Introduction
Cancer is one of the leading causes of mortality
worldwide (1). Although progress in
cancer treatment has been achieved in previous decades, the
prognosis for the majority of cancer types remains poor. Early
diagnosis and treatment are important in improving the prognosis of
patients with cancer. However, the sensitivity and specificity of
the cancer markers used currently are not satisfactory (2). Therefore, novel molecular markers for
predicting advanced cancer status and prognosis are required.
Although 70–80% of the human genome is transcribed
into RNA, protein-coding sequences account for a small fraction of
the total transcripts, indicating that the number of non-coding
RNAs is increased compared with that of protein-coding genes
(3–6). Specifically, long noncoding RNAs
(lncRNAs) constitute a class of non-coding RNAs measuring >200
nucleotides in length, with no protein-coding capacity. These
lncRNAs serve key roles in chromatin regulation, gene expression,
growth, differentiation and development (7). Although the existence of lncRNAs has
been known for some time, the term ‘lncRNA’ was not adopted until
more recently. There are a large number of lncRNAs included in
numerous databases, including LNCipedia (http://www.lncipedia.org) and the NONCODE database
(http://www.noncode.org). As this number continues
to increase, there are more opportunities to investigate the
functions of these noncoding elements, particularly in association
with cancer treatment and prognosis (8).
Accumulating evidence has suggested an oncogenic or
tumor suppressive role for lncRNAs during tumorigenesis, in which
numerous lncRNAs have been revealed to be dysregulated (9,10).
lncRNA-X-inactive specific transcript (XIST) was the first
functional lncRNA identified to be responsible for X-chromosome
inactivation (11). A growing body
of data has revealed that lncRNA-XIST behaves in an oncogenic
manner in colorectal (12), bladder
(13) and gastric cancer (14), in addition to nasopharyngeal cancer
(15) and osteosarcoma (OS)
(16).
By contrast, XIST has been demonstrated to serve a
tumor suppressor role in other studies (17–19).
However, the majority of the data described so far is limited by
discrete outcomes and small sample sizes. In recent years, several
meta-analyses were conducted to evaluate the prognostic value of
lncRNA-XIST in patients with cancer, and it was identified that the
expression level of XIST was associated with overall survival,
lymph node metastasis, distant metastasis and tumor stage (20–22).
However, following these meta-analyses, a number of studies
concerning XIST and cancer were published, with some describing
contradicting conclusions (23,24).
Therefore, we propose that an updated meta-analysis is required. In
order to assess the value of lncRNA-XIST in predicting the
progression of clinicopathological features in patients with
cancer, a meta-analysis was conducted and a case series of 45
patients with OS was described.
Patients and methods
Search strategy
To identify the incidence of lncRNA-XIST expression
in cancer, PubMed (https://www.ncbi.nlm.nih.gov/pubmed), Web of Science
(www.webofknowledge.com/), Embase
(https://www.embase.com/) and the Cochrane Library
(https://www.cochranelibrary.com/)
databases were searched for articles published prior to January
2019, using the search terms ‘long non-coding RNA’ OR ‘lncRNA,’ AND
‘cancer’ OR ‘sarcoma’ OR ‘carcinoma’ OR ‘neoplasm’ OR ‘malignancy’,
AND ‘XIST’. Additionally, reference lists of associated reviews
were searched to identify any potentially relevant studies. The
inclusion criteria were as follows: i) The publication explored the
relevance of lncRNA-XIST expression in human tumor tissues; ii)
high and low lncRNA-XIST expression groups were defined, or the
relevant data to categorize patients into these groups was present;
and iii) the publication language was confined to English. The
following articles were excluded from the study i) Reviews,
editorials, meetings, abstracts and commentaries; ii) publications
with no target data or relevant outcomes; and iii) duplicate
studies.
The following basic information was extracted from
each study using a standardized data collection method: i) First
authors; ii) publication year; iii) study population; iv) sample
size; v) tumor type; and vi) lncRNA-XIST detection method. If any
essential information was not available from the original article,
best efforts were made to contact the corresponding author to
obtain the missing data. As summarized in Table I, all of the included publications
were evaluated based on the critical checklist of the Dutch
Cochrane Centre, as proposed by Meta-analysis of Observational
Studies in Epidemiology (25).
 | Table I.Characteristics of the studies
included. |
Table I.
Characteristics of the studies
included.
| First author | Year | Country | Sample size | Ethnicity | Cancer type | Detection
method | Quality assessment
score | (Refs.) |
|---|
| Tantai | 2015 | China | 32 | Asian | NSCLC | qPCR | 6 | (32) |
| Kobayashi | 2016 | Japan | 49 | Asian | Cervical squamous
cell carcinoma | qPCR | 5 | (19) |
| Fang | 2016 | China | 53 | Asian | NSCLC | qPCR | 7 | (33) |
| Chen | 2016 | China | 106 | Asian | Gastric cancer | qPCR | 5 | (34) |
| Li | 2017 | China | 145 | Asian | Osteosarcoma | qPCR | 6 | (35) |
| Chen | 2017 | China | 115 | Asian | Colorectal
cancer | qPCR | 8 | (36) |
| Wei | 2017 | China | 64 | Asian | Pancreatic
cancer | qPCR | 7 | (37) |
| Du | 2017 | China | 69 | Asian | Glioma | qPCR | 7 | (38) |
| Wang | 2017 | China | 30 | Asian | Glioma | qPCR | 7 | (39) |
| Song | 2017 | China | 50 | Asian |
Colorectal cancer | qPCR | 6 | (12) |
| Ma | 2017 | China | 98 | Asian | Gastric cancer | qPCR | 8 | (14) |
| Mo | 2017 | China | 88 | Asian | Hepatocellular
carcinoma | qPCR | 7 | (40) |
| Sun | 2017 | China | 47 | Asian | Cervical
cancer | qPCR | 6 | (41) |
| Xiong | 2017 | China | 67 | Asian | Bladder cancer | qPCR | 6 | (42) |
| Sun | 2017 | China | 50 | Asian | NSCLC | qPCR | 6 | (43) |
| Du | 2017 | China | 62 | Asian | Prostate
cancer | qPCR | 7 | (18) |
| Hu | 2017 | China | 52 | Asian | Bladder cancer | qPCR | 7 | (13) |
| Wu | 2017 | China | 127 | Asian |
Esophageal squamous cell
carcinoma | qPCR | 6 | (44) |
| Kong | 2018 | China | 52 | Asian | Hepatocellular
carcinoma | qPCR | 7 | (45) |
| Yi | 2018 | China | 140 | Asian | Esophageal squamous
cell carcinoma | qPCR | 7 | (46) |
| Liang | 2017 | China | 73 | Asian | Pancreatic
carcinoma | qPCR | 7 | (47) |
| Sun | 2018 | China | 120 | Asian | Colon cancer | qPCR | 7 | (48) |
| Liu | 2018 | China | 77 | Asian | Thyroid cancer | qPCR | 7 | (49) |
| Hu | 2018 | China | 30 | Asian | Retinoblastoma | qPCR | 6 | (50) |
| Zhu | 2018 | China | 52 | Asian | Cervical
cancer | qPCR | 7 | (51) |
Evaluation of clinical samples
A total of 45 (25 females and 20 males) paired OS
tissues and adjacent healthy tissues from patients aged from 9–14
years (from January 2009 to September 2017), with no preoperative
history of radiotherapy and/or chemotherapy were obtained from the
Children's Hospital of Chongqing Medical University (Chongqing,
China). The tumor and adjacent healthy tissues were obtained during
biopsy/resection prior to chemotherapy. Staging was performed based
on the Musculoskeletal Tumor Society staging system (26). Following resection, the tissues were
immediately frozen in liquid nitrogen. The present study was
performed with the approval of the Institutional Review Board of
Children's Hospital of Chongqing Medical University. Written
informed consent was obtained from the parents of all patients.
Total RNA was isolated with TRIzol®
reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and subjected to reverse transcription (RT)
reactions using hexamers, dNTPs and M-MuLV Reverse Transcriptase
(supplied with 10X Reaction Buffer) (New England BioLabs, Inc.).
The thermocycling conditions of the RT polymerase chain reaction
(RT-PCR) were as follows: 37°C for 60 min, then at 95°C for 1 min,
followed by holding at 4°C. The resultant cDNA products were
diluted 10- to 100-fold and used as templates. RT-quantitative PCR
(RT-qPCR) analysis was performed using the optimized touchdown qPCR
protocol described previously by Zhang et al (27). Briefly, the SYBR Green qPCR reactions
(Bio-Rad Laboratories, Inc.) were performed in triplicate,
according to manufacturer's protocol, under the following
thermocycler conditions: 95°C for 3 min, followed by 95°C for 20
sec and 66°C for 10 sec for 4 cycles (decreasing by 3°C per cycle);
then 95°C for 20 sec, 55°C for 10 sec and 70°C for 1 sec, for 40
cycles. The 2−ΔΔCq method was used to determine the
relative quantitation of lncRNA-XIST expression levels (28). The primer sequences used in were as
followed: GAPDH forward, 5′-GTCAAGGCTGAGAACGGGAA-3′; GAPDH reverse,
5′-AAATGAGCCCCAGCCTTCTC-3′; lncRNA-XIST forward,
5′-GGTGGACATGTGCGGTCA-3′; and lncRNA-XIST reverse,
5′-CCTGCGGCAAAACCCAAC−3′.
Statistical analysis
All statistical analyses were performed using Stata
12.0 (StataCorp LP) and Revman5.2 software (Cochrane). The combined
odds ratio (OR) and 95% confidence interval (CI) were used to
determine the association between lncRNA-XIST expression level and
clinical risk. The combined effect size was statistically
significant when it did not overlap with 1. Heterogeneity across
the studies was quantified using the I2 statistic. A
fix-effects model with the inverse variance method was conducted
when the calculated I2 <50% (29,30). If
I2 >50%, subgroup analysis was performed. Potential
publication bias was assessed using Egger's bias indicator test
with the linear regression method, respectively, and sensitivity
analysis was also conducted. For analysis of the clinical samples,
the patients were divided into two groups according to the
expression level of lncRNA-XIST (the median expression level 2.4
was used as the cut-off). The χ2 test was used to
identify the differences between categorical variables, and the
two-tailed Student's t-test was used for comparisons between
groups. Overall survival was estimated with Kaplan-Meier method.
P<0.05 was considered to indicate a statistically significant
difference.
Quality assessment
The quality of each study was assessed using the
Newcastle-Ottawa Scale (31),
consisting of three parts: Selection; outcome; and comparability,
with a score range of 0–9. A score ≥6 was considered to indicate
high quality.
Results
Literature analysis
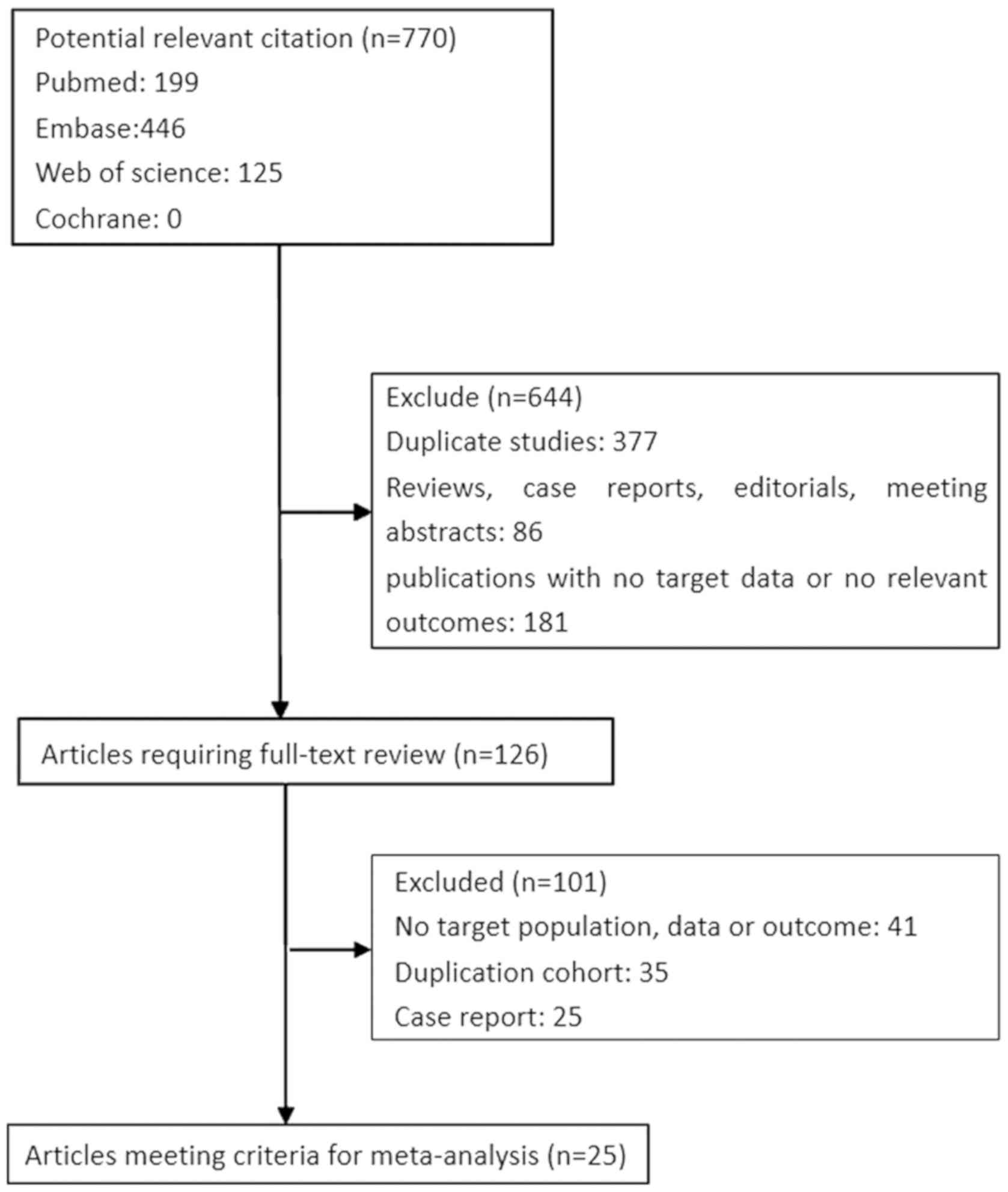
A total of 770 citations were retrieved from an
initial online search for literature associated with lncRNA-XIST
expression in cancer. A total of 644 citations were excluded
following initial screening of titles and abstracts; of the
remaining 125 candidate studies (which were reviewed in their
entirety), 100 were also excluded. A total of 25 studies were
included in the final analysis (Fig.
1).
Study characteristics
The characteristics of the final 25 articles are
presented in Table I (12–14,18,19,32–51).
These studies were published between 2015 and 2018, with sample
sizes of between 31–146 patients. A total of 1,869 patients were
divided into two groups (high and low expression of lncRNA-XIST)
according to RT-qPCR results. The majority of the studies were
conducted in China and the patients presented with the following 12
cancer types: Hepatocellular carcinoma; gastric cancer; pancreatic
cancer; osteosarcoma; cervical cancer; bladder cancer; esophageal
squamous cell carcinoma; glioma; colorectal cancer; non-small-cell
lung cancer (NSCLC); thyroid cancer; retinoblastoma; and cervical
cancer.
lncRNA-XIST expression and patient
outcome
As outlined in Table
II, the results demonstrated that high expression levels of
lncRNA-XIST were associated with lymphatic metastasis (OR =2.32;
95% CI 1.81–3.00; P=0.028; Fig. 2),
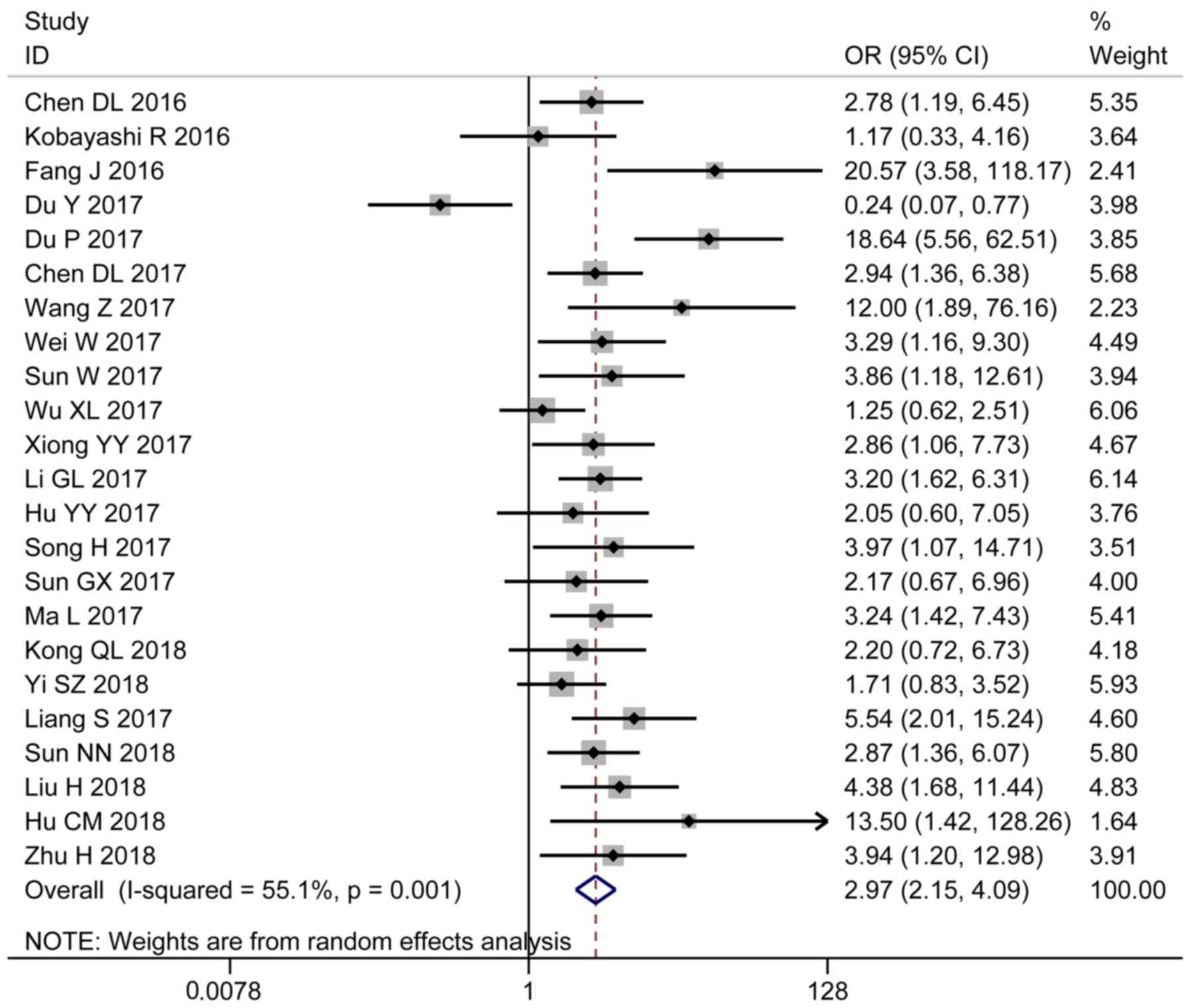
larger tumor size (OR=2.60; 95% CI 1.91–3.56; P=0.001; Fig. 3), advanced cancer stage (OR=2.97; 95%
CI 2.15–4.09; P=0.001; Fig. 4) and
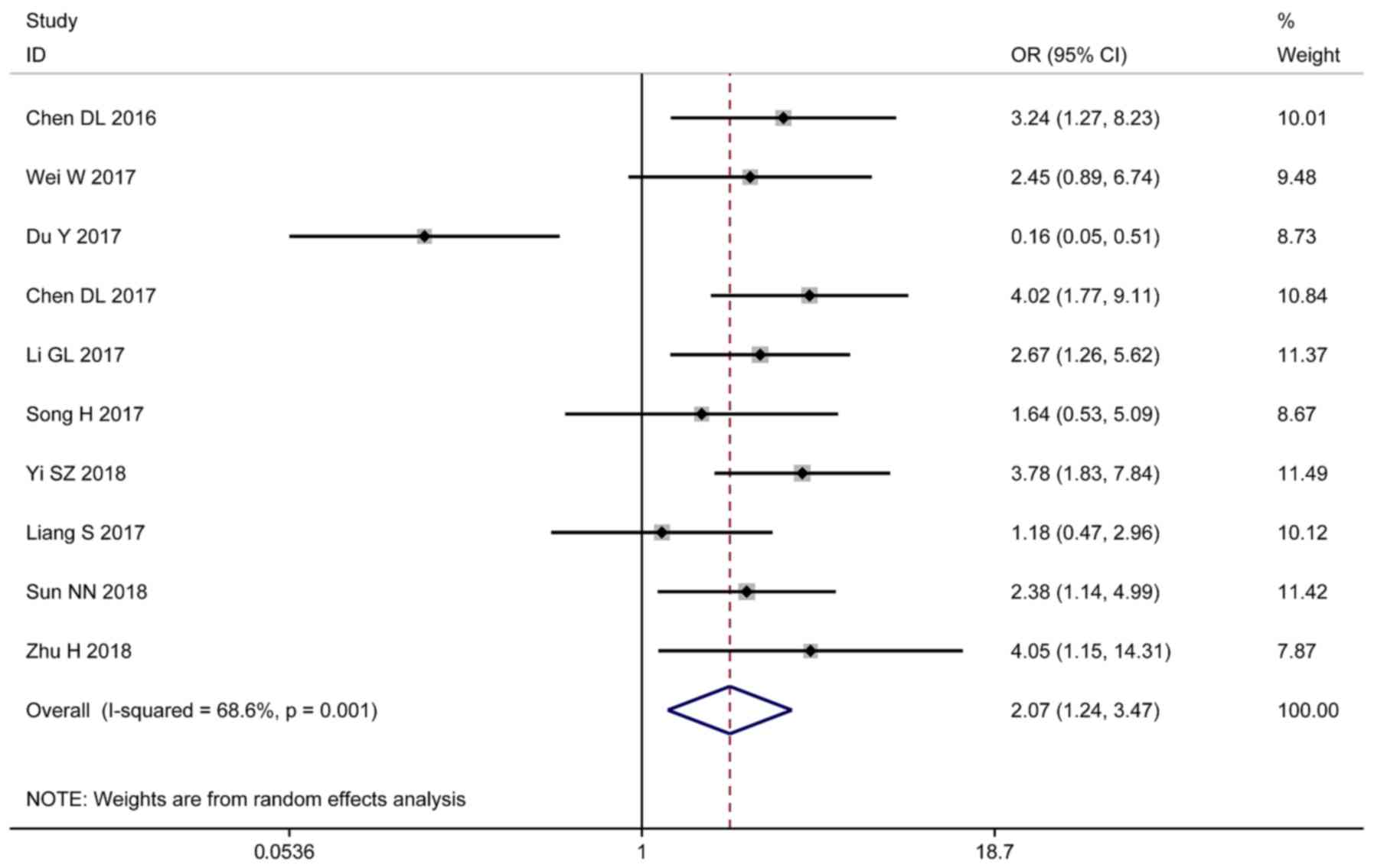
positive distant metastasis (OR=2.07; 95% CI 1.24–3.47; P=0.001;
Fig. 5). However, sex (OR=0.96; 95%
CI 0.79–1.18; P=0.916) was not associated with lncRNA-XIST
expression level (Fig. 6).
 | Table II.Primary outcome of X-inactive
specific transcript expression on disease characteristics. |
Table II.
Primary outcome of X-inactive
specific transcript expression on disease characteristics.
|
|
|
|
|
|
| Egger's test |
|---|
|
|
|
|
|
|
|
|
|---|
| Disease
characteristics | OR (95% CI) | Z-value | P-value |
Phet | I2
(%) | t | P-value |
|---|
| Lymph node
metastasis (yes vs. no) | 2.93 (2.09,
4.10) | 6.27 | <0.001 | 0.37 | 7 | −0.47 | 0.65 |
| Tumour size (bigger
vs. smaller) | 3.07
(2.40,3.92) | 8.98 | <0.001 | 0.16 | 26 | −0.57 | 0.58 |
| Stage (III/IV vs.
I/II) | 2.86
(1.85,4.42) | 4.72 | <0.001 | 0.00 | 64 |
1.89 | 0.08 |
| Distant metastasis
(yes vs. no) | 1.76
(0.76,4.08) | 3.59 | <0.001 | 0.64 | 79 | −1.77 | 0.15 |
| Sex (female vs.
male) | 1.02
(0.79,1.30) | 0.13 |
0.89 | 0.57 | 0 |
0.71 | 0.49 |
Publication bias and sensitivity
analysis
The Egger linear regression test indicated a
potential publication bias in the stage category (Table II). Next, sensitivity analysis was
performed to evaluate the stability of the present study. Each
parameter was excluded from the sensitivity analysis, and the
results of our meta-analysis were consistent, indicating that the
combined results were stable (Figs.
S1-S5). Due to the marked heterogeneity observed, subgroup
analysis was performed for distant metastasis, stage and tumor
size. From the Figs. S6-S9,
subgroup analysis results of cancer types, publication year, sample
size and quality assessment for distant metastasis demonstrated
that the levels of heterogeneity had not decreased, indicating that
cancer types, publication year, sample size and quality assessment
were not the source of heterogeneity. The same results were
identified in tumor size or stage subgroup analyses (Figs. S10-S17).
Quality assessment
The scores of the included studies ranged between
5–8, as determined using the Newcastle-Ottawa Scale. The majority
of the studies scored >7; 15 studies scored 7 and 2 studies
scored 8 (Table I).
Clinical sample confirmation
To determine the role of lncRNA-XIST in patients
with OS, expression levels were determined in the cancerous and
adjacent healthy tissues of patients using RT-qPCR (Fig. 7A). The results revealed that
lncRNA-XIST expression level was significantly upregulated in OS
(Fig. 7A), and that high expression
levels were inversely associated with patient overall survival
(Fig. 7B). The potential association
between lncRNA-XIST expression level and patient
clinicopathological features was also evaluated (Table III). The results revealed that high
lncRNA-XIST expression level correlated with advanced clinical
stage and tumor size.
 | Table III.Correlations between
clinicopathological features and the expression of XIST in
osteosarcoma tissues. |
Table III.
Correlations between
clinicopathological features and the expression of XIST in
osteosarcoma tissues.
|
|
| XIST
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Number of
patients | High | Low | P-value |
|---|
| Sex |
|
|
|
|
|
Male | 22 | 12 | 10 | 0.87 |
|
Female | 23 | 12 | 11 |
|
| Ages, years [Mean
(SD)] |
| 13.2 (2.4) | 12.7 (2.9) | 0.29 |
| Histological
type |
|
|
|
|
|
Osteoblastic | 31 | 17 | 14 | 0.76 |
|
Chondroblastic | 14 | 7 | 7 |
|
| TNM stage |
|
|
|
|
| I,
II | 23 | 8 | 15 | 0.01 |
|
III | 22 | 16 | 6 |
|
| Tumor size, cm |
|
|
|
|
|
<5 | 18 | 6 | 12 | 0.02 |
|
>5 | 27 | 18 | 9 |
|
Discussion
Cancer is one of the leading causes of mortality
worldwide (52). The identification
of novel biomarkers is necessary for early diagnosis, and to
improve the prognosis of patients with cancer. An increasing number
of studies have suggested that lncRNAs are aberrantly expressed in
various types of human cancer; furthermore, an association between
lncRNA expression, pathophysiological features and patient survival
has also been indicated, making lncRNAs promising biomarkers for
cancer prognosis (53). It has also
been demonstrated that lncRNAs may exert their functions via
transcription and epigenetic regulation, as they also serve as
scaffolds in the formation of ribonucleoprotein complexes (54). Various lncRNAs, including
lncRNA-maternally expressed 3 (55),
lncRNA-colon cancer associated transcript 2 (56) and lncRNA-HOX transcript antisense RNA
(57) have been identified as novel
indicators of poor prognosis in a number of types of human
cancer.
Accumulating evidence implies a regulatory role for
lncRNA-XIST, the earliest identified lncRNA, in various malignant
tumors. Wei et al (37)
demonstrated that lncRNA-XIST was involved in the proliferation,
invasion, and epithelial-mesenchymal transition of cancer cells.
Additionally, Wang et al (39) suggested that lncRNA-XIST promoted
glioma cell proliferation by targeting microRNA-137. A study by
Chen et al (34) indicated
that high lncRNA-XIST expression levels were positively correlated
with aggressive tumor phenotypes and prognosis in gastric cancer,
and enhanced the functions of enhancer of zeste homolog 2. In
nasopharyngeal carcinoma, abnormal expression of lncRNA-XIST was
revealed to promote cell proliferation, partially by suppressing
miRNA-34a-5p, and subsequently activating E2F transcription factor
3 (15). Small interfering RNA
inhibition of lncRNA-XIST suppressed the proliferation, migration
and invasion of NSCLC cells in vitro, and suppressed tumor
growth in vivo (33). Based
on these data, lncRNA-XIST may be a potential prognostic marker for
patients with cancer (35–38).
Conversely, lncRNA-XIST may serve as a tumor
suppressor in specific types of cancer. Kobayashi et al
(19) revealed that increased
expression levels of lncRNA-XIST were positively associated with
favorable prognosis in cervical squamous cell carcinoma. In breast
cancer, a decreased expression of lncRNA-XIST upregulated the
phosphorylation of protein kinase B and inhibited tumor growth
(17). Based on these contradictory
results, the true value of lncRNA-XIST as a tumor marker remains to
be determined. It is necessary to comprehensively evaluate the
clinical significance of lncRNA-XIST. Therefore, meta-analyses were
conducted to evaluate the prognostic value of lncRNA-XIST in
patients with cancer. The study conducted by Hu et al
(20) examined 9 studies with 853
patients with cancer, and identified that the expression level of
lncRNA-XIST was markedly associated with overall survival, disease
free survival, tumor type, lymph node metastasis, distant
metastasis and tumor stage. Mao et al (21) identified 15 eligible studies
containing 1,209 patients for inclusion in their meta-analysis;
they observed that increased lncRNA-XIST expression levels in
cancer tissues were associated with a poorer overall survival. In
the study conducted by Liu et al (22), 15 studies with a total of 920
patients were included in the meta-analysis, and the results
suggested that high lncRNA-XIST expression levels were associated
with distant metastasis, tumor stage and poor prognosis. However, a
number of contrasting studies were published concerning the role of
lncRNA-XIST in cancer: In a recent study by Du et al
(18), lncRNA-XIST served as a tumor
suppressor in prostate cancer, its expression correlating with
prognosis and tumor stage. In addition, Sun et al (23) identified that lncRNA-XIST regulated
the microRNA-106b-5p/cyclin-dependent kinase inhibitor 1 axis to
suppress tumor progression in renal cell carcinoma. Consequently,
we proposed that an updated meta-analysis was required. In the
present study, a comprehensive and detailed meta-analysis was
conducted to investigate the association between lncRNA-XIST
expression and the clinicopathological characteristics of patients
with cancer. The analysis of 25 studies, including 1,869 cancer
patients, indicated that high lncRNA-XIST expression level was
significantly associated with lymphatic metastasis, larger tumor
size, advanced stage and positive distant metastasis, suggesting
that increased lncRNA-XIST levels may be associated with advanced
disease presentation. It should also be noted that the prognostic
value of lncRNA-XIST may vary between different types of cancer.
For example, the expression level of lncRNA-XIST was significantly
associated with tumor size in all types of cancer in the included
studies, with the exception of NSCLC and cervical squamous cell
carcinoma.
As a number of the clinicopathological
characteristics of cancer may be associated with the sex of the
patients, and lncRNA-XIST is responsible for X-chromosome
inactivation, it was speculated that there may be a sex-specific
association between the expression of lncRNA-XIST and the features
of different types of cancer. Although the results of the present
study revealed no significant association between lncRNA-XIST
expression and sex, this should be considered in future
studies.
Following a review of the current literature,
potential associations between clinicopathological features and the
expression of lncRNA-XIST in OS tissues were determined. It was
confirmed that lncRNA-XIST expression was upregulated in OS
tissues, and that high expression levels were inversely correlated
with the overall survival of patients with OS. Advanced staging and
increased tumor size were closely associated with increased
lncRNA-XIST expression levels. The results of the present study
were consistent with the conclusions of meta-analysis, additionally
highlighting the importance of lncRNA-XIST in human
malignancies.
In the present study, a number of limitations should
be considered. Only papers written in English were included.
Additionally, the majority of the studies originated from China,
therefore the results are largely representative of the Chinese
population. Furthermore, the cut-off values for high and low
expression levels of lncRNA-XIST differed between the studies.
Also, due to the small number of studies comparing the expression
of lncRNA-XIST with distant metastasis, the significance of certain
data may have been limited by population size. Positive results
were described in the majority of studies, whilst those with
negative results were less likely to be published, which may be
suggestive of a publication bias. Also, the association between
lncRNA-XIST expression and the survival rates of patients was not
determined. However, recent studies revealed that high expression
levels were correlated with decreased overall and disease-free
survival (20,22,58). The
subgroup analysis did not identify a source of heterogeneity,
although significant heterogeneity in distant metastasis, tumor
size and stage was observed. More high-quality original studies are
required to confirm the conclusions of the present study.
Given the aforementioned limitations, the present
study supports the hypothesis that the upregulation of lncRNA-XIST
expression may be considered a credible predictive factor for
advanced clinicopathological features in human cancer. In the
future, large-scale and multicenter studies are required to confirm
these results, and to validate the clinical significance of
lncRNA-XIST expression in human cancer.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr T.-C. He and Ms
Mia Spezia of The University of Chicago Medical Center for their
critical analysis of the manuscript.
Funding
The present study was supported by a research grant
from Chongqing Science and Technology Commission (grant no.
cstc2015jcyjA10046) and the Chengdu Women's and Children's Central
Hospital of Chongqing Medical University Research Projects (Project
No.: 1712).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CY and JT conceived and designed the study, acquired
data, and drafted the manuscript. CD and CY performed the
acquisition of data and analyzed the data. XH and KW acquired
data.
Ethics approval and consent to
participate
The present study was performed with the approval of
the Institutional Review Board of Children's Hospital of Chongqing
Medical University. Written informed consent was obtained from the
parents of all patients.
Patient consent for publication
Written informed consent was obtained from the
parents of all patients.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cui Z, Chen Y, Xiao Z, Hu M, Lin Y, Chen Y
and Zheng Y: Long noncoding RNAs as auxiliary biomarkers for
gastric cancer screening: A pooled analysis of individual studies.
Oncotarget. 7:25791–800. 2016.PubMed/NCBI
|
|
3
|
ENCODE Project Consortium, Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kapranov P, Willingham AT and Gingeras TR:
Genome-wide transcription and the implications for genomic
organization. Nat Rev Genet. 8:413–423. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
ENCODE Project Consortium: An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Blythe AJ, Fox AH and Bond CS: The ins and
outs of lncRNA structure: How, why and what comes next. Biochim
Biophys Acta. 1859:46–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brown CJ, Ballabio A, Rupert JL,
Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF: A gene from
the region of the human X inactivation centre is expressed
exclusively from the inactive X chromosome. Nature. 349:38–44.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song H, He P, Shao T, Li Y, Li J and Zhang
Y: Long non-coding RNA XIST functions as an oncogene in human
colorectal cancer by targeting miR-132-3p. J BUON. 22:696–703.
2017.PubMed/NCBI
|
|
13
|
Hu Y, Deng C, Zhang H, Zhang J, Peng B and
Hu C: Long non-coding RNA XIST promotes cell growth and metastasis
through regulating miR-139-5p mediated Wnt/β-catenin signaling
pathway in bladder cancer. Oncotarget. 8:94554–94568. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma L, Zhou Y, Luo X, Gao H, Deng X and
Jiang Y: Long non-coding RNA XIST promotes cell growth and invasion
through regulating miR-497/MACC1 axis in gastric cancer.
Oncotarget. 8:4125–4135. 2017.PubMed/NCBI
|
|
15
|
Song P, Ye LF, Zhang C, Peng T and Zhou
XH: Long non-coding RNA XIST exerts oncogenic functions in human
nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 592:8–14.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang C, Wu K, Wang S and Wei G: Long
non-coding RNA XIST promotes osteosarcoma progression by targeting
YAP via miR-195-5p. J Cell Biochem. 119:5646–5656. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang YS, Chang CC, Lee SS, Jou YS and
Shih HM: Xist reduction in breast cancer upregulates AKT
phosphorylation via HDAC3-mediated repression of PHLPP1 expression.
Oncotarget. 7:43256–43266. 2016.PubMed/NCBI
|
|
18
|
Du Y, Weng XD, Wang L, Liu XH, Zhu HC, Guo
J, Ning JZ and Xiao CC: LncRNA XIST acts as a tumor suppressor in
prostate cancer through sponging miR-23a to modulate RKIP
expression. Oncotarget. 8:94358–94370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kobayashi R, Miyagawa R, Yamashita H,
Morikawa T, Okuma K, Fukayama M, Ohtomo K and Nakagawa K: Increased
expression of long non-coding RNA XIST predicts favorable prognosis
of cervical squamous cell carcinoma subsequent to definitive
chemoradiation therapy. Oncol Lett. 12:3066–3074. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu S, Chang J, Li Y, Wang W, Guo M, Zou
EC, Wang Y and Yang Y: Long non-coding RNA XIST as a potential
prognostic biomarker in human cancers: A meta-analysis. Oncotarget.
9:13911–13919. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mao H, Wang K, Feng Y, Zhang J, Pan L,
Zhan Y, Sheng H and Luo G. Prognostic role of long non-coding RNA
XIST expression in patients with solid tumors: A meta-analysis.
Cancer Cell Int. 18:342018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu X, Ming X, Jing W, Luo P, Li N, Zhu M,
Yu M, Liang C and Tu J: Long non-coding RNA XIST predicts worse
prognosis in digestive system tumors: A systemic review and
meta-analysis. Biosci Rep. 38:BSR201801692018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun K, Jia Z, Duan R, Yan Z, Jin Z, Yan L,
Li Q and Yang J: Long non-coding RNA XIST regulates miR-106b-5p/P21
axis to suppress tumor progression in renal cell carcinoma. Biochem
Biophys Res Commun. 510:416–420. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang R and Xia T: Long non-coding RNA
XIST regulates PDCD4 expression by interacting with miR-21-5p and
inhibits osteosarcoma cell growth and metastasis. Int J Oncol.
51:1460–1470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stroup DF, Berlin JA, Morton SC, Olkin I,
Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA and Thacker
SB: Meta-analysis of observational studies in epidemiology: A
proposal for reporting. Meta-analysis of Observational Studies in
Epidemiology (MOOSE) group. JAMA. 283:2008–2012. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Enneking WF: A system of staging
musculoskeletal neoplasms. Clin Orthop Relat Res. 9–24.
1986.PubMed/NCBI
|
|
27
|
Zhang Q, Wang J, Deng F, Yan Z, Xia Y,
Wang Z, Ye J, Deng Y, Zhang Z, Qiao M, et al: TqPCR: A touchdown
qPCR assay with significantly improved detection sensitivity and
amplification efficiency of SYBR green qPCR. PLoS One.
10:e01326662015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bowden J, Tierney JF, Copas AJ and Burdett
S: Quantifying, displaying and accounting for heterogeneity in the
meta-analysis of RCTs using standard and generalised Q statistics.
BMC Med Res Methodol. 11:412011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wells GA, Shea B, O'Connell D, Peterson J,
Welch V, Losos M and Tugwell P: The Newcastle–Ottawa Scale (NOS)
for assessing the quality of nonrandomized studies in
meta-analyses. http://www.ohri.ca/programs/clinical_epidemiology/oxford.aspJune
15–2019
|
|
32
|
Tantai J, Hu D, Yang Y and Geng J:
Combined identification of long non-coding RNA XIST and HIF1A-AS1
in serum as an effective screening for non-small cell lung cancer.
Int J Clin Exp Pathol. 8:7887–7895. 2015.PubMed/NCBI
|
|
33
|
Fang J, Sun CC and Gong C: Long noncoding
RNA XIST acts as an oncogene in non-small cell lung cancer by
epigenetically repressing KLF2 expression. Biochem Biophys Res
Commun. 478:811–817. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL,
Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al: Long non-coding
RNA XIST regulates gastric cancer progression by acting as a
molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin
Cancer Res. 35:1422016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li GL, Wu YX, Li YM and Li J: High
expression of long non-coding RNA XIST in osteosarcoma is
associated with cell proliferation and poor prognosis. Eur Rev Med
Pharmacol Sci. 21:2829–2834. 2017.PubMed/NCBI
|
|
36
|
Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng
ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ and Xu RH: Long
noncoding RNA XIST expedites metastasis and modulates
epithelial-mesenchymal transition in colorectal cancer. Cell Death
Dis. 8:e30112017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wei W, Liu Y, Lu Y, Yang B and Tang L:
LncRNA XIST promotes pancreatic cancer proliferation through
miR-133a/EGFR. J Cell Biochem. 118:3349–3358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Du P, Zhao H, Peng R, Liu Q, Yuan J, Peng
G and Liao Y: LncRNA-XIST interacts with miR-29c to modulate the
chemoresistance of glioma cell to TMZ through DNA mismatch repair
pathway. Biosci Rep. 37:BSR201706962017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Z, Yuan J, Li L, Yang Y, Xu X and
Wang Y: Long non-coding RNA XIST exerts oncogenic functions in
human glioma by targeting miR-137. Am J Transl Res. 9:1845–1855.
2017.PubMed/NCBI
|
|
40
|
Mo Y, Lu Y, Wang P, Huang S, He L, Li D,
Li F, Huang J, Lin X, Li X, et al: Long non-coding RNA XIST
promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in
hepatocellular carcinoma. Tumour Biol. 39:10104283176909992017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun G, Wang C and Zhang H: Long non-coding
RNA XIST promotes cervical cancer cell epithelial-mesenchymal
transition through the Wnt/β-catenin pathway. Int J Clin Exp
Pathol. 10:2333–2339. 2017.
|
|
42
|
Xiong Y, Wang L, Li Y, Chen M, He W and Qi
L: The long non-coding RNA XIST interacted with MiR-124 to modulate
bladder cancer growth, invasion and migration by targeting androgen
receptor (AR). Cell Physiol Biochem. 43:405–418. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun W, Zu Y, Fu X and Deng Y: Knockdown of
lncRNA-XIST enhances the chemosensitivity of NSCLC cells via
suppression of autophagy. Oncol Rep. 38:3347–3354. 2017.PubMed/NCBI
|
|
44
|
Wu X, Dinglin X, Wang X, Luo W, Shen Q, Li
Y, Gu L, Zhou Q, Zhu H, Li Y, et al: Long noncoding RNA XIST
promotes malignancies of esophageal squamous cell carcinoma via
regulation of miR-101/EZH2. Oncotarget. 8:76015–76028.
2017.PubMed/NCBI
|
|
45
|
Kong Q, Zhang S, Liang C, Zhang Y, Kong Q,
Chen S, Qin J and Jin Y: LncRNA XIST functions as a molecular
sponge of miR-194-5p to regulate MAPK1 expression in hepatocellular
carcinoma cell. J Cell Biochem. 119:4458–4468. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yi SZ, Zhang W, Zhou QY and Cheng GR:
lncRNA XIST promotes aggressive tumor phenotype and is associated
with poor prognosis in esophageal squamous cell carcinoma. Int J
Clin Exp Med. 11:8317–8323. 2018.
|
|
47
|
Liang S, Gong X, Zhang G, Huang G, Lu Y
and Li Y: The lncRNA XIST interacts with miR-140/miR-124/iASPP axis
to promote pancreatic carcinoma growth. Oncotarget.
8:113701–113718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sun N, Zhang G and Liu Y: Long non-coding
RNA XIST sponges miR-34a to promotes colon cancer progression via
Wnt/β-catenin signaling pathway. Gene. 665:141–148. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu H, Deng H, Zhao Y, Li C and Liang Y:
LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor
growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin
Cancer Res. 37:2792018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hu C, Liu S, Han M, Wang Y and Xu C:
Knockdown of lncRNA XIST inhibits retinoblastoma progression by
modulating the miR-124/STAT3 axis. Biomed Pharmacother.
107:547–554. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhu H, Zheng T, Yu J, Zhou L and Wang L:
LncRNA XIST accelerates cervical cancer progression via
upregulating Fus through competitively binding with miR-200a.
Biomed Pharmacother. 105:789–797. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yarmishyn AA and Kurochkin IV: Long
noncoding RNAs: A potential novel class of cancer biomarkers. Front
Genet. 6:1452015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cui X, Jing X, Long C, Tian J and Zhu J:
Long noncoding RNA MEG3, a potential novel biomarker to predict the
clinical outcome of cancer patients: A meta-analysis. Oncotarget.
8:19049–19056. 2017.PubMed/NCBI
|
|
56
|
Fan YH, Fang H, Ji CX, Xie H, Xiao B and
Zhu XG: Long noncoding RNA CCAT2 can predict metastasis and poor
prognosis: A meta-analysis. Clin Chim Acta. 466:120–126. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang S, Chen S, Yang G, Gu F, Li M, Zhong
B, Hu J, Hoffman A and Chen M: Long noncoding RNA HOTAIR as an
independent prognostic marker in cancer: A meta-analysis. PLoS One.
9:e1055382014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhu J, Kong F, Xing L, Jin Z and Li Z:
Prognostic and clinicopathological value of long noncoding RNA XIST
in cancer. Clin Chim Acta. 479:43–47. 2018. View Article : Google Scholar : PubMed/NCBI
|