Introduction
Cholangiocarcinoma (CCA) is a malignant tumor of the
extrahepatic bile duct, extending from the hilar area to the lower
end of the common bile duct (1). The
etiology of CCA may be associated with cholelithiasis, primary
sclerosing cholangitis and other types of disease (2). Surgical treatment, radiotherapy and
chemotherapy can be used clinically; however, the prognosis is poor
(3,4). Consequently, it is crucial to
investigate efficient therapies and identify sensitive biomarkers
for early diagnosis of CCA. Molecular-targeted therapy has
exhibited significant effects against cell proliferation,
recurrence and metastasis in CCA, with few adverse effects
(5). Therefore, it is important to
understand the molecular mechanisms of carcinogenesis and
progression in CCA, which may contribute to identifying novel
diagnostic markers and sensitive therapeutic targets.
Long non-coding RNAs (lncRNAs) are a class of RNA
molecules that are >200 nucleotides and exhibit no protein
coding function (6). Recent studies
have demonstrated that lncRNAs are closely associated with the
development of diverse human diseases, including cancer (7,8). lncRNAs
are involved in the regulation of gene expression at the
epigenetic, transcriptional and post-transcriptional levels and
affect the occurrence, development and invasion of tumors (9,10).
Investigations regarding the association between lncRNAs and
digestive system tumors may provide a new strategy for the
prevention, diagnosis and treatment of these tumors (11,12).
Recently, the biological roles and mechanisms of several lncRNAs,
including AFAP1 antisense RNA 1 (AS1), ASAP1 intronic transcript 1,
colon cancer associated transcript 1, nuclear enriched abundant
transcript 1 and sprouty4-intron transcript 1, have been reported
to be involved in CCA occurrence and metastasis (13–17). A
previous study demonstrated that the expression of lncRNA
FLVCR1-AS1 is significantly increased in hepatocellular carcinoma.
Additionally, it was identified to promote cancer cell
proliferation, migration and invasion by sponging microRNA
(miR)-513c in hepatocellular carcinoma (18). However, to the best of our knowledge,
the role and function of FLVCR1-AS1 in CCA remains unclear.
The present study identified an increased expression
level of FLVCR1-AS1 in CCA tumor tissues and cell lines. The
biological effect of FLVCR1-AS1 on cell proliferation, migration
and invasion was investigated. It was revealed that downregulation
of FLVCR1-AS1 inhibited these processes. Furthermore,
bioinformatics analysis and luciferase reporter assay demonstrated
that FLVCR1-AS1 serves an oncogenic role in CCA by sponging
miR-485-5p.
Materials and methods
Patients and tissue samples
In total, 22 paired CCA and normal tissue samples
were collected from patients (age range, 42–77 years; mean age, 59
years; 13 males and 9 females) who underwent surgical treatment at
Shanghai Seventh People's Hospital Affiliated to Shanghai
University of Traditional Chinese Medicine (Shanghai, China)
between January 2010 and December 2016. The clinical
characteristics of patients with CCA are presented in Table I; the Tumor-Node-Metastasis (TNM)
staging system was used as previously described (19). Tissue samples were snap frozen in
liquid nitrogen immediately after surgical resection and stored at
−80°C. Patients who received radiotherapy and/or immunotherapy
before or following surgery were excluded from the study. Samples
were collected after written informed consent was obtained from all
patients. The study protocol conformed to the ethical guidelines of
the 1975 Declaration of Helsinki and was approved by the Ethics
Committee of Shanghai Seventh People's Hospital Affiliated to
Shanghai University of Traditional Chinese Medicine (Shanghai,
China).
 | Table I.Clinical characteristics of patients
with cholangiocarcinoma. |
Table I.
Clinical characteristics of patients
with cholangiocarcinoma.
| Characteristic | Patients (n=22) |
|---|
| Sex |
|
| Male | 13 |
|
Female | 9 |
| Age, years |
|
|
>60 | 12 |
| ≤60 | 10 |
| Tumor size, cm |
|
|
>3 | 13 |
| ≤3 | 9 |
| Lymph node
metastasis |
|
| No | 15 |
| Yes | 7 |
| TNM stage |
|
| I/II | 14 |
|
III/IV | 8 |
Cell culture and CCA cell
transfection
Human cell lines, including the noncancerous
cholangiocyte cell line HIBEC and the CCA cell lines RBE, CCLP1,
HuCCT1 and HCCC-9810 were purchased from the Chinese Academy of
Sciences Cell Bank (Shanghai, China). All cells were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc.) and grown in humidified 5% CO2
at 37°C. Short hairpin RNA (shRNA) targeting FLVCR1-AS1
(shFLVCR1-AS1) and the relative control shRNA (shNC) were obtained
from Shanghai Genepharma Co., Ltd., (Shanghai, China). The sequence
of shFLVCR1-AS1 was 5′-GGTAAGCAGTGGCTCCTCTAA-3′ and the sequence of
shNC was 5′-AATTCTCCGAACGTGTCACGT-3′. The transfection was
performed using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) in 5×106 cells at a final
concentration of 50 nM, according to the manufacturer's protocol.
After transfection for 24 h, expression of FLVCR1-AS1 was validated
by reverse transcription-quantitative PCR (RT-qPCR).
RT-qPCR
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Qiagen GmbH, Hilden, Germany),
according to the manufacturer's protocol. Following quantification
using Nanodrop 2000 (Thermo Fisher Scientific, Inc.), the extracted
total RNA was reverse-transcribed using Reverse Transcription kit
(Takara Biotechnology Co., Ltd., Dalian, China). qPCR with
SYBR® Green RT-PCR Master mix (Takara Biotechnology,
Co., Ltd., Dalian, China) was performed on an Applied Biosystems
7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Primer specific sequences (FLVCR1-AS1, forward,
5′-GTGGCTCTCTCGTTCCC-3′ and reverse, 5′-CCGTCCTTCGGTAGTGTC-3′;
miR-485-5p, forward, 5′-AGAGGCTGGCCGTGAT-3′ and reverse,
5′-ATGTGTTGCTGTGTTTGTCG-3′) were synthesized by Shanghai Sangon
Biological Engineering Technology & Services Co., Ltd.
(Shanghai, China). The PCR conditions were as follows: Initial
denaturation at 95°C for 10 min, followed by 40 cycles of
denaturation at 95°C for 5 sec, annealing at 60°C for 40 sec and
extension at 72°C for 10 sec. Universal miRNA RT-qPCR primer,
5′-AACGAGACGACGACAGAC-3′; 5′-GCAAATTCGTGAAGCGTTCCATA-3′ for U6 was
used as an endogenous control to normalize miR-485-5p expression
level. GAPDH (forward, 5′-TCCTCTGACTTCAACAGCGACAC-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAATTC-3′) was used as an endogenous control
to normalize lncRNA FLVCR1-AS1 expression level. The relative
expression level was calculated using the 2−ΔΔCq method
(20). All the experiments were
performed in triplicate.
Cell proliferation (MTT) assay
CCA cells transfected either with shNC or
shFLVCR1-AS1 were seeded into 96 well plates at 5,000 cells/well.
At 12, 24, 48 and 72 h after transfection, the medium was removed
by suction and replaced with serum-free RPMI-1640 medium containing
1 mg/ml MTT. The cells were then incubated at 37°C for 4 h.
Following removal of the MTT solution, the formazan precipitate was
dissolved with 100 µl DMSO and the absorbance was measured at 570
and 600 nm in a microplate reader. shNC cells were used as the
normal controls.
Colony formation assay
CCA cells were trypsinized, counted and seeded into
12-well plates at 100 cells/well. The medium was replaced every 3
days during colony growth. Following incubation in Dulbecco's
modified Eagle's medium (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) for 12 days, the colonies were fixed with methanol for 20
min at room temperature and stained with 0.2% crystal violet for 15
min (Sigma-Aldrich; Merck KGaA) at room temperature. The colonies
were counted under an inverted microscope (Nikon Corporation,
Tokyo, Japan) at magnification, ×100.
In vivo xenograft experiments
All animal experiments were performed in accordance
with institutional and international animal regulations. The animal
protocols were approved by the Animal Care and Use Committee at
Shanghai Seventh People's Hospital Affiliated to Shanghai
University of Traditional Chinese Medicine (Shanghai, China). Male
6-week old BALB/c nude mice (N=8) were purchased from Beijing HFK
Bioscience Co. Ltd. (Beijing, China). All mice were kept in
pathogen-free conditions in 26–28°C and 50% humidity on a 12 h
light/dark cycle. Mice had ad libitum access to food and
water. For tumor propagation analysis, HuCCT1 cells, transfected
either with shNC or shFLVCR1-AS1, were subcutaneously injected into
the flanks of BALB/c nude mice at 1×106 cells/mouse. The
length and width of tumors were measured using a caliper every 7
days for a period of 28 days. All mice were euthanized at day 28
post-injection, and the tumor nodules of the mice were removed and
weighed. The tumor volume was calculated using the following
formula: Tumor volume (mm3)=length (mm) × width
(mm)2/2.
Wound-healing assay
A wound-healing assay was performed to detect cell
migration. In brief, 48 h after transfection, CCA cells were
cultured in 6-well plates (5×104 cells/well). The
monolayer of cells was scratched using a sterile plastic
micropipette tip and cells were cultured under standard conditions
for 24 h. Following several washes, recovery of the wound was
observed and images were obtained using a light microscope
(magnification, ×100; Olympus Corporation, Tokyo, Japan).
Cell invasion assay
After transfection for 48 h, CCA cells
(1×105) transfected with shNC or shFLVCR1-AS1 were
seeded into the upper chamber of Matrigel-coated inserts with
serum-free medium. RPMI-1640 medium with 10% FBS was added to the
lower chamber as a chemoattractant. The cells were allowed to
invade for 48 h at 37°C with 5% CO2. Migrating cells
were fixed in 70% ethanol for 30 min and stained with 0.1% crystal
violet for 10 min at 25°C. The number of cells that migrated to the
lower side were counted in five randomly selected fields under an
inverted microscope (Olympus Corporation) at ×100
magnification.
Luciferase reporter assay
Wild-type FLVCR1-AS1 (5′-AUACACGGCCCUC-3′) and
mutant FLVCR1-AS1 (5′-AUACUGCCGCCUC-3′) sequences were cloned into
a pmirGLO reporter vector (Promega Corporation, Madison, WI, USA).
HuCCT1 cells were co-transfected with NC mimic
(5′-GCAUGAUGGUCAUAGGUCC-3′) or miR-485-5p mimic
(5′-AGAGGCUGGCCGUGAUGAAUUC-3′), and wild-type FLVCR1-AS1 or mutant
FLVCR1-AS1 (1 µg per well) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Relative luciferase
activity was measured on a dual-luciferase reporter assay system
(Promega Corporation) at 48 h post-transfection. Data are presented
as the ratio of Renilla luciferase activity to firefly
luciferase activity.
Western blot analysis
CCA cells were lysed using RIPA lysis buffer
(Beyotime Biotechnology, Shanghai, China) and sonicated. Lysates
containing soluble proteins were collected and stored at 80°C.
Protein concentration was determined using a Bradford assay
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Total protein
lysates (10 µl per lane) were resolved on 10% SDS-PAGE gels and
transferred to polyvinyldifluoride membranes (EMD Millipore,
Billerica, MA, USA). Following blocking in TBS containing 0.1%
Tween-20 (TBS-T) with 5% non-fat dry milk for 30 min at room
temperature, membranes were washed four times in TBS-T and
incubated with primary antibodies overnight at 4°C. All primary
antibodies were obtained from Abcam (Cambridge, MA, USA) and used
at the following dilutions: Anti-Twist (1:500; catalog no.
ab49254), anti-matrix metalloproteinase (MMP)-2 (1:500; catalog no.
ab92536), anti-MMP-9 (1:500; ab38898), anti-β-actin (1:1,000;
catalog no. ab8226). Following extensive washing, membranes were
incubated with horseradish peroxidase-linked goat polyclonal
anti-rabbit IgG secondary antibodies at a dilution of 1:2,000 for 1
h at room temperature. Immunoreactivity was detected by enhanced
chemiluminescence using an ECL kit (Pierce; Thermo Fisher
Scientific, Inc.) and exposure to radiography film. GAPDH served as
the loading control. Western blots were quantified by densitometry
with Labworks Software (version 4.0; UVP BioImaging Systems,
Upland, CA, USA).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analysis was performed using SPSS 16.0 software (SPSS,
Inc., Chicago, IL, USA). Two-tailed Student's t-test was applied to
compare the differences between two groups and one-way analysis of
variance followed by Dunnett's multiple comparison test was used to
compare the differences among three independent groups. Correlation
between FLVCR1-AS1 and miR-485-5p expression in CCA tissues was
identified using Pearson's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference. Each
experiment was repeated in triplicate.
Results
Expression of FLVCR1-AS1 in tissues
from patients with CCA and cells
To investigate the role of FLVCR1-AS1 in CCA, the
expression of FLVCR1-AS1 in 22 CCA and normal tissue samples was
analyzed by RT-qPCR. The clinical characteristics of patients with
CCA are presented in Table I. It was
identified that the expression level of FLVCR1-AS1 was
significantly increased in CCA tissues compared with normal tissues
(P<0.001; Fig. 1A). In addition,
RT-qPCR demonstrated that the expression of FLVCR1-AS1 was higher
in CCA cell lines (RBE, HCCC-9810, HuCCT1 and CCLP1) compared with
the HIBEC noncancerous cholangiocyte cell line (P<0.01; Fig. 1B). These results indicated that
FLVCR1-AS1 may act as an oncogene in CCA. HuCCT1 and CCLP1 cells
exhibited the highest expression of FLVCR1-AS1 and were used for
subsequent experiments.
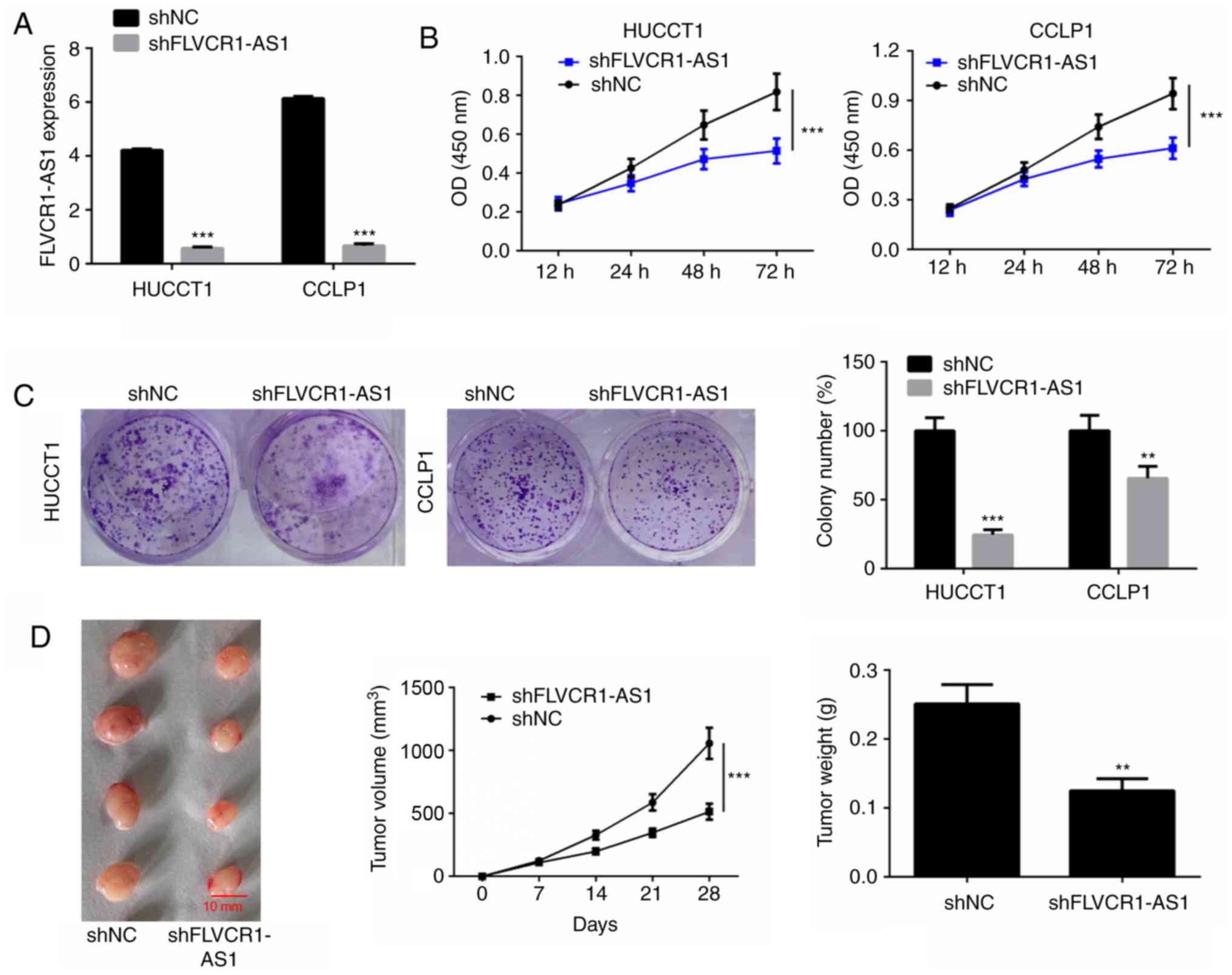
Downregulation of FLVCR1-AS1 inhibits
cell proliferation in vitro and in vivo
To investigate whether FLVCR1-AS1 influences cell
viability of HuCCT1 and CCLP1 cells, FLVCR1-AS1 was silenced by
transfection with shFLVCR1-AS1. The transfection efficiency in
HuCCT1 and CCLP1 cells was determined by measuring the expression
level of FLVCR1-AS1 using RT-qPCR (Fig.
2A). MTT and colony formation assays were performed to detect
cell proliferation and the results revealed that silencing of
FLVCR1-AS1 significantly suppressed cell viability and
proliferation of HuCCT1 and CCLP1 cells compared with the
respective shNC groups (P<0.01; Fig.
2B and C).
The biological role of FLVCR1-AS1 on CCA tumor
growth was evaluated in a xenograft mouse model. HuCCT1 cells,
transfected with shNC or shFLVCR1-AS1, were implanted
subcutaneously into mice. Subsequently, tumor growth was measured
every 7 days for a period of 28 days. FLVCR1-AS1-knockdown
significantly delayed tumor growth in vivo (Fig. 2D). At 4 weeks post-implantation, mice
were sacrificed and tumors were harvested and weighed. Silencing of
FLVCR1-AS1 significantly decreased the tumor weight (Fig. 2D).
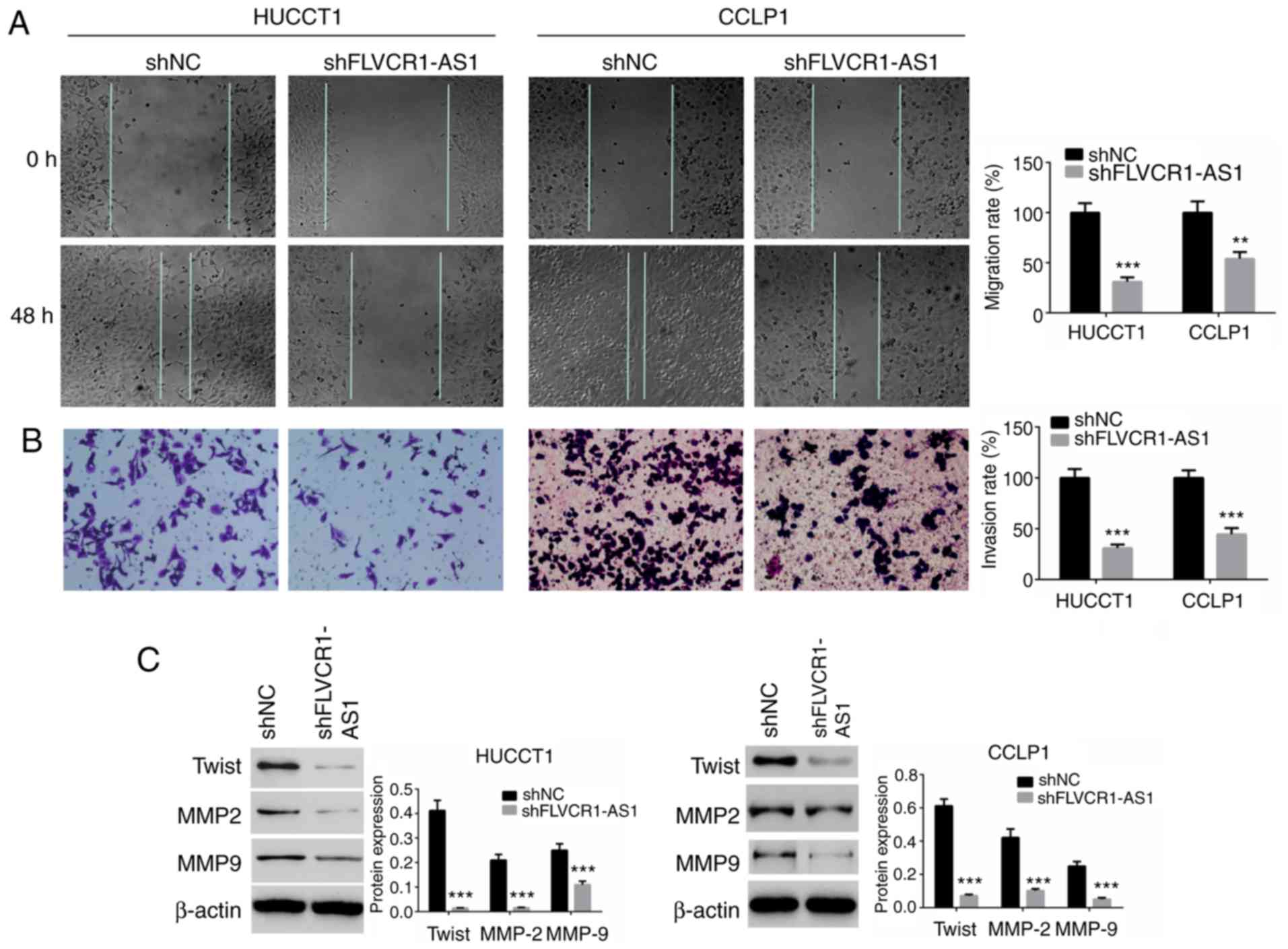
FLVCR1-AS1 suppresses cell migration
and invasion
To identify the effect of FLVCR1-AS1 on cell
migration and invasion, wound-healing and Transwell assays were
performed. As presented in Fig. 3A,
the migration ability of cells transfected with shFLVCR1-AS1 was
significantly suppressed compared with the shNC groups (P<0.01).
The invasion of HuCCT1 and CCLP1 cells transfected with
shFLVCR1-AS1 was also significantly reduced compared with the shNC
groups (P<0.01; Fig. 3B).
Expression levels of migration and invasion-associated proteins,
including Twist, MMP-2 and MMP-9, were determined by western blot
analysis. The expression levels of Twist, MMP-2 and MMP-9 in the
shFLVCR1-AS1 groups were significantly decreased compared with the
shNC groups (P<0.01; Fig.
3C).
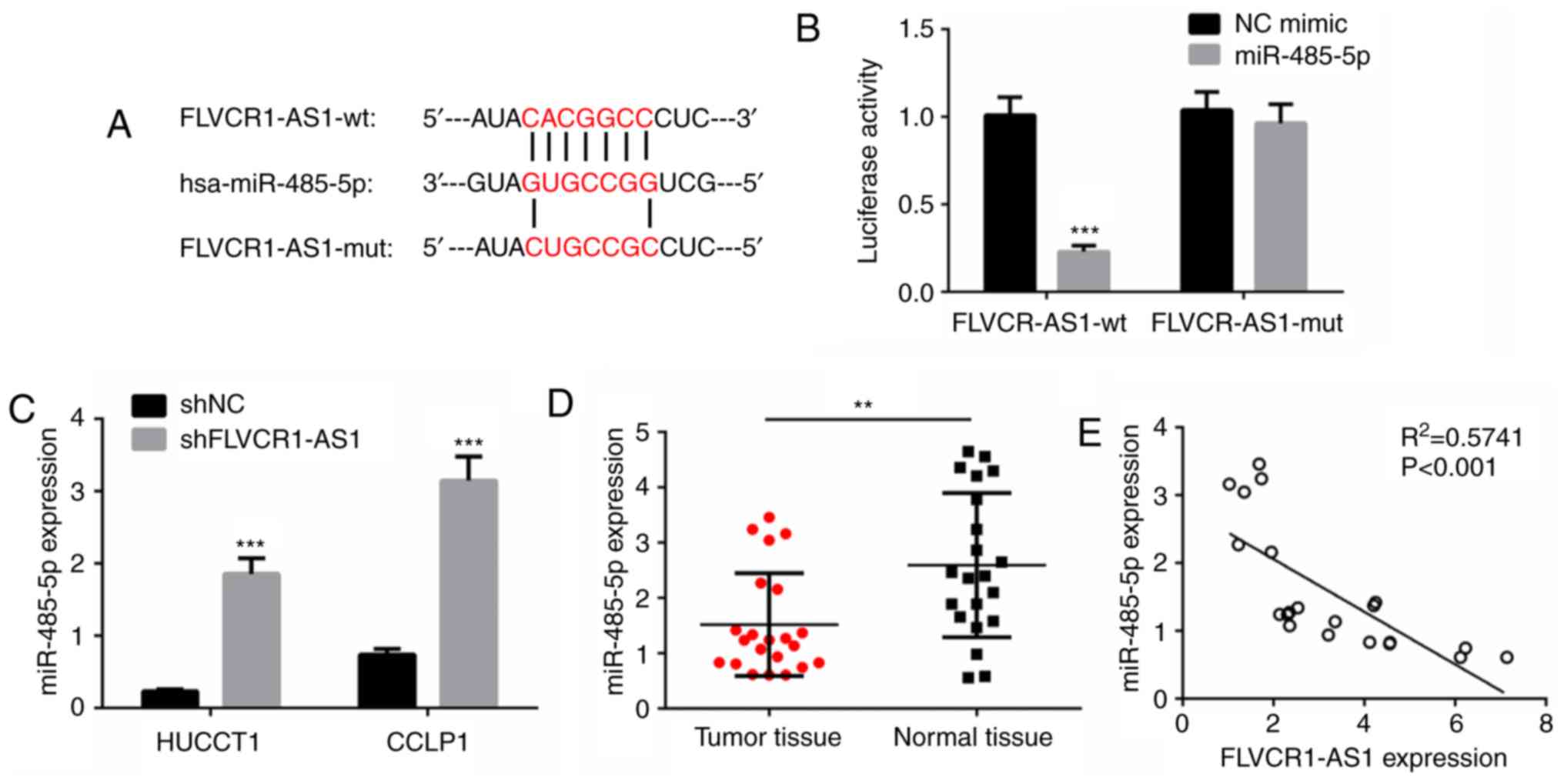
FLVCR1-AS1 interacts with miR-485-5p
in HuCCT1 and CCLP1 cells
To investigate the molecular mechanism by which
FLVCR1-AS1 promotes CCA cell proliferation, migration and invasion,
target miRNAs of FLVCR1-AS1 were predicted with miRanda.
miR-485-5p, which is commonly reported to be involved in many types
of human tumor (21,22), was selected as a candidate target of
FLVCR1-AS1 (Fig. 4A). Furthermore,
luciferase reporter vectors carrying either the predicted
miR-485-5p binding site (wild-type FLVCR1-AS1) or its corresponding
mutant fragment (mutant FLVCR1-AS1) were constructed. As presented
in Fig. 4B, co-transfection of
wild-type FLVCR1-AS1 and miR-485-5p mimic significantly reduced the
luciferase activity, while co-transfection of mutant FLVCR1-AS1 and
miR-485-5p mimic did not affect luciferase activity. The present
study further evaluated the expression of miR-485-5p in HuCCT1 and
CCLP1 cells transfected with shFLVCR1-AS1 or shNC. As presented in
Fig. 4C, the expression of
miR-485-5p was significantly increased in the shFLVCR1-AS1 groups
compared with the shNC groups (P<0.001). Furthermore, the
expression of miR-485-5p in CCA samples and normal tissues was
determined by RT-qPCR. A significant decrease was observed in CCA
tumor samples compared with the normal tissue samples (P<0.001;
Fig. 4D). Pearson's correlation
analysis revealed that FLVCR1-AS1 expression was negatively
correlated with miR-485-5p expression in CCA tissues (P<0.001;
Fig. 4E). These data indicated that
FLVCR1-AS1 may interact with miR-485-5p in CCA cells.
Discussion
A number of studies have reported that lncRNAs serve
a crucial role in the occurrence, development and prognosis of
malignant neoplasms, including CCA (12,23). The
present study determined the expression of FLVCR1-AS1 in CCA
tissues and cell lines, and examined the effect of FLVCR1-AS1 on
CCA cell growth, migration and invasion. In addition, the mechanism
by which FLVCR1-AS1 functions as an oncogene in CCA was
evaluated.
FLVCR1-AS1 is understood to exhibit tumor-promoting
activity in hepatocellular carcinoma (24). Initially, the present study
identified that the expression of FLVCR1-AS1 was significantly
increased in CCA tissues compared with the adjacent normal tissues.
The expression of FLVCR1-AS1 was further upregulated in CCA cell
lines. These results indicate that FLVCR1-AS1 may serve a role in
the development and progression of CCA. CCA cell lines (HuCCT1 and
CCLP1) stably transfected with shFLVCR1-AS1 or shNC were
established, and cell proliferation and colony formation assays
were performed. Silencing of FLVCR1-AS1 significantly inhibited
cell proliferation and colony formation. Furthermore,
FLVCR1-AS1-knockdown significantly inhibited tumor growth in a
xenograft model, which supports the in vitro data.
Therefore, silencing FLVCR1-AS1 was demonstrated to inhibit tumor
growth in vitro and in vivo. Immoderate tumor cell
migration and invasion promote cancer metastasis (25). The effects of FLVCR1-AS1-knockdown on
cell migration and invasion were evaluated by wound-healing and
Transwell assays, respectively. Knockdown of FLVCR1-AS1
significantly weakened the migration and invasion abilities of CCA
cells. Additionally, increased expression levels of Twist, MMP-2
and MMP-9 serve important roles in the development of malignant
tumors (26,27), and the expression levels of these
proteins were identified to be significantly reduced following
silencing of FLVCR1-AS1.
A previous study indicated that FLVCR1-AS1 promotes
cancer cell proliferation, migration and invasion by sponging
miR-513c in hepatocellular carcinoma (24). To elucidate the underlying molecular
mechanisms by which FLVCR1-AS1 serves a tumor-promoting role in
CCA, bioinformatics analysis and a luciferase reporter assay were
performed. The analysis and reporter assay revealed that FLVCR1-AS1
competitively bound to miR-485-5p in CCA cells. miR-485-5p has been
reported to act as a negative regulator in the progression of
gastric and breast cancer (21,22).
Previous studies have indicated that miR-485-5p exerts its
antitumor effect by targeting nucleoside diphosphate linked moiety
X, peroxisome proliferator-activated receptor-g coactivator-1α,
flotillin-1 and survivin (28–30). In
the present study, the expression of miR-485-5p in CCA tissues was
significantly decreased compared with the normal tissues.
Furthermore, FLVCR1-AS1 expression was identified to be inversely
correlated with miR-485-5p expression in the CCA tissues.
In summary, lncRNA FLVCR1-AS1 was revealed to be
significantly upregulated in CCA tissues and cell lines, and was
demonstrated to function as an oncogene in CCA by sponging
miR-485-5p. The present study provided novel insights that may
promote understanding of the molecular mechanism of lncRNA
FLVCR1-AS1 in CCA tumorigenesis and metastasis. Investigating other
downstream targets of lncRNA FLVCR1-AS1 may be the focus of future
studies. lncRNA FLVCR1-AS1 may serve as a potential therapeutic
target and a novel diagnostic marker for CCA.
Acknowledgments
Not applicable.
Funding
This study was supported by the Shanghai Scientific
Research Project Fund (grant no. 16411967000).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
All authors (WB, FC, SN, HL, JY, ZS and BZ) were
involved in the conception and design of this study. SN and HL
performed the literature search and data extraction. SN, HL, FC, JY
and ZS analyzed the data. BZ drafted the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The use of human tissues was approved by the Ethics
Committee of Shanghai Seventh People's Hospital affiliated to
Shanghai University of Traditional Chinese Medicine (Shanghai,
China) and written informed consent was obtained from all patients.
All animal experiments were performed in accordance with
institutional and international animal regulations. The animal
experimental protocol was approved by the Animal Care and Use
Committee of Shanghai Seventh People's Hospital affiliated to
Shanghai University of Traditional Chinese Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang J, Liu Y, Wang B, Lan H, Liu Y, Chen
F, Zhang J and Luo J: Sumoylation in p27kip1 via RanBP2 promotes
cancer cell growth in cholangiocarcinoma cell line QBC939. BMC Mol
Biol. 18:232017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sha M, Jeong S and Xia Q: Antiviral
therapy improves survival in patients with HBV infection and
intrahepatic cholangiocarcinoma undergoing liver resection: Novel
concerns. J Hepatol. 68:1315–1316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jin Y, Gao J, Weng Q, Xiong F, Gu S,
Shivaram G, Zhang F and Yang X: Cholangiocarcinoma: Molecular
imaging-guided radiofrequency hyperthermia-enhanced intratumoral
herpes simplex virus thymidine kinase gene therapy. Am J Cancer
Res. 8:502–513. 2018.PubMed/NCBI
|
|
4
|
Suksawat M, Techasen A, Namwat N, Boonsong
T, Titapun A, Ungarreevittaya P, Yongvanit P and Loilome W:
Inhibition of endothelial nitric oxide synthase in
cholangiocarcinoma cell lines-a new strategy for therapy. FEBS Open
Bio. 8:513–522. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yoshikawa D, Ojima H, Kokubu A, Ochiya T,
Kasai S, Hirohashi S and Shibata T: Vandetanib (ZD6474), an
inhibitor of VEGFR and EGFR signalling, as a novel
molecular-targeted therapy against cholangiocarcinoma. Br J Cancer.
100:1257–1266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chiu HS, Somvanshi S, Patel E, Chen TW,
Singh VP, Zorman B, Patil SL, Pan Y, Chatterjee SS; Cancer Genome
Atlas Research Network, ; et al: Pan-cancer analysis of lncRNA
regulation supports their targeting of cancer genes in each tumor
context. Cell Rep. 23:297–312.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vrba L and Futscher BW: Epigenetic
silencing of lncRNA MORT in 16 TCGA cancer types. F1000Res.
7:2112018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang G, Pian C, Chen Z, Zhang J, Xu M,
Zhang L and Chen Y: Identification of cancer-related miRNA-lncRNA
biomarkers using a basic miRNA-lncRNA network. PLoS One.
13:e1966812018.
|
|
9
|
Lou Y, Jiang H, Cui Z, Wang X, Wang L and
Han Y: Gene microarray analysis of lncRNA and mRNA expression
profiles in patients with high-grade ovarian serous cancer. Int J
Mol Med. 42:91–104. 2018.PubMed/NCBI
|
|
10
|
Sun T, Du SY, Armenia J, Qu F, Fan J, Wang
X, Fei T, Komura K, Liu SX, Lee GM and Kantoff PW: Expression of
lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter
facilitates the development of castration-resistant prostate
cancer. Oncogenesis. 7:302018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lv L, Wei M, Lin P, Chen Z, Gong P, Quan Z
and Tang Z: Integrated mRNA and lncRNA expression profiling for
exploring metastatic biomarkers of human intrahepatic
cholangiocarcinoma. Am J Cancer Res. 7:688–699. 2017.PubMed/NCBI
|
|
13
|
Guo L, Zhou Y, Chen Y, Sun H, Wang Y and
Qu Y: LncRNA ASAP1-IT1 positively modulates the development of
cholangiocarcinoma via hedgehog signaling pathway. Biomed
Pharmacother. 103:167–173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu Y, Yao Y, Jiang X, Zhong X, Wang Z, Li
C, Kang P, Leng K, Ji D, Li Z, et al: SP1-induced upregulation of
lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding
EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J
Exp Clin Cancer Res. 37:812018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi X, Zhang H, Wang M, Xu X, Zhao Y, He
R, Zhang M, Zhou M, Li X, Peng F, et al: LncRNA AFAP1-AS1 promotes
growth and metastasis of cholangiocarcinoma cells. Oncotarget.
8:58394–58404. 2017.PubMed/NCBI
|
|
16
|
Zhang S, Xiao J, Chai Y, Du YY, Liu Z,
Huang K, Zhou X and Zhou W: LncRNA-CCAT1 promotes migration,
invasion, and EMT in intrahepatic cholangiocarcinoma through
suppressing miR-152. Dig Dis Sci. 62:3050–3058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang C, Li JY, Tian FZ, Zhao G, Hu H, Ma
YF and Yang YL: LncRNA NEAT1 promotes growth and metastasis of
cholangiocarcinoma cells. Oncol Res. 26:879–888. 2018. View Article : Google Scholar
|
|
18
|
Jiang XM, Li ZL, Li JL, Zheng WY, Li XH,
Cui YF and Sun DJ: LncRNA CCAT1 as the unfavorable prognostic
biomarker for cholangiocarcinoma. Eur Rev Med Pharmacol Sci.
21:1242–1247. 2017.PubMed/NCBI
|
|
19
|
Igami T, Ebata T, Yokoyama Y, Sugawara G,
Takahashi Y and Nagino M: Staging of peripheral-type intrahepatic
cholangiocarcinoma: Appraisal of the new TNM classification and its
modifications. World J Surg. 35:2501–2509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y
and Li Z: MiR-485-3p and miR-485-5p suppress breast cancer cell
metastasis by inhibiting PGC-1α expression. Cell Death Dis.
7:e21592016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jing LL and Mo XM: Reduced miR-485-5p
expression predicts poor prognosis in patients with gastric cancer.
Eur Rev Med Pharmacol Sci. 20:1516–1520. 2016.PubMed/NCBI
|
|
23
|
Zheng B, Jeong S, Zhu Y, Chen L and Xia Q:
miRNA and lncRNA as biomarkers in cholangiocarcinoma(CCA).
Oncotarget. 8:100819–100830. 2017.PubMed/NCBI
|
|
24
|
Zhang K, Zhao Z, Yu J, Chen W, Xu Q and
Chen L: LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate
cancer cell proliferation, migration, and invasion in
hepatocellular carcinoma. J Cell Biochem. 119:6045–6056. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu B, Yan S, Jia Y, Ma J, Wu S, Xu Y,
Shang M and Mao A: TLR2 promotes human intrahepatic
cholangiocarcinoma cell migration and invasion by modulating NF-κB
pathway-mediated inflammatory responses. FEBS J. 283:3839–3850.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ince AT, Yildiz K, Gangarapu V, Kayar Y,
Baysal B, Karatepe O, Kemik AS and Şentürk H: Serum and biliary
MMP-9 and TIMP-1 concentrations in the diagnosis of
cholangiocarcinoma. Int J Clin Exp Med. 8:2734–2740.
2015.PubMed/NCBI
|
|
27
|
Wang Q, Tang H, Yin S and Dong C:
Downregulation of microRNA-138 enhances the proliferation,
migration and invasion of cholangiocarcinoma cells through the
upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol Rep. 29:2046–2052.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang M, Cai WR, Meng R, Chi JR, Li YR,
Chen AX, Yu Y and Cao XC: miR-485-5p suppresses breast cancer
progression and chemosensitivity by targeting survivin. Biochem
Biophys Res Commun. 501:48–54. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duan J, Zhang H, Li S, Wang X, Yang H,
Jiao S and Ba Y: The role of miR-485-5p/NUDT1 axis in gastric
cancer. Cancer Cell Int. 17:922017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kang M, Ren MP, Zhao L, Li CP and Deng MM:
miR-485-5p acts as a negative regulator in gastric cancer
progression by targeting flotillin-1. Am J Transl Res. 7:2212–2222.
2015.PubMed/NCBI
|