents the null GSTM1 alleles lack the respective enzyme
function (13).
A number of studies have been performed to assess
the association between gene polymorphisms and cancer
susceptibility (14–18). One meta-analysis showed no
association between CYP1A1 rs4646903 polymorphism and
digestive tract cancers risk (14),
while another meta-analysis confirmed association existed between
CYP1A1 rs4646903 and gastric cancer (15). Zhang et al (16) indicated that CYP2E1 rs2031920
polymorphisms revealed no association with the risk of GC, however
when GSTM1 was null, the association became significant. GSTM1/T1
null genotype was reported to increase GC risk, and combination of
the CYP1A1 rs4646422 variant allele and GSTM1/T1 null
genotypes was also associated with a statistically significant
increased risk (17). A recent
meta-analysis suggested the association between GSTM1 and digestive
cancers, and two potential gene-smoking interactions were also
found (18). The results from these
studies have not always been consistent. In addition, to the best
of our knowledge, the evaluation of gene-gene and gene-environment
interactions regarding upper digestive cancer risk is insufficient
at present. To clarify the combined effects of CYP1A1
rs4646903, CYP2E1 rs2031920, GSTM1 null polymorphisms
and smoking or alcohol consumption on upper digestive tract cancer
risk, a population-based case-control study was performed in
Anyang, a typical high-incidence area of upper digestive cancer in
Northern China (19,20).
Materials and methods
Patient and control selection
This case-control study included 194 patients with
EC, 212 patients with GCC, 135 patients with gastric antral
carcinoma (GAC), and 212 controls. The mean ages ± standard
deviation of these four groups were 63±7.179, 64±9.070, 63±6.852
and 63±4.646 years. The sex ratio (male vs. female) of these four
groups were 65.5 vs. 34.5%, 67.9 vs. 32.1%, 67.4 vs. 32.6% and 66.5
vs. 33.5%. All subjects were recruited from Anyang Cancer Hospital
(Henan, China) between July 2015 and July 2017, with the study
conducted during the same period. Inclusion criteria were as
follows: Age between 30–79 years old with Han ethnicity;
pathological diagnosis confirming ECC, GCC or GAC and no
simultaneous malignancies. Patients who had undergone chemotherapy
or radiotherapy prior to surgery were excluded from the present
study. The cancer diagnoses were confirmed histologically. Subjects
with no sign of a tumor based on gastroscopy were recruited from a
cancer screening program for early detection of upper digestive
tract cancers in the same area. All subjects underwent a personal
interview and provided information on sociodemographic
characteristics, recent and prior tobacco or alcohol use, and
family history of cancer. Smoking status was stratified into three
levels: Never smoked, smoking for <30 years and smoking for ≥30
years; alcohol consumption status was stratified into three levels:
Never to occasional; ≥1 day/week and <150 g/week; ≥1 day/week
and >150 g/week. The Anyang Tumor Hospital Institutional Review
Board approved the present study. All patients and controls signed
a study-specific written informed consent form.
PCR analysis of gene
polymorphisms
DNA was extracted from the buffy coat of blood
samples from the patients and controls using a FlexiGene DNA kit
(cat. no. 51206; Qiagen China Co., Ltd.) for PCR or PCR-restriction
fragment length polymorphism (RFLP) experiments. The polymorphisms
of CYP2E1 rs2031920 C>T and GSTM1 (21) were detected by PCR using the Thermal
Cycler K640 (Hangzhou Jingle Scientific Instrument Co., Ltd.).
Nested PCR (22) was used to amplify
the CYP1A1 rs4646903 T>C. The PCR thermocycling
conditions included initial denaturation at 95°C for 15 min
followed by 35 cycles of denaturation at 95°C for 1 min, annealing
for 1 min (annealing temperatures are presented in Table I), and extension at 75°C for 1 min;
and a final extension at 72°C for 10 min. The amplified products
were digested and examined using 1.5% agarose gel electrophoresis,
and were visualized using a UV transilluminator (Beijing Liuyi
Biotechnology Co., Ltd.). Table I
presents the primer sequences, annealing temperatures, and
digestion enzymes used. A total of 15% of the PCR products were
selected for direct sequencing to confirm the RFLP results. The
primers used for CYP1A1 and CYP2E1 sequencing were
the same as the primers used in PCR. For GSTM1, the primers used
for sequencing were cited from Khabaz et al (23). No deviation was found between the
RFLP results and the sequencing data.
 | Table I.PCR primers and restriction
conditions used in the present study. |
Table I.
PCR primers and restriction
conditions used in the present study.
| Gene | Primer | Annealing
temperature | Restriction
enzyme | Fragment
length |
|---|
| CYP1A1 | Forward
5′-TCACTCGTCTAAATACTCACCCTG-3′ (C1F) | 60°C | MspI | 298 bp
(wild-type) |
| rs4646903
T>C | Reverse
5′-TAGGAGTCTTGTCTCATGCCT-3′ (C1R) |
|
| 298, 135 and 160 bp
(heterozygous) |
|
| Forward
5′-CAGTGAAGAGGTGTAGCCGCT-3′ (C2F) |
|
| 135 and 160 bp
(homozygous) |
|
| Reverse
5′-GAGGCAGGTGGATCACTTGAGCTC3′ (C2R) |
|
|
|
| CYP2E1 | Forward
5′-AACGCCCCTTCTTGGTTCAG-3′ | 60°C | RsaI | 265 and 150 bp
(wild-type); |
| rs2031920
C>T | Reverse
5′-CATACAGACCCTCTTCCACCTT-3′ |
|
| 416, 265 and 150 bp
(heterozygous) |
|
|
|
|
| 416 bp
(homozygous) |
| GSTM1 | Forward
5′-GAACTCCCTGAAAAGCTAAAGC-3′ | 60°C | – | 215 bp
(present) |
|
| Reverse
5′-GTTGGGCTCAAATATACGGTGG-3′ |
|
| No fragment
(null) |
Statistical analysis
SPSS 19.0 software (IBM Corp.) was used for
statistical analysis, and all tests were repeated three times.
Pearson's χ2 test or Fisher's exact test were used to
examine differences between groups and unpaired t-tests to compare
means. All tests were two-sided. Hardy-Weinberg equilibrium test
was used to confirm the CYP1A1 and CYP2E1 genotype distributions.
The Bonferroni correction was used to evaluate the associations
found and a P-value of <0.05/m was considered statistically
significant (m=the total comparison times). Cancer risk associated
with genotype or environmental exposure factors was estimated by
calculating odds ratios (OR) and 95% confidence intervals (CI)
using unconditional logistic regression. After adjusting for
potential confounding factors, multivariate logistic regression was
used to assess the association between smoking, alcohol, and the
metabolic gene polymorphisms.
Results
Patient and control
characteristics
Table II presents
the demographic profiles of the 541 patients and 212 controls.
There were no significant differences between the cases and
controls in sex, mean age, marital status, education level, labor
type and economic income. Upper digestive tract cancer and family
history of cancer were significantly associated for EC (P=0.017),
GCC (P=0.002) and GAC (P=0.001).
 | Table II.Demographic characteristics of
patients in the current study. |
Table II.
Demographic characteristics of
patients in the current study.
|
|
| EC | GCC | GAC |
|---|
|
|
|
|
|
|
|---|
|
Characteristics | Controls n=212 | n=194 | χ2 | P-value | n=212 | χ2 | P-value | n=135 | χ2 | P-value |
|---|
| Sex |
|
Male | 141 | 127 | 0.049 | 0.824 | 144 | 0.096 | 0.756 | 91 | 0.030 | 0.862 |
|
Female | 71 | 67 |
|
| 68 |
|
| 44 |
|
|
| Mean age ±
SDa, years | 63±4.646 | 63±7.179 | – | 0.874 | 64±9.070 | – | 0.396 | 63±6.852 | – | 0.456 |
| Marital
statusb |
|
Yes | 209 | 190 | – | 0.836 | 208 | – | 0.685 | 134 | – | 0.147 |
| No | 3 | 4 |
|
| 4 |
|
| 1 |
|
|
|
Educationb |
|
≤Primary school | 136 | 130 | – | 0.320 | 134 | – | 0.974 | 73 | – | 0.127 |
| Junior
or senior | 73 | 64 |
|
| 75 |
|
| 58 |
|
|
|
≥College | 3 | 0 |
|
| 3 |
|
| 4 |
|
|
| Occupation |
|
Labor | 22 | 18 | 3.793 | 0.285 | 25 | 2.567 | 0.463 | 19 | 1.475 | 0.688 |
|
Farmers | 175 | 170 |
|
| 178 |
|
| 105 |
|
|
| Civil
jobs | 7 | 2 |
|
| 6 |
|
| 6 |
|
|
| Other
jobs | 8 | 4 |
|
| 3 |
|
| 5 |
|
|
| Incomec, yuan |
|
≤1,999 | 130 | 122 | 5.705 | 0.058 | 125 | 0.627 | 0.731 | 76 | 0.939 | 0.625 |
|
2,000-3,999 | 71 | 70 |
|
| 78 |
|
| 52 |
|
|
|
≥4,000 | 11 | 2 |
|
| 9 |
|
| 7 |
|
|
| Family history |
|
Yes | 37 | 141 | 5.716 | 0.017 | 64 | 9.475 | 0.002 | 45 | 11.526 | 0.001 |
| No | 175 | 53 |
|
| 148 |
|
| 90 |
|
|
Detection of CYP1A1, CYP2E1 and GSTM1
variants in upper digestive tract cancers
A total of 194 EC, 212 GCC and 135 GAC cases, and
212 controls were examined to detect CYP1A1 rs4646903,
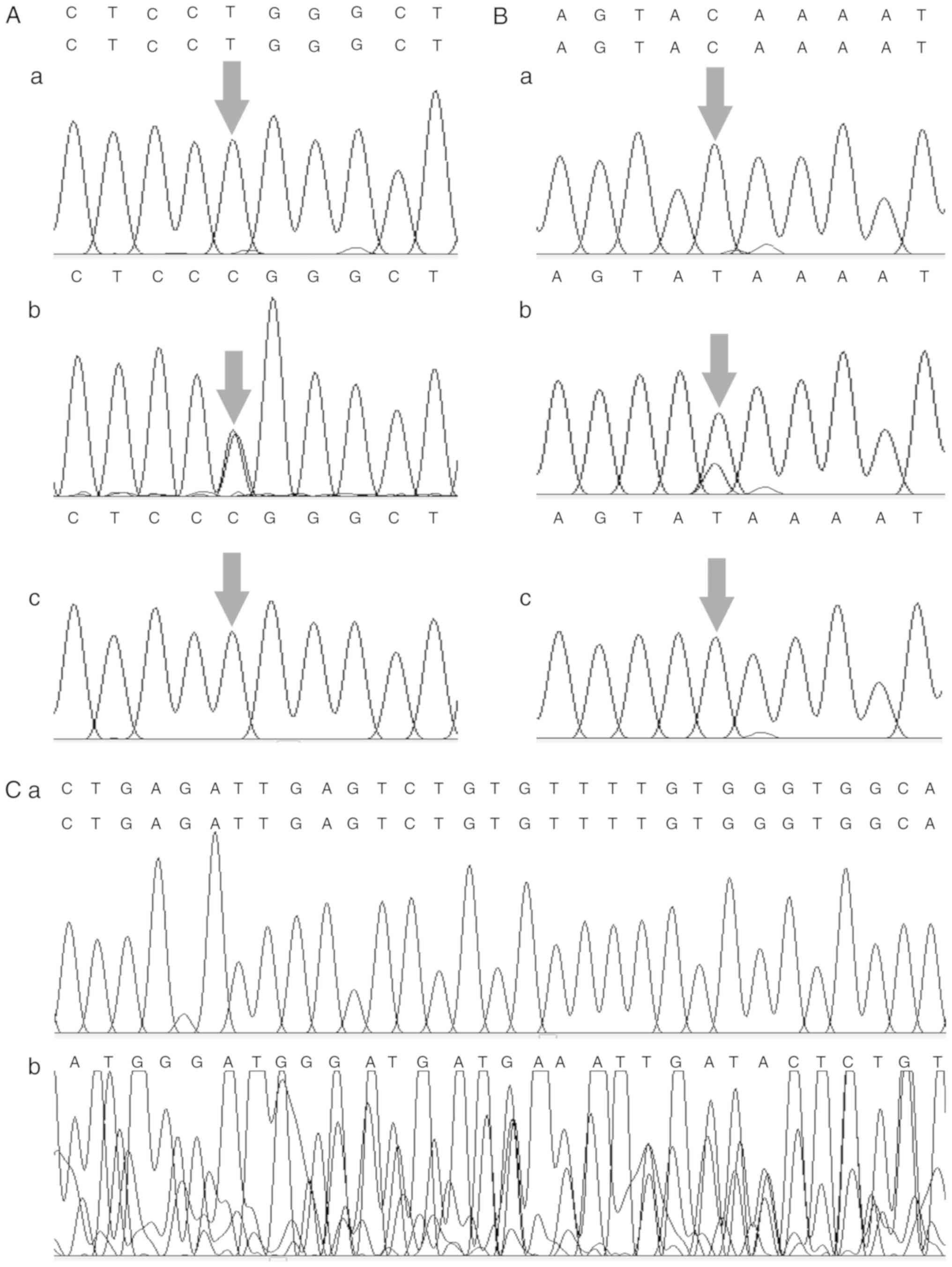
CYP2E1 rs2031920 and GSTM1 polymorphisms. Fig. 1 shows examples of gene polymorphisms
in PCR-amplified fragments or digestion fragments. Fig. 2 shows the sequencing chromatogram of
CYP1A1 rs4646903 and CYP2E1 rs2031920. Among the
controls, both the CYP1A1 and CYP2E1 genotype
distributions were in Hardy-Weinberg equilibrium.
 | Figure 1.PCR analysis of polymorphisms of
CYP1A1 rs4646903, CYP2E1 rs2031920 and GSTM1.
(A) CYP1A1 rs4646903 polymorphism: Lane 3, 6 and 7: Wild
genotype (298 bp); lane 5 and 8: Homozygous variant (160 and 135
bp), and lane 1, 2 and 4: Heterozygous variant (298 and 160 bp and
135 bp). (B) CYP2E1 rs2031920 polymorphism: Lane 4, 6 and 8:
Wild genotype (416 and 265 and 150 bp); lane 2 and 7: Homozygous
variant (416 bp), and lane 1, 3 and 5: Heterozygous variant (265
and 150 bp). (C) GSTM1 genotypes: Lane 3, 5 and 6: Null
genotype (no band) and lane 1, 2, 4 and 7: Wild genotype (215 bp
band). CYP1A1, Cytochrome P4501A1; CYP2E1, Cytochrome
P4502E1; GSTM1, Glutathione S-transferase mu 1. |
Association between smoking, alcohol
consumption, CYP1A1, CYP2E1, GSTM1 and upper digestive tract
cancers
Smoking and alcohol consumption were confirmed to be
main risk factors for upper digestive tract cancers (Table III). After adjusting for matching
variables and potential confounders, smoking increased EC, GCC and
GAC risk compared with non-smoking status: EC [OR (95% CI)=3.594
(2.077–6.221); P<0.001]; GCC [OR (95% CI)=4.658 (2.654–8.174);
P<0.001] and GAC [OR (95% CI)=3.999 (2.131–7.505); P<0.001],
as did alcohol consumption: EC [OR (95% CI)=1.953 (1.210–3.151);
P=0.006]; GCC [OR (95% CI)=2.442 (1.523–3.914); P<0.001] and GAC
[OR (95% CI)=1.765 (1.030–3.025); P=0.039]. Dose-dependent trends
were observed with these two risk factors, with ORs increasing as
the total smoking years or alcohol consumption amount increased
(Table III). It was indicated that
the GSTM1 null genotype had protective effects against EC,
decreasing EC risk [OR (95% CI)=0.510 (0.340–0.765); P=0.001].
 | Table III.Odds ratios and 95% Confidence
Intervals of smoking, alcohol and GSTM1 genotypes in upper
digestive tract cancer. |
Table III.
Odds ratios and 95% Confidence
Intervals of smoking, alcohol and GSTM1 genotypes in upper
digestive tract cancer.
|
|
| EC | GCC | GAC |
|---|
|
|
|
|
|
|
|---|
| Factors | Controls n=212 | n=194 | ORc (95% CI) | P-value | n=212 | ORc (95% CI) | P-value | n=135 | ORc (95% CI) | P-value |
|---|
| Smoking |
|
Non-smokers | 136 | 92 | 1.00
(reference) |
| 89 | 1.00
(reference) |
| 59 | 1.00
(reference) |
|
|
Smokers | 76 | 102 | 3.594b (2.077–6.221) | <0.001 | 123 | 4.658b (2.654–8.174) | <0.001 | 76 | 3.999b (2.131–7.505) | <0.001 |
| Smoking years |
|
<30 | 22 | 28 | 3.225b (1.570–6.626) | 0.001 | 28 | 3.500b (1.672–7.327) | 0.001 | 21 | 3.700b (1.657–8.264) | 0.001 |
|
≥30 | 54 | 74 | 3.773b (2.096–6.790) | <0.001 | 95 | 5.185b (2.866–9.382) | <0.001 | 55 | 4.153b (2.115–8.156) | <0.001 |
| Alcohol |
| Never
to occasional | 135 | 103 | 1.00
(reference) |
| 99 | 1.00
(reference) |
| 70 | 1.00
(reference) |
|
|
Frequent drinkers | 77 | 91 | 1.953b (1.210–3.151) | 0.006 | 113 | 2.442b (1.523–3.914) | <0.001 | 65 | 1.765a (1.030–3.025) | 0.039 |
| Alcohol
consumption |
| ≥1 day
and <150 g/week | 40 | 43 | 1.872a (1.044–3.355) | 0.035 | 40 | 1.687a (0.933–3.051) | 0.084 | 21 | 1.080
(0.535–2.182) | 0.830 |
| ≥1 day
and ≥150 g/week | 37 | 48 | 2.024a (1.158–3.538) | 0.013 | 73 | 3.139b (1.832–5.378) | <0.001 | 44 | 2.398b (1.310–4.389) | 0.005 |
| GSTM1 |
|
Present | 74 | 100 | 1.00
(reference) |
| 84 | 1.00
(reference) |
| 55 | 1.00
(reference) |
|
|
Null | 138 | 94 | 0.510b (0.340–0.765) | 0.001 | 128 | 0.862
(0.575–1.290) | 0.470 | 80 | 0.823
(0.518–1.306) | 0.408 |
CYP1A1 rs4646903 polymorphism was
significantly associated with GCC risk [CC vs. TT: OR (95%
CI)=1.936 (1.035–3.620), P=0.039; CC vs. CT+TT: OR (95% CI)=2.263
(1.272–4.026), P=0.005]; CYP2E1 rs2031920 was significantly
associated with EC risk [c1/c2 vs. c1/c1: OR (95% CI)=1.673
(1.111–2.520), P=0.014; c1/c2+c2/c2 vs. c1/c1: OR (95% CI)=1.595
(1.071–2.375), P=0.022] (Tables IV
and V).
 | Table IV.Adjusted odds ratios and 95%
confidence intervals of the CYP1A1 rs4646903 genotype in
upper digestive tract cancer. |
Table IV.
Adjusted odds ratios and 95%
confidence intervals of the CYP1A1 rs4646903 genotype in
upper digestive tract cancer.
|
| Number (%) |
| Adjusted
ORsc of different
modes of inheritance (95% CIs) |
|---|
|
|
|
|
|
|---|
| Factors | TT | CT | CC | P-value | ③ vs. ① | P-value | ② vs. ① | P-value | ②+③ vs. ① | P-value | ③ vs. ①+② | P-value |
|---|
| Controls
(n=212) | 74 (34.9) | 116 (54.7) | 22 (10.4) |
| 1.00
(reference) |
| 1.00
(reference) |
| 1.00
(reference) |
| 1.00
(reference) |
|
| EC (n=194) | 28 (14.4) | 90 (46.4) | 76 (39.2) | 0.198 | 1.175
(0.607–2.274) | 0.633 | 0.693
(0.449–1.069) | 0.097 | 0.768
(0.507–1.162) | 0.212 | 1.453
(0.790–2.674) | 0.230 |
| GCC (n=212) | 76 (35.8) | 96 (45.3) | 40 (18.9) | 0.028 | 1.936a (1.035–3.620) | 0.039 | 0.760
(0.494–1.169) | 0.212 | 0.940
(0.626–1.410) | 0.764 | 2.263b (1.272–4.026) | 0.005 |
| GAC (n=135) | 54 (40.0) | 63 (46.7) | 18 (13.3) | 0.326 | 1.295
(0.617–2.721) | 0.495 | 0.735
(0.451–1.199) | 0.217 | 0.820
(0.515–1.307) | 0.405 | 1.543
(0.774–3.076) | 0.218 |
 | Table V.Adjusted odds ratios and 95%
confidence intervals of CYP2E1 rs2031920 genotypes in upper
digestive tract cancer. |
Table V.
Adjusted odds ratios and 95%
confidence intervals of CYP2E1 rs2031920 genotypes in upper
digestive tract cancer.
|
| Number (%) |
| Adjusted
ORsb of different
modes of inheritance (95% CIs) |
|---|
|
|
|
|
|
|---|
| Factors | c1/c1 | c1/c2 | c2/c2 | P-value | ③ vs. ① | P-value | ② vs. ① | P-value | ②+③ vs. ① | P-value | ③ vs. ①+② | P-value |
|---|
| Controls
(n=212) | 118 (55.7) | 84 (39.6) | 10 (4.7) |
| 1.00
(reference) |
| 1.00
(reference) |
| 1.00
(reference) |
| 1.00
(reference) |
|
| EC (n=194) | 86 (44.3) | 100 (51.5) | 8 (4.1) | 0.054 | 0.993
(0.367–2.686) | 0.990 | 1.673a (1.111–2.520) | 0.014 | 1.595a (1.071–2.375) | 0.022 | 0.789
(0.297–2.094) | 0.634 |
| GCC (n=212) | 115 (54.2) | 87 (41.0) | 10 (4.7) | 0.955 | 0.974
(0.383–2.475) | 0.956 | 1.051
(0.702–1.575) | 0.808 | 1.043
(0.705–1.541) | 0.834 | 0.955
(0.381–2.392) | 0.921 |
| GAC (n=135) | 82 (60.7) | 48 (35.6) | 5 (3.7) | 0.630 | 0.752
(0.238–2.380) | 0.628 | 0.827
(0.517–1.321) | 0.426 | 0.819
(0.520–1.288) | 0.387 | 0.806
(0.258–2.519) | 0.710 |
Gene-gene and gene-environment
association between smoking, alcohol consumption, and CYP1A1 or
CYP2E1
Gene-gene and gene-environment association between
cigarette smoking, alcohol consumption, and CYP1A1 rs4646903
or CYP2E1 rs2031920 polymorphisms are presented in Table VI. An association existed between
CYP1A1 and smoking in EC, GCC and GAC; CYP1A1 and
alcohol drinking in EC and GCC; CYP2E1 and smoking in EC,
GCC and GAC; and CYP2E1 and alcohol drinking in EC and GCC.
No association was observed between CYP1A1 and
CYP2E1. Compared with non-smokers with wild-type
CYP1A1 (TT), smokers with a CYP1A1 heterozygous
variant genotype had a 2.597, 4.359 and 3.503-fold increased risk
of EC, GCC and GAC, respectively. Smokers with a CYP1A1
homozygous variant genotype had a 5.125, 8.618 and 6.070-fold
increased risk of EC, GCC and GAC, respectively. Compared with
non-drinkers with wild-type CYP1A1 (TT), alcohol drinkers
with a CYP1A1 homozygous variant genotype had a 4.124, 6.820
and 4.489-fold increased risk of EC, GCC and GAC, respectively.
Compared with non-smokers with wild-type CYP2E1 (c1/c1),
smokers with a CYP2E1 heterozygous variant genotype had a
6.345, 5.318 and 3.300-fold increased risk of EC, GCC and GAC,
respectively. In addition, smokers with a CYP2E1 homozygous
variant genotype had 6.661 and 7.621-fold increased risk for GCC
and GAC. Compared with non-drinkers with wild-type CYP2E1
(c1/c1), alcohol drinkers with a CYP2E1 heterozygous variant
genotype had a 3.820 and 3.070-fold increased risk of EC and GCC,
respectively. These results indicated the association between
smoking or alcohol consumption and CYP1A1 rs4646903 or
CYP2E1 rs2031920 in UDTC. No associations were observed
between CYP1A1 rs4646903 and CYP2E1 rs2031920.
 | Table VI.Association of smoking, alcohol, and
CYP1A1 rs4646903, CYP2E1 rs2031920 variants in upper
digest tract cancers. |
Table VI.
Association of smoking, alcohol, and
CYP1A1 rs4646903, CYP2E1 rs2031920 variants in upper
digest tract cancers.
|
|
|
| EC | GCC | GAC |
|---|
|
|
|
|
|
|
|
|---|
|
Factorse | Variant | Controls n=212 | n=194 | ORc (95% CI) | P-value | n=212 | ORc (95% CI) | P-value | n=135 | ORc (95% CI) | P-value |
|---|
| Smoking | rs4646903 |
|
|
| 0.011d |
|
| 0.001d |
|
| 0.049d |
| No | TT | 41 | 32 | 1.00
(reference) |
| 29 | 1.00
(reference) |
| 19 | 1.00
(reference) |
|
|
| CT | 79 | 44 | 0.700
(0.378–1.296) | 0.257 | 37 | 0.704
(0.368–1.346) | 0.288 | 29 | 0.907
(0.440–1.871) | 0.791 |
|
| CC | 16 | 16 | 1.175
(0.487–2.834) | 0.719 | 23 | 2.494a (1.072–5.800) | 0.033 | 11 | 1.722
(0.639–4.637) | 0.282 |
|
Yes | TT | 33 | 44 | 3.188b (1.482–6.857) | 0.003 | 47 | 4.193b (1.863–9.438) | 0.001 | 35 | 4.439b (1.846–10.674) | 0.001 |
|
| CT | 37 | 46 | 2.597a (1.225–5.505) | 0.013 | 59 | 4.359b (1.979–9.601) | <0.001 | 34 | 3.503a (1.447–8.478) | 0.005 |
|
| CC | 6 | 12 | 5.125b (1.551–16.943) | 0.007 | 17 | 8.618b (2.710–27.403) | <0.001 | 7 | 6.070b (1.580–23.325) | 0.009 |
| Smoking | rs2031920 |
|
|
| 0.002d |
|
| 0.001d |
|
| 0.017d |
| No | c1/c1 | 73 | 44 | 1.00
(reference) |
| 49 | 1.00
(reference) |
| 40 | 1.00
(reference) |
|
|
| c1/c2 | 55 | 43 | 1.336
(0.756–2.361) | 0.319 | 36 | 1.046
(0.584–1.874) | 0.880 | 19 | 0.646
(0.291–1.101) | 0.204 |
|
| c2/c2 | 8 | 5 | 0.998
(0.289–3.439) | 0.997 | 4 | 0.809
(0.224–2.922) | 0.746 | NA | NA | NA |
|
Yes | c1/c1 | 45 | 42 | 2.834b (1.430–5.613) | 0.003 | 66 | 4.236b (2.147–8.359) | <0.001 | 42 | 2.818b (1.345–5.904) | 0.006 |
|
| c1/c2 | 29 | 57 | 6.345b (3.113–12.930) | <0.001 | 51 | 5.318b (2.546–11.106) | <0.001 | 29 | 3.300b (1.465–7.434) | 0.004 |
|
| c2/c2 | 2 | 3 | 3.185
(0.467–21.740) | 0.237 | 6 | 6.661a (1.202–36.901) | 0.030 | 5 | 7.621b (1.277–45.480) | 0.026 |
| Alcohol | rs4646903 |
|
|
| 0.037d |
|
| 0.002d |
|
| 0.136d |
| No | TT | 41 | 38 | 1.00
(reference) |
| 34 | 1.00
(reference) |
| 29 | 1.00
(reference) |
|
|
| CT | 76 | 48 | 0.633
(0.353–1.135) | 0.137 | 41 | 0.625
(0.340–1.149) | 0.139 | 32 | 0.639
(0.332–1.230) | 0.188 |
|
| CC | 18 | 17 | 0.920
(0.406–2.088) | 0.842 | 24 | 1.641
(0.749–3.593) | 0.217 | 9 | 0.762
(0.292–1.991) | 0.578 |
|
Yes | TT | 33 | 38 | 1.579 (0.
786–3.172) | 0.204 | 42 | 1.822
(0.892–3.722) | 0.102 | 25 | 1.220
(0.550–2.705) | 0.631 |
|
| CT | 40 | 42 | 1.280
(0.649–2.522) | 0.486 | 55 | 1.877
(0.948–3.714) | 0.072 | 31 | 1.116
(0.518–2.402) | 0.785 |
|
| CC | 4 | 11 | 4.124a (1.122–15.155) | 0.033 | 16 | 6.820b (1.974–23.561) | 0.002 | 9 | 4.489a (1.185–17.002) | 0.028 |
| Alcohol | rs2031920 |
|
|
| 0.020d |
|
| 0.016d |
|
| 0.178d |
| No | c1/c1 | 71 | 46 | 1.00
(reference) |
| 51 | 1.00
(reference) |
| 45 | 1.00
(reference) |
|
|
| c1/c2 | 58 | 54 | 1.545
(0.901–2.651) | 0.114 | 43 | 1.109
(0.638–1.928) | 0.713 | 22 | 0.624
(0.330–1.181) | 0.147 |
|
| c2/c2 | 6 | 3 | 0.782
(0.180–3.398) | 0.743 | 5 | 1.271
(0.361–4.479) | 0.709 | 3 | 0.972
(0.222–4.263) | 0.970 |
|
Yes | c1/c1 | 47 | 40 | 1.789
(0.944–3.390) | 0.075 | 64 | 2.467b (1.343–4.532) | 0.004 | 37 | 1.380
(0.702–2.714) | 0.351 |
|
| c1/c2 | 26 | 46 | 3.820b (1.913–7.629) | <0.001 | 44 | 3.070b (1.537–6.134) | 0.001 | 26 | 1.801
(0.834–3.890) | 0.134 |
|
| c2/c2 | 4 | 5 | 1.796
(0.444–7.272) | 0.412 | 5 | 1.679
(0.415–6.797) | 0.468 | 2 | 0.710
(0.118–4.273) | 0.708 |
| rs2031920 | rs4646903 |
|
|
| 0.060d |
|
| 0.976d |
|
| 0.998d |
|
c1/c1 | TT | 46 | 37 | 1.00
(reference) |
| 49 | 1.00
(reference) |
| 29 | 1.00
(reference) |
|
|
c1/c1 | CT | 25 | 42 | 2.256
(1.150–4.424) | 0.018 | 24 | 0.857
(0.422–1.740) | 0.669 | 23 | 1.604
(0.747–3.442) | 0.225 |
|
c1/c1 | CC | 3 | 2 | 0.664
(0.100–4.389) | 0.670 | 3 | 0.715
(0.128–4.004) | 0.703 | 2 | 1.195
(0.181–7.905) | 0.853 |
|
c1/c2 | TT | 66 | 39 |
0.678(0.372–1.237) | 0.205 | 48 | 0.620
(0.353–1.090) | 0.097 | 40 | 1.007
(0.530–1.911) | 0.984 |
|
c1/c2 | CT | 46 | 41 | 1.055
(0.567–1.961) | 0.866 | 44 | 0.833
(0.459–1.510) | 0.547 | 20 | 0.687
(0.329–1.432) | 0.316 |
|
c1/c2 | CC | 4 | 5 | 1.328
(0.321–5.497) | 0.696 | 4 | 0.861
(0.197–3.766) | 0.842 | 3 | 1.228
(0.239–6.318) | 0.806 |
|
c2/c2 | TT | 6 | 10 | 2.193
(0.713–6.746) | 0.171 | 18 | 3.222
(1.149–9.039) | 0.026 | 13 | 4.359
(1.433–13.260) | 0.010 |
|
c2/c2 | CT | 13 | 17 | 1.477
(0.620–3.517) | 0.378 | 19 | 1.398
(0.609–3.209) | 0.430 | 5 | 0.682
(0.210–2.212) | 0.524 |
|
c2/c2 | CC | 3 | 1 | 0.409
(0.039–4.330) | 0.458 | 3 | 0.991
(0.183–5.364) | 0.991 | NA | NA | NA |
Discussion
In the present study, it was confirmed that smoking
and alcohol consumption were the main risk factors of upper
digestive tract cancers. In addition, it was indicated that
CYP1A1 rs4646903 polymorphisms increased GCC risk,
CYP2E1 rs2031920 increased EC risk, while the GSTM1
null genotype decreased EC risk. Regarding the gene-gene or
gene-environment associations in this study, associations between
CYP1A1 rs4646903, CYP2E1 rs2031920 and smoking or
alcohol were detected in UDTC.
To date, an increasing number of studies have
investigated the associations between CYP1A1 rs4646903
polymorphisms and digestive cancer risk (15,18,24,25). In
a recent meta-analysis, seven articles reported on CYP1A1
rs4646903 polymorphisms in four digestive cancers, and no
associations were found in stratified analysis and subgroup
analyses (18). In addition, in
another meta-analysis, CYP1A1 rs4646903 polymorphisms were
confirmed to be associated with an increased susceptibility to
colorectal cancer, however not to esophageal cancer or gastric
cancer (24). In the present study,
no association between the CYP1A1 rs4646903 CC genotype and
EC or GAC were detected, which was consistent with the
aforementioned studies. However, in another meta-analysis, 11
studies about CYP1A1 rs4646903 polymorphisms and GC were
included, and significant results were found among a large
sample-size subgroup (15).
Furthermore, evidence was also found to support an association
between CYP1A1 rs4646903 polymorphisms and digestive tract
cancer in the subgroups of Caucasian and mixed individuals
(24). This suggested that the
associations may vary across different sample sizes and
ethnicities. This study found associations between CYP1A1
rs4646903 polymorphisms and GCC. To the best of our knowledge, a
limited number of studies have been performed in GCC. One report in
Linzhou found an association between the CYP1A1 rs4646903
variant allele, and a reduced risk of GCC in people with Dysplasia,
who were at high risk for the development of GCC (25). However, the study only included 90
cases of GCC, decreasing the reliability of the results.
One meta-analysis in China suggested that the
CYP2E1 rs2031920 polymorphism was a risk factor for EC, and
the c2 allele was demonstrated to be a factor that decreases the
risk of EC in the mainland Chinese population (26). However, in this research,
CYP2E1 rs2031920 genotypes tended to increase EC risk. One
report in Guangzhou Chinese population and another report in a
Northern Jiangsu Chinese population also showed that the
CYP2E1 rs2031920 polymorphisms could be risk factors for the
development of gastric cancer (27,28).
Molecular biological evidence has shown that the CYP2E1
rs2031920 variant in the CYP2E1 promoter enhances gene
transcriptional activity by altering its binding to its
transcription factor, particularly, hepatocyte nuclear factor-1
(29), and influencing its
susceptibility to N-nitrosamine-linked carcinogenesis (30), indicating that the CYP2E1
rs2031920 variant may be associated with an increased cancer risk.
The present study's results supported the aforementioned
findings.
It was indicated that the GSTM1 null genotype
had protective effects against EC, decreasing EC risk. However,
increased upper digestive tract cancer risk was associated with
GSTM1 non-null genotypes. To the best of our knowledge, this
is not consistent with most other studies (17,18). A
most recent meta-analysis on four digestive cancers showed that the
GSTM1 polymorphism was associated with the risk of the four
digestive cancers among the Asian population, as subgroup analyses
by cancer site showed that the GSTM1 null genotype increased
the total gastric cancer risk in the population (18). Another meta-analysis in a Japanese
population showed that GSTM1 null, GSTT1 null and
GSTM1/T1 both or either null genotypes were associated with
increased risk, though this was not statistically significantly
(15). However, there are a number
of reports showing that cancer risk is associated with GSTM1
non-null genotypes (30–33). There are several possible reasons for
this observation. One is that the loss of one GST enzyme may be
negligible compared with the large extended GST family (23). Even if the GSTM1
detoxification function is lost, other GST family members can still
act to decrease cancer risk. Furthermore, some carcinogens,
including N-hydroxy-Trp-P-2, have enhanced genotoxicity and
carcinogenicity after binding to glutathione (34). Furthermore, it appears that
GSTM1 null individuals have higher DNA adduct levels than
GSTM1-expressing individuals (35).
Regarding the gene-gene or gene-environment
associations in this study, an association between CYP1A1
rs4646903, CYP2E1 rs2031920 and smoking or alcohol was
detected. Two meta-analyses showed that CYP2E1 rs2031920 may
modify the susceptibility to gastric cancer among individuals who
have a smoking history, or when GSTM1 or GSTT1 are
null, or CYP2E1 rs2031920 is homozygous wild-type (16,36). An
increased risk was seen in CYP1A1 rs4646422 variant subjects
whose smoking was categorized as ≤30 pack-years, or whose
GSTM1/T1 were both null genotypes, or who were null
for either GSTM1/T1 individually (17). These studies suggested that tumor
incidence is often due to a combination of exposure to external
environmental factors and internal gene aberrance. These
interactions have a greater impact on cancer susceptibility
compared with single genes.
Associations between metabolic gene polymorphisms
and human cancers have been debated. The differences stem from
several factors, including ethnic or geographic differences, as
Asian populations have been reported to be more prone compared with
Caucasian populations to show significant associations between
metabolic gene polymorphisms and carcinogenesis (18,37,38).
Even in populations containing the same ethnic group, the
associations vary by region (14).
It is believed that these inconsistent results across ethnicity and
geographic areas derive mainly from the unequal frequency of
genetic polymorphisms (30,39). Another factor is the different host
habits and environmental factor exposure levels, including tobacco
use and alcohol consumption (4,5), family
history of cancer and Helicobacter pylori infection
(40), which have been identified as
risk factors for upper digestive tract cancers. Other environmental
factors include low socioeconomic status (41), poor oral hygiene (42), nutritional deficiencies, diet
(43) and high salt intake (44). It has been hypothesized that various
living environments lead to different degrees of cancer
susceptibility (45). Specific
associations are easily found in subgroups with exposure to
negative factors, including smoking, H. pylori infection, or
low consumption of fruit. A lack of statistical power has also been
identified as a contributing factor, as the number of subjects who
carry the ‘unfavorable’ gene polymorphism combinations becomes
visible and can be assessed only if sufficient subjects are
available with the specific genetic profile required (46). Furthermore, the ‘Berkson bias’ is
typically present in hospital-based studies, as the controls may
only represent a sample of an ill-defined reference population and
may not be representative of the general population (47). In addition, in terms of gene-gene and
gene-environment interaction, tumor incidence is often a
combination of multiple factors (48). A negative association between a gene
and cancer susceptibility does not mean that the gene has no impact
on cancer risk. In terms of methodological differences, the most
popular method in previous studies has been PCR-RFLP (21,30).
Although PCR-RFLP is a simple, specific and efficient method of SNP
detection, it has obvious limitations with respect to accuracy,
particularly for subjects who carry a heterozygous mutation
(49). With the development of
molecular detection technology, a number of researchers have begun
to use TaqMan assays (25,50), which may be faster and more accurate
compared with PCR-RFLP. A superior new method is genome sequencing
(23,51), particularly genome-wide associated
studies, which can assay huge amounts of SNPs in a large number of
samples and facilitates rapid detection.
In conclusion, it was indicated that smoking/alcohol
consumption are upper digestive tract cancer risk factors. The
CYP1A1 rs4646903 and CYP2E1 rs2031920 genotypes may
contribute to higher GCC and EC susceptibility, respectively. The
GSTM1 null genotype may serve a protective effect against
EC. The gene-environment associations present increase the cancer
risk. In the future, the present study may be improved by
increasing the sample size and applying more advanced SNP detection
methods, including a TaqMan assay or genome sequencing.
Acknowledgments
We are also grateful to Dr. Xiang Yuan and Dr Jin-yu
Kong at the First Affiliated Hospital of Henan University of
Science and Technology University (Xinxiang, China), for their kind
assistance in editing this manuscript.
Funding
This work was supported by grants from the National
Natural Science Foundations (grant no. U1504814) and Major Projects
of Science and Technology Department in Henan Province (grant nos.
161100311200 and 161100311300) and Xinxiang Science and Technology
Project (grant no. CXGG17032).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FYZ, FZ and SML designed the experiment. FZ, JFS,
SML, YJH, LJD, ZWG, JL, XJD, FFS, YWZ and NCW collected the data
and performed the experiments. JFS analyzed and interpreted the
data. FZ and JFS were major contributors in writing the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The Anyang Tumor Hospital Institutional Review Board
approved the present study (no. AZLL022015005150701). All patients
and controls signed a study-specific written informed consent
form.
Patient consent for publication
All patients and controls have provided consent for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Piazuelo MB and Correa P: Gastric cáncer:
Overview. Colomb Med (Cali). 44:192–201. 2013.PubMed/NCBI
|
|
2
|
Global Burden of Disease Cancer
Collaboration, ; Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional, and national cancer incidence, mortality, years
of life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miao Y, Liu R, Pu Y and Yin L: Trends in
esophageal and esophagogastric junction cancer research from 2007
to 2016: A bibliometric analysis. Medicine (Baltimore).
96:e69242017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haas SL, Ye W and Löhr JM: Alcohol
consumption and digestive tract cancer. Curr Opin Clin Nutr Metab
Care. 15:457–467. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dong J and Thrift AP: Alcohol, smoking and
risk of oesophago- gastric cancer. Best Pract Res Clin
Gastroenterol. 31:509–517. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sitarz R, Skierucha M, Mielko J, Offerhaus
GJA, Maciejewski R and Polkowski WP: Gastric cancer: Epidemiology,
prevention, classification, and treatment. Cancer Manag Res.
10:239–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matejcic M and Iqbal Parker M:
Gene-environment interactions in esophageal cancer. Crit Rev Clin
Lab Sci. 52:211–231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miller EC: Some current perspectives on
chemical carcinogenesis in humans and experimental animals:
Presidential address. Cancer Res. 38:1479–1496. 1978.PubMed/NCBI
|
|
10
|
Kiyohara C, Wakai K, Mikami H, Sido K,
Ando M and Ohno Y: Risk modification by CYP1A1 and GSTM1
polymorphisms in the association of environmental tobacco smoke and
lung cancer: A case-control study in Japanese nonsmoking women. Int
J Cancer. 107:139–144. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Landi MT, Bertazzi PA, Shields PG, Clark
G, Lucier GW, Garte SJ, Cosma G and Caporaso NE: Association
between CYP1A1 genotype, mRNA expression and enzymatic activity in
humans. Pharmacogenetics. 4:242–246. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marchand LL, Wilkinson GR and Wilkens LR:
Genetic and dietary predictors of CYP2E1 activity: A phenotyping
study in Hawaii Japanese using chlorzoxazone. Cancer Epidemiol
Biomarkers Prev. 8:495–500. 1999.PubMed/NCBI
|
|
13
|
Strange RC and Fryer AA: The glutathione
S-transferases: Influence of polymorphism on cancer susceptibility.
IARC Sci Publ. 231–249. 1999.PubMed/NCBI
|
|
14
|
Liu C, Jiang Z, Deng QX and Zhao YN:
Meta-analysis of association studies of CYP1A1 genetic
polymorphisms with digestive tract cancer susceptibility in
Chinese. Asian Pac J Cancer Prev. 15:4689–4695. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xue H, Lu Y, Xue Z, Lin B, Chen J, Tang F
and Huang G: The effect of CYP1A1 and CYP1A2 polymorphisms on
gastric cancer risk among different ethnicities: A systematic
review and meta-analysis. Tumour Biol. 35:4741–4756. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang MX, Liu K, Wang FG, Wen XW and Song
XL: Association between CYP2E1 polymorphisms and risk of gastric
cancer: An updated meta-analysis of 32 case-control studies. Mol
Clin Oncol. 4:1031–1038. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hidaka A, Sasazuki S, Matsuo K, Ito H,
Charvat H, Sawada N, Shimazu T, Yamaji T, Iwasaki M, Inoue M, et
al: CYP1A1, GSTM1 and GSTT1 genetic polymorphisms and gastric
cancer risk among Japanese: A nested case-control study within a
large-scale population-based prospective study. Int J Cancer.
139:759–768. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du L, Lei L, Zhao X, He H, Chen E, Dong J,
Zeng Y and Yang J: The interaction of smoking with gene
polymorphisms on four digestive cancers: A systematic review and
meta-analysis. J Cancer. 9:1506–1517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li JY, Liu BQ, Li GY, Chen ZJ, Sun XI and
Rong SD: Atlas of cancer mortality in the People's Republic of
China. An aid for cancer control and research. Int J Epidemiol.
10:127–133. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou MG, Wang XF, Hu JP, Li GL, Chen WQ,
Zhang SW, Wan X, Wang LJ, Xiang C, Hu YS and Yang GH: Geographical
distribution of cancer mortality in China, 2004–2005. Zhonghua Yu
Fang Yi Xue Za Zhi. 44:303–308. 2010.(In Chinese). PubMed/NCBI
|
|
21
|
Choudhury JH, Singh SA, Kundu S, Choudhury
B, Talukdar FR, Srivasta S, Laskar RS, Dhar B, Das R, Laskar S, et
al: Tobacco carcinogen-metabolizing genes CYP1A1, GSTM1, and GSTT1
polymorphisms and their interaction with tobacco exposure influence
the risk of head and neck cancer in Northeast Indian population.
Tumour Biol. 36:5773–5783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu X, Kelsey KT, Wiencke JK, Wain JC and
Christiani DC: Cytochrome P450 CYP1A1 MspI polymorphism and lung
cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 5:687–692.
1996.PubMed/NCBI
|
|
23
|
Khabaz MN, Nedjadi T, Gari MA, Al-Maghrabi
JA, Atta HM, Bakarman M and Gazzaz ZJ: GSTM1 gene polymorphism and
the risk of colorectal cancer in a Saudi Arabian population. Genet
Mol Res. 15:2016. View Article : Google Scholar
|
|
24
|
Ren A, Qin T, Wang Q, Du H, Zhong D, Hua Y
and Zhu L: Cytochrome P450 1A1 gene polymorphisms and digestive
tract cancer susceptibility: A meta-analysis. J Cell Mol Med.
20:1620–1631. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Roth MJ, Abnet CC, Johnson LL, Mark SD,
Dong ZW, Taylor PR, Dawsey SM and Qiao YL: Polymorphic variation of
Cyp1A1 is associated with the risk of gastric cardia cancer: A
prospective case-cohort study of cytochrome P-450 1A1 and GST
enzymes. Cancer Causes Control. 15:1077–1083. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Leng WD, Zeng XT, Chen YJ, Duan XL, Niu
YM, Long RP and Luo ZX: Cytochrome P450 2E1 RsaI/PstI polymorphism
and risk of esophageal cancer: A meta-analysis of 17 case-control
studies. Exp Ther Med. 4:938–948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Elingarami S, Liu H, Kalinjuma AV, Hu W,
Li S and He N: Polymorphisms in NEIL-2, APE-1, CYP2E1 and mdm2
genes are independent predictors of gastric cancer Risk in a
Northern Jiangsu Population (China). J Nanosci Nanotechnol.
15:4815–4828. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen ZH, Xian JF and Luo LP: Analysis of
ADH1B Arg47His, ALDH2 Glu487Lys, and CYP4502E1 polymorphisms in
gastric cancer risk and interaction with environmental factors.
Genet Mol Res. 15:2016. View Article : Google Scholar
|
|
29
|
Hayashi S, Watanabe J and Kawajiri K:
Genetic polymorphisms in the 5′-flanking region change
transcriptional regulation of the human cytochrome P450IIE1 gene. J
Biochem. 110:559–565. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tan W, Song N, Wang GQ, Liu Q, Tang HJ,
Kadlubar FF and Lin DX: Impact of genetic polymorphisms in
cytochrome P450 2E1 and glutathione S-transferases M1, T1, and P1
on susceptibility to esophageal cancer among high-risk individuals
in China. Cancer Epidemiol Biomarkers Prev. 9:551–556.
2000.PubMed/NCBI
|
|
31
|
Chen C, Madeleine MM, Lubinski C, Weiss
NS, Tickman EW and Daling JR: Glutathione S-transferase M1
genotypes and the risk of anal cancer: A population-based
case-control study. Cancer Epidemiol Biomarkers Prev. 5:985–991.
1996.PubMed/NCBI
|
|
32
|
Li D, Dandara C and Parker MI: The 341C/T
polymorphism in the GSTP1 gene is associated with increased risk of
oesophageal cancer. BMC Genet. 11:472010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lewis SJ, Cherry NM, Niven RM, Barber PV
and Povey AC: GSTM1, GSTT1 and GSTP1 polymorphisms and lung cancer
risk. Cancer Lett. 180:165–171. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saito K, Yamazoe Y, Kamataki T and Kato R:
Glutathione transferase-mediated and non-enzymatic activation and
detoxication of the N-hydroxy derivative of Trp-P-2, a potent
pyrolysate promutagen. Xenobiotica. 14:545–548. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Houlston RS: Glutathione S-transferase M1
status and lung cancer risk: A meta-analysis. Cancer Epidemiol
Biomarkers Prev. 8:675–682. 1999.PubMed/NCBI
|
|
36
|
Zhuo W, Zhang L, Wang Y, Ling J, Zhu B and
Chen Z: CYP2E1 RsaI/PstI polymorphism and gastric cancer
susceptibility: Meta-analyses based on 24 case-control studies.
PLoS One. 7:e482652012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu B, Liu K, Huang H, Yuan J, Yuan W, Wang
S, Chen T, Zhao H and Yin C: MspI and Ile462Val polymorphisms in
CYP1A1 and overall cancer risk: A meta-analysis. PLoS One.
8:e851662013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meng X, Liu Y and Liu B: Glutathione
S-transferase M1 null genotype meta-analysis on gastric cancer
risk. Diagn Pathol. 9:1222014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Garte S, Gaspari L, Alexandrie AK,
Ambrosone C, Autrup H, Autrup JL, Baranova H, Bathum L, Benhamou S,
Boffetta P, et al: Metabolic gene polymorphism frequencies in
control populations. Cancer Epidemiol Biomarkers Prev.
10:1239–1248. 2001.PubMed/NCBI
|
|
40
|
Choi YJ and Kim N: Gastric cancer and
family history. Korean J Intern Med. 31:1042–1053. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Uthman OA, Jadidi E and Moradi T:
Socioeconomic position and incidence of gastric cancer: A
systematic review and meta-analysis. J Epidemiol Community Health.
67:854–860. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gupta B and Johnson NW: Emerging and
established global life-style risk factors for cancer of the upper
aero-digestive tract. Asian Pac J Cancer Prev. 15:5983–5991. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Johnson IT: Understanding the association
between diet and nutrition in upper gastrointestinal cancer. Expert
Rev Gastroenterol Hepatol. 9:1347–1349. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Goral V: Etiopathogenesis of gastric
cancer. Asian Pac J Cancer Prev. 17:2745–2750. 2016.PubMed/NCBI
|
|
45
|
Yoshida T, Ono H, Kuchiba A, Saeki N and
Sakamoto H: Genome-wide germline analyses on cancer susceptibility
and GeMDBJ database: Gastric cancer as an example. Cancer Sci.
101:1582–1589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Taioli E: Gene-environment interaction in
tobacco-related cancers. Carcinogenesis. 29:1467–1474. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Conn HO, Snyder N and Atterbury CE: The
Berkson bias in action. Yale J Biol Med. 52:141–147.
1979.PubMed/NCBI
|
|
48
|
Weinstein IB: Cell culture systems for
studying multifactor interactions in carcinogenesis. Dev Toxicol
Environ Sci. 8:149–64. 1980.PubMed/NCBI
|
|
49
|
Jin YW, Qu YJ, Wang H, Bai JL and Song F:
Limitation of PCR-RFLP method for the detection of genetic
mutations in spinal muscular atrophy. Zhonghua Yi Xue Yi Chuan Xue
Za Zhi. 29:34–37. 2012.(In Chinese). PubMed/NCBI
|
|
50
|
Blakely T, Barendregt JJ, Foster RH, Hill
S, Atkinson J, Sarfati D and Edwards R: The association of active
smoking with multiple cancers: National census-cancer registry
cohorts with quantitative bias analysis. Cancer Causes Control.
24:1243–1255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tan YH, Sidik SM, Syed Husain SN, Lye MS
and Chong PP: CYP1A1 MspI polymorphism and cervical carcinoma risk
in the multi-ethnic population of malaysia: A case-control study.
Asian Pac J Cancer Prev. 17:57–64. 2016. View Article : Google Scholar : PubMed/NCBI
|