Introduction
Nasopharyngeal carcinoma (NPC), which is a malignant
tumor, is the most prevalent head and neck cancer in south China
(1–3). The pathogenesis of NPC is complicated,
including the Epstein-Barr virus (EBV) infection, carcinogen
hazards and individual susceptibility (1,4). In
clinic, NPC exhibits low differentiation and high rates of
metastasis compared with other types of cancers (5–7).
Although chemotherapy and radiotherapy are used, the treatment
efficacy for NPC is still unsatisfactory due to the local
recurrence and distant metastasis (5,8).
Therefore, it is important to understand the molecular mechanisms
of NPC.
The kinesin superfamily proteins (KIFs) are a
conserved class of microtubule-dependent motor proteins that serve
the role of oncogenes in cancer cells (9,10).
Microtubules (MTs) are the important components of cytoskeleton are
vital for mitotic activity of tumor and for invading tissues
(9). Kinesin family member 2A
(KIF2A), which is a member of the kinesin-13 family, is associated
with the process of mitosis and mitotic spindle assembly in cancer
cells (11,12). KIF2A is able to inhibit the process
of microtubule polymerization and is vital for the tumor growth and
invasion (11). Recent studies have
demonstrated that KIF2A is involved in carcinogenesis and prognosis
in different types of human cancer, such as prostate, breast and
ovarian cancer (13–15). However, the role of KIF2A in human
NPC is still unknown.
The aim of the present study was to investigate the
function of KIF2A in the prognosis of nasopharyngeal carcinoma and
nasopharyngeal carcinoma cells. The expression of KIF2A was
examined in 97 human NPC tumor samples; the prognostic value of
KIF2A for patients NPC was evaluated, and the biological functions
of KIF2A in the 5-8F NPC cell line were analyzed.
Materials and methods
Patient specimens
A total of 97 patients (47 male, 50 female; age
range, 25–78 years; median age, 44 years) diagnosed with NPC by
biopsy between January 2017 and June 2018 at the Sixth Affiliated
Hospital of Guangzhou Medical University (Guangdong, China) were
included in the present study. All specimens had confirmed
pathological diagnosis and were classified according to the World
Health Organization (WHO) Tumor-Node-Metastasis staging system
(16,17). The patients with a history of
radiotherapy or chemotherapy were excluded. Sample collection
protocols were approved by the Ethics Committee of the Sixth
Affiliated Hospital of Guangzhou Medical University (approval no.
ERC-2016-33). Written informed consent was obtained from all
patients.
Immunohistochemistry
Immunohistochemical staining assays were performed
on NPC tissue sections using the EnVision Detection system
according to the manufacturer's protocol. Briefly, the tissues were
fixed in 4% paraformaldehyde for 24 h at 4°C and embedded in
paraffin, and then six consecutive sections (4 µm) were obtained
from each tissue. The sections were incubated with a KIF2A primary
antibody (1:500; catalog no. PA1-640; Thermo Fisher Scientific,
Inc.) overnight at 4°C. The slides were treated with horseradish
peroxidase-conjugated secondary antibodies (1:1,000; catalog no.
GP016029; Gene Tech Co, Ltd.). The negative controls included the
sections incubated without primary antibodies. The sections were
evaluated and scored by three independent pathologists according
the percentage of positive-stained cells (0, 0%; 1, 1–10%; 2,
11–60%; 3, 61–75%; and 4, 76–100%). The average staining intensity
was calculated, and an average score <3 was considered as weak
KIF2A protein expression, and an average score ≥3 was considered
strong.
Cell culture and RNA interference
The human NPC cell line 5-8F was purchased from the
Cancer Institute, Southern Medical University (Guangdong, China).
Cells were maintained in RPMI-1640 medium (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
µg/ml streptomycin in a humidified atmosphere with 5%
CO2 at 37°C. For RNA interference, 5-8F cells were
transfected with 50 nM KIF2A small-interfering (si)RNA
(5′-GGCAAAGAGAUUGACCUGG-3′; Invitrogen; Thermo Fisher Scientific,
Inc.) with Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Scrambled siRNA (5′-AUCAGCAUACCUCA-3′) and mock treatment (using
Lipofectamine) were used as controls.
Western blotting
Cells were collected and treated with RIPA buffer
(Beyotime Institute of Biotechnology) on ice and the extracted
protein was quantified by a bicinchoninic acid kit (Beyotime
Institute of Biotechnology). A total of 30 µg protein sample was
separated by 10% SDS-PAGE and subsequently transferred onto a PVDF
membrane (18). Following blocking
with 5% BSA in TBS + 0.1% Tween-20 for 1 h, the membrane was
incubated with antibodies against KIF2A (1:500; catalog no.
PA1-640; Thermo Fisher Scientific, Inc.) or β-actin (1:1,000;
catalog no. 3700; Cell Signaling Technology, Inc.) overnight at
4°C. The membrane was subsequently incubated with horseradish
peroxidase-conjugated goat anti-mouse secondary antibodies
(1:1,000; catalog no. GP016029; Gene Tech Co., Ltd.) for 1 h. The
results were analyzed using the fluorescence imaging system
(Bio-Rad Laboratories, Inc.). The density of the protein bands was
analyzed using ImageJ software (version 1.8.0; National Institutes
of Health).
Reverse transcription-quantitative
PCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Following quantitation, 5 µg total RNA was
reverse-transcribed into cDNA using a PrimeScript™ RT reagent kit
(Takara Bio, Inc.) according to manufacturer's protocol. The mRNA
expression levels were detected with the 2−∆∆Cq method
using SYBR Green kit (Takara Bio, Inc.) with specific primers
(19). The thermocycling conditions
were as follows: 45 cycles of 95°C for 5 min, 95°C for 30 sec, 57°C
for 30 sec and 72°C for 1 min. The gene-specific primers were
synthesized by Sangon Biotech Co., Ltd., and the sequences are
presented in Table I.
 | Table I.Reverse transcription-quantitative PCR
primer sequences. |
Table I.
Reverse transcription-quantitative PCR
primer sequences.
| Gene | Sequence (5′→3′) | Size (bp) |
|---|
| KIF2A | F:
AAGGCAAAGAGATTGACCTGG | 151 |
|
| R:
GAAGCTACAGTCCGTCGATTC |
|
| Bcl-2 | F:
GGTGGGGTCATGTGTGTGG | 89 |
|
| R:
CGGTTCAGGTACTCAGTCATCC |
|
| Bax | F:
CCCGAGAGGTCTTTTTCCGAG | 155 |
|
| R:
CCAGCCCATGATGGTTCTGAT |
|
| VEGF | F:
AGGGCAGAATCATCACGAAGT | 75 |
|
| R:
AGGGTCTCGATTGGATGGCA |
|
| GAPDH | F:
CTGGGCTACACTGAGCACC | 101 |
|
| R:
AAGTGGTCGTTGAGGGCAATG |
|
Cell proliferation assay
Cells were seeded at the density of 1×103
cells/well in a 96-well plate and were cultured for 24, 48 and 72
h. Cell viability was assessed by MTT assay. MTT reagent (5 mg/ml;
Amresco, LLC) was added to each well and incubated in a dark
environment for 3 h at 37°C with 5% CO2. DMSO (100
µl/well) was added into each well. Absorbance at 490 nm was
recorded using a microplate reader (Thermo Fisher Scientific,
Inc.).
Flow cytometry assay
Following cell collection (1×106), the
cell cycle was evaluated by a Cell Cycle Analysis kit (Beyotime) in
the dark for 30 min at 37°C. Apoptosis was evaluated by
phycoerythrin Annexin V Apoptosis Detection kit (BD Biosciences)
following the manufacturer's protocol. After staining, the cells
were analyzed by flow cytometry (BD Biosciences).
Cell invasion assay
The invasive ability of 5-8F cells was determined by
Matrigel assay using Transwell chambers (BD Biosciences). The
filters were pre-coated with 50 µl Matrigel and incubated in a
humidified incubator for polymerization. The lower compartment was
filled with DMEM medium (Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS. Cells with 0.2 ml serum-free DMEM were
seeded in the upper compartment of the chamber (2×105
cells/chamber). Following incubation at 37°C for 24 h, the cells on
the upper surface of the filter were removed using a cotton swab.
Cells that traversed the filter were fixed with 10% methanol for 5
min at 4°C, stained with 0.5% eosin for 30 min at 37°C and counted
using a light microscope.
Statistical analysis
All experiments were performed in triplicate.
Statistical analyses were performed using SPSS version 20.0
software package (IBM Corp.). One-way analysis of variance with
Bonferroni correction was used for multiple comparisons. P<0.05
was considered to indicate a statistically significant
difference.
Results
Association between of KIF2A
expression and clinicopathological parameters in NPC
The protein expression level of KIF2A in NPC tissue
was detected by immunohistochemistry. The results demonstrated that
the expression levels of KIF2A were not associated with the sex and
age of patients. However, KIF2A expression levels were associated
with the size, location and stage of the primary tumor, the nodal
status and the tumor stage. High KIF2A expression was significantly
associated with T3-T4 (P=0.005), N2-N3 (P=0.037) and stage III–IV
(P=0.014) (Table II).
 | Table II.Association between KIF2A expression
and the clinicopathological features in patients. |
Table II.
Association between KIF2A expression
and the clinicopathological features in patients.
|
|
| KIF2A expression
(%) |
|
|---|
|
|
|
|
|
|---|
| Variable | Patients, n | Low, n=57 | High, n=40 | P-value |
|---|
| Sex |
|
|
| 0.315 |
| Male | 47 | 29 (50.9) | 24 (60.0) |
|
|
Female | 50 | 28 (49.1) | 16 (40.0) |
|
| Age, years |
|
|
| 0.827 |
|
<60 | 78 | 47 (82.5) | 30 (75.0) |
|
|
≥60 | 19 | 10 (17.5) | 10 (25.0) |
|
| Primary tumor |
|
|
| 0.005a |
|
T1-T2 | 68 | 48 (84.2) | 15 (37.5) |
|
|
T3-T4 | 29 | 9
(15.8) | 25 (62.5) |
|
| Nodal status |
|
|
| 0.037a |
|
N0-N1 | 75 | 52 (91.2) | 25 (62.5) |
|
|
N2-N3 | 22 | 5 (8.8) | 15 (37.5) |
|
| Stage |
|
|
| 0.014a |
|
I–II | 73 | 50 (87.7) | 15 (37.5) |
|
|
III–IV | 24 | 7
(12.3) | 25 (62.5) |
|
KIF2A silencing decreases 5-8F cell
viability
To identify the functions of KIF2A in nasopharyngeal
carcinoma, siRNA knockdown of KIF2A was performed. Western blotting
and RT-qPCR results confirmed that the mRNA and protein expression
of KIF2A was inhibited in siRNA-transfected cells compared with the
control group and the scrambled siRNA group (P<0.05; Fig. 1A and B). The effects of KIF2A siRNA
on the viability of 5-8F cells were investigated by MTT assay; the
results demonstrated that the viability of KIF2A-knockdown cells
was decreased at 48 and 72 h compared with the control groups
(Fig. 1C).
Effects of KIF2A silencing on the
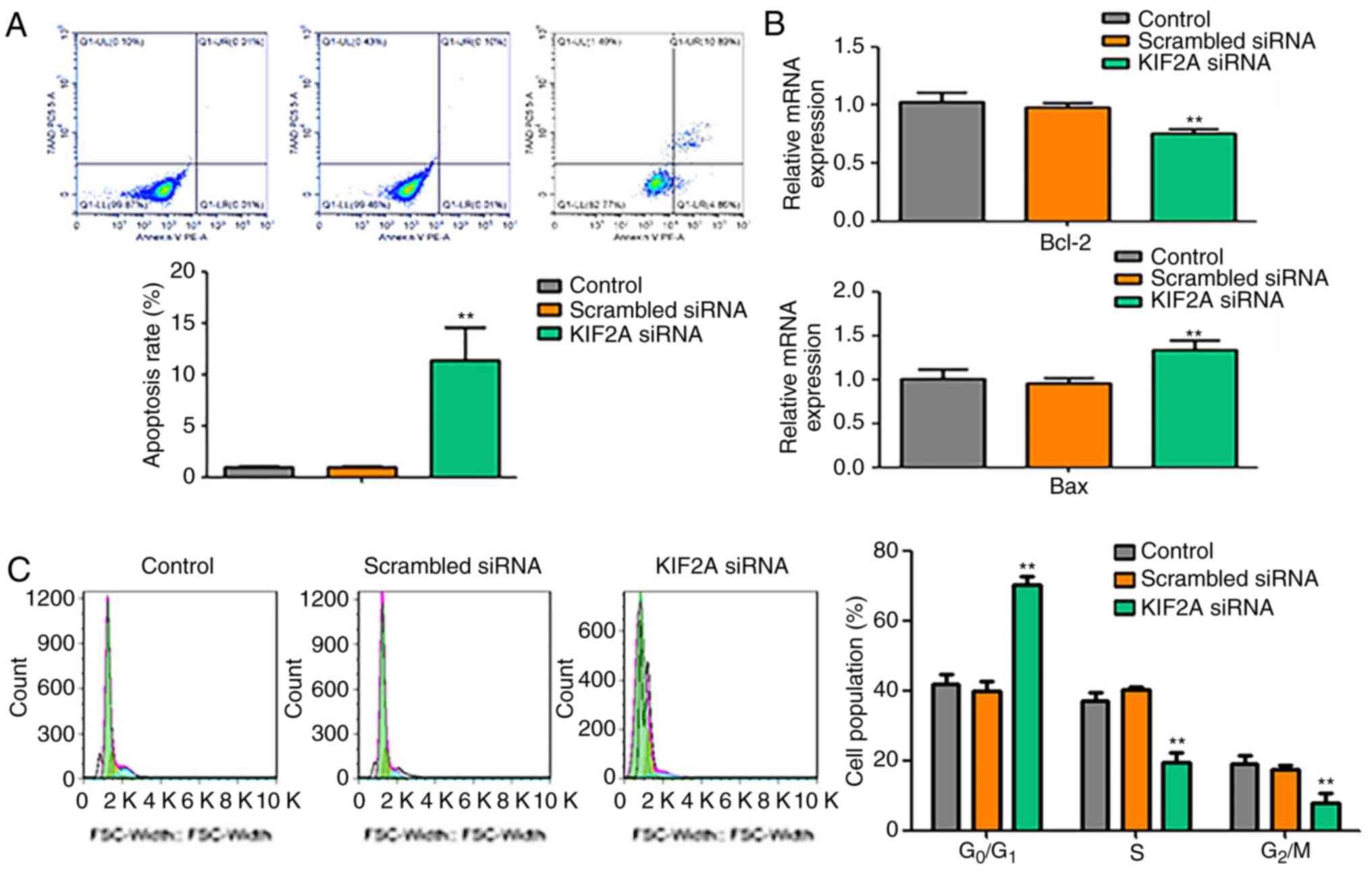
apoptosis and cell cycle of 5-8F cells
To assess whether the decrease of cell proliferation
induced by KIF2A siRNA was associated with apoptosis and cell cycle
arrest, apoptotic rate and cell cycle of 5-8F cells were examined.
The results demonstrated that the number of apoptotic cells in the
KIF2A siRNA group was higher compared with those in the control and
scrambled siRNA groups (Fig. 2A). In
the KIF2A siRNA group, the mRNA expression level of Bcl-2 was
decreased, whereas the expression level of Bax was increased
compared with the control groups (Fig.
2B). The cell cycle assay revealed that, compared with the
control and scrambled siRNA groups, the percentage of cells in the
G0/G1 phase of the cell cycle was significantly increased, whereas
the percentages of cells in the S and G2/M phases were
significantly decreased in the KIF2A siRNA group (Fig. 2C).
KIF2A knockdown inhibits the invasive
ability and vascular endothelial growth factor (VEGF) expression in
5-8F cells
To evaluate the invasive ability of 5-8F cells in
the KIF2A siRNA group, Matrigel cell invasion assay was performed.
The results demonstrated that the invasive capacity of 5-8F cells
was significantly decreased in the KIF2A siRNA group compared with
the control and scrambled siRNA groups (P<0.05; Fig. 3A). In addition, the expression levels
of the angiogenesis marker VEGF of 5-8F were evaluated by RT-qPCR.
Compared with the control and scrambled siRNA groups, VEGF
expression levels were significantly decreased in the KIF2A siRNA
group (P<0.05; Fig. 3B).
Discussion
Nasopharyngeal carcinoma (NPC), a common malignant
tumor, derives from the nasopharyngeal epithelium (20–22). The
long-term survival rate for NPC patients is currently
unsatisfactory. Therefore, it is important to investigate the
molecular mechanisms of NPC tumorigenesis and progression to
improve the therapy of NPC.
KIF2A, which is a type of motor protein with an
oncogenic function, has been identified to be overexpressed in a
number of human cancers (15,23).
KIF2A is a member of the kinesin-13 family of proteins, and serves
the function of depolymerizing the ends of microtubules (MTs)
(24,25). MTs are vital components of the
cytoskeleton and are involved in the invasion and metastasis of
cancer (26,27). Therefore, the aim of the present
study was to evaluate the expression of KIF2A in NPC and explore
the relationship between KIF2A expression and the basic
characteristics of 5-8F cells.
In patients with NPC, the expression levels of KIF2A
were not related with the sex and age of the patients; however, the
levels of KIF2A were notably associated with the primary tumor, the
nodal status and the tumor stage, which suggested that high levels
of KIF2A may be associated with the malignant degree of NPC.
Similarly, in a previous study, the mRNA and protein expression
levels of KIF2A were significantly higher in grade III–IV glioma
tissues compared with grade I–II and healthy tissues (28). Therefore, the results of the present
study indicated that KIF2A may be involved in the malignant
development of NPC.
A previous study demonstrated that the silencing of
KIF2A significantly inhibits the growth and invasion of oral tongue
squamous cell carcinoma, which suggested that KIF2A may regulate
the viability and invasion of cancer cells (29). In the present study, 5-8F cell
proliferation decreased and apoptosis increased in the KIF2A siRNA
group compared with the control groups. The Bax/Bcl-2 ratio is an
effective method to identify apoptotic cells (30,31). In
the KIF2A siRNA group, the mRNA level of anti-apoptotic Bcl-2
decreased, whereas the pro-apoptotic Bax increased, which
demonstrated that the ratio of apoptotic cells increased.
The cell cycle analysis in the present study
demonstrated that the number of cells arrested at the G0/G1 phase
was increased in the KIF2A siRNA group compared with the control
groups. A recent study has revealed that the MRE11-RAD50-NBS1
complex activates KIF2A, which leads to spindle turnover and DNA
damage (11); therefore, KIF2A may
be associated with the DNA structure and spindle dynamics.
In addition, the results of the present study
demonstrated that the invasive ability and the expression of the
angiogenesis marker VEGF in 5-8F cells were inhibited in the KIF2A
siRNA group compared with the control groups. A previous study has
reported that KIF2A is associated with the prognosis of ovarian
cancer, and the inhibition of KIF2A decreases invasion in ovarian
cancer in vitro (15).
Downregulation of KIF2A inhibits gastric cancer cell invasion by
suppressing membrane type I-matrix metallopeptidase (32). Therefore, KIF2A may be associated
with the invasion and angiogenesis of 5-8F cells.
In conclusion, the present study demonstrated that
the expression of KIF2A was correlated with poor
clinicopathological features in NPC. In addition, KIF2A may serve
an important role in the progression of NPC and the proliferation
5-8F cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Qingyuan
Science and Technology Project (grant no. 2016B030).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
CC and QZ designed the study. QZ, TL and CC guided
the experiments. QZ, DL and WL performed the experiments. QZ, SY
and HG collected and processed the clinical data. QZ, HG and TL
analyzed and interpreted the patient data. QZ wrote the paper, and
TL and CC reviewed and edited the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Sample collection protocols were approved by the
Ethics Committee of the Sixth Affiliated Hospital of Guangzhou
Medical University (approval no. ERC-2016-33). Written informed
consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wei KR, Zheng RS, Zhang SW, Liang ZH, Li
ZM and Chen WQ: Nasopharyngeal carcinoma incidence and mortality in
China, 2013. Chin J Cancer. 36:902017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tang XR, Li YQ, Liang SB, Jiang W, Liu F,
Ge WX, Tang LL, Mao YP, He QM, Yang XJ, et al: Development and
validation of a gene expression-based signature to predict distant
metastasis in locoregionally advanced nasopharyngeal carcinoma: A
retrospective, multicentre, cohort study. Lancet Oncol. 19:382–393.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang F, Li J, Xiao H, Zou Y, Liu Y and
Huang W: AFAP1-AS1: A novel oncogenic long non-coding RNA in human
cancers. Cell Prolif. 51:2018. View Article : Google Scholar
|
|
4
|
Tsao SW, Tsang CM and Lo KW: Epstein-Barr
virus infection and nasopharyngeal carcinoma. Philos Trans R Soc
Lond B Biol Sci. 19:3722017.
|
|
5
|
He JY, Han P, Zhang Y, Liu YD, Song SJ,
Feng GK, An Y, Zhou AJ, Wang HB, Yuan L, et al: Overexpression of
Nogo receptor 3 (NgR3) correlates with poor prognosis and
contributes to the migration of epithelial cells of nasopharyngeal
carcinoma patients. J Mol Med (Berl). 96:265–279. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu JF, Huang W, Yi HM, Xiao T, Li JY,
Feng J, Yi H, Lu SS, Li XH, Lu RH, et al: Annexin A1-suppressed
autophagy promotes nasopharyngeal carcinoma cell invasion and
metastasis by PI3K/AKT signaling activation. Cell Death Dis.
9:11542018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
You R, Cao YS, Huang PY, Chen L, Yang Q,
Liu YP, Zou X, Zhang YN, Jiang R, Zhang MX, et al: The changing
therapeutic role of chemo-radiotherapy for loco-regionally advanced
nasopharyngeal carcinoma from two/three-dimensional radiotherapy to
intensity-modulated radiotherapy: A network meta-analysis.
Theranostics. 7:4825–4835. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xie P, Yang JP, Cao Y, Peng LX, Zheng LS,
Sun R, Meng DF, Wang MY, Mei Y, Qiang YY, et al: Promoting
tumorigenesis in nasopharyngeal carcinoma, NEDD8 serves as a
potential theranostic target. Cell Death Dis. 8:e28342017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lucanus AJ and Yip GW: Kinesin
superfamily: Roles in breast cancer, patient prognosis and
therapeutics. Oncogene. 37:833–838. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen J, Li S, Zhou S, Cao S, Lou Y, Shen
H, Yin J and Li G: Kinesin superfamily protein expression and its
association with progression and prognosis in hepatocellular
carcinoma. J Cancer Res Ther. 13:651–659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu R, Xu Y, Huo W, Lv Z, Yuan J, Ning S,
Wang Q, Hou M, Gao G, Ji J, et al: Mitosis-specific MRN complex
promotes a mitotic signaling cascade to regulate spindle dynamics
and chromosome segregation. Proc Natl Acad Sci USA.
115:E10079–E10088. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen MH, Liu Y, Wang YL, Liu R, Xu BH,
Zhang F, Li FP, Xu L, Lin YH, He SW, et al: KIF2A regulates the
spindle assembly and the metaphase I-anaphase I transition in mouse
oocyte. Sci Rep. 6:393372016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tao T, Chen M, Jiang R, Guan H, Huang Y,
Su H, Hu Q, Han X and Xiao J: Involvement of EZH2 in aerobic
glycolysis of prostate cancer through miR-181b/HK2 axis. Oncol Rep.
37:1430–1436. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao P, Lan F, Zhang H, Zeng G and Liu D:
Down-regulation of KIF2A inhibits gastric cancer cell invasion via
suppressing MT1-MMP. Clin Exp Pharmacol Physiol. 45:1010–1018.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sheng N, Xu YZ, Xi QH, Jiang HY, Wang CY,
Zhang Y and Ye Q: Overexpression of KIF2A is suppressed by miR-206
and associated with poor prognosis in ovarian cancer. Cell Physiol
Biochem. 50:810–822. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Palazzi M, Guzzo M, Tomatis S, Cerrotta A,
Potepan P, Quattrone P and Cantù G: Improved outcome of
nasopharyngeal carcinoma treated with conventional radiotherapy.
Int J Radiat Oncol Biol Phys. 60:1451–1458. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
OuYang PY, You KY, Zhang LN, Xiao Y, Zhang
XM and Xie FY: External validity of a prognostic nomogram for
locoregionally advanced nasopharyngeal carcinoma based on the 8th
edition of the AJCC/UICC staging system: A retrospective cohort
study. Cancer Commun (Lond). 38:552018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anufrieva KS, Shender VC, Arapidi GP,
Pavlyukov MS, Shakhparonov MI, Shnaider PV, Butenko IO, Lagarkova
MA and Govorun VM: Therapy-induced stress response is associated
with downregulation of pre-mRNA splicing in cancer cells. Genome
Med. 10:492018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xing H, Chen X and Han Y: Role of
regenerating gene IA expression on local invasion and survival in
nasopharyngeal carcinoma. Biol Res. 50:372017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zuo LL, Zhang J, Liu LZ, Zhou Q, Du SJ,
Xin SY, Ning ZP, Yang J, Yu HB, Yue WX, et al: Cadherin 6 is
activated by Epstein-Barr virus LMP1 to mediate EMT and metastasis
as an interplay node of multiple pathways in nasopharyngeal
carcinoma. Oncogenesis. 6:4022017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xing H, Chen X and Han Y: Role of
regenerating gene IA expression on local invasion and survival in
nasopharyngeal carcinoma. Biol Res. 50:372017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Watanabe T, Kakeno M, Matsui T, Sugiyama
I, Arimura N, Matsuzawa K, Shirahige A, Ishidate F, Nishioka T,
Taya S, et al: TTBK2 with EB1/3 regulates microtubule dynamics in
migrating cells through KIF2A phosphorylation. J Cell Biol.
210:737–751. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ali A, Veeranki SN, Chinchole A and Tyagi
S: MLL/WDR5 complex regulates Kif2A localization to ensure
chromosome congression and proper spindle assembly during mitosis.
Dev Cell. 41:605–622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kwon HJ, Park JE, Song H and Jang CY: DDA3
and Mdp3 modulate Kif2a recruitment onto the mitotic spindle to
control minus-end spindle dynamics. J Cell Sci. 129:2719–2725.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yan C, Wang F, Peng Y, Williams CR,
Jenkins B, Wildonger J, Kim HJ, Perr JB, Vaughan JC, Kern ME, et
al: Microtubule acetylation is required for mechanosensation in
drosophila. Cell Rep. 25:1051–1065.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sandi MJ, Marshall CB, Balan M, Coyaud É,
Zhou M, Monson DM, Ishiyama N, Chandrakumar AA, La Rose J, Couzens
AL, et al: MARK3-mediated phosphorylation of ARHGEF2 couples
microtubules to the actin cytoskeleton to establish cell polarity.
Sci Signal. 10(pii): eaan32862017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang X, Ma C, Wang Q, Liu J, Tian M, Yuan
Y, Li X and Qu X: Role of KIF2A in the progression and metastasis
of human glioma. Mol Med Rep. 13:1781–1787. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang CQ, Li YJ, Wei ZM, Zhu CJ, Qu X, Wei
FC, Xing XM and Yu WJ: Stable gene-silence of Kif2a synergistic
with 5-fluorouracil suppresses oral tongue squamous cell carcinoma
growth in vitro and in vivo. Oral Surg Oral Med Oral Pathol Oral
Radiol. 116:49–54. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang P, Zhang Y, Liu K, Liu B, Xu W, Gao
J, Ding L and Tao L: Ivermectin induces cell cycle arrest and
apoptosis of HeLa cells via mitochondrial pathway. Cell Prolif.
52:e125432019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou S, Wang Y and Zhu JJ: Simultaneous
detection of tumor cell apoptosis regulators Bcl-2 and bax through
a dual-signal-marked electrochemical immunosensor. ACS Appl Mater
Interfaces. 8:7674–7682. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao P, Lan F, Zhang H, Zeng G and Liu D:
Down-regulation of KIF2A inhibits gastric cancer cell invasion via
suppressing MT1-MMP. Clin Exp Pharmacol Physiol. 45:1010–1018.
2018. View Article : Google Scholar : PubMed/NCBI
|