Introduction
Gastric cancer (GC), the second most common
malignant cancer worldwide, gives rise to a number of
cancer-associated fatalities annually (1). Despite improvements in the diagnosis
and treatment of the disease, however, the prognosis and the
five-year survival rate of GC remains poor (2). Most patients with GC are diagnosed at
an advanced stage, at which point GC treatment becomes difficult
due to extensive tumor invasion and lymphatic metastasis (3,4).
Consequently, to provide improved GC interventions, a greater
understanding of the mechanism of GC and accurate identification of
novel therapeutic targets are necessary.
MicroRNAs (miRs/miRNAs) are a class of short
non-coding RNAs of ~22 nucleotides in length, with the ability to
regulate gene expression by directing target mRNAs for degradation
(5,6). Several studies have shown that miRNAs
are involved in regulating several cellular functions, including
cell proliferation, apoptosis, motility and differentiation
(6). In recent years, a great number
of miRNAs have been identified and characterized as tumor
suppressors or oncogenes in GC, and many of these molecules have
been associated with GC metastasis, invasion and drug resistance
(7,8). miR-761 was recently reported to play
important roles in human cancer, including breast cancer,
hepatocellular carcinoma, non-small cell lung cancer, glioma and
colorectal cancer (9–14). miR-761 serves as an oncogene or tumor
suppressor in different types of cancer, depending on the tissue in
which it is expressed. However, the expression, biological function
and molecular mechanism of miR-761 in the progression of GC have
not been well elucidated. Therefore, the determination of the
function and mechanism of miR-761 in GC is essential.
In the present study, the expression level of
miR-761 was decreased in GC tissues and cell lines compared with
that in normal cells. Overexpression of miR-761 decreased, whereas
knockdown of miR-761 promoted, the proliferation and migration of
GC cells. The molecular mechanism study also demonstrated that RIN1
is a direct downstream target of miR-761 in GC cells, and that
overexpression of RIN1 could rescue the inhibitory effect of the
miR-761 mimic on the biological function of GC cells.
Materials and methods
Cell culture
Human GC cell lines MGC-803 (well differentiated
adenocarcinoma), SGC-7901 (moderately differentiated
adenocarcinoma), AGS (poorly differentiated adenocarcinoma) and
BGC-823 (poorly differentiated adenocarcinoma), immortal human
gastric epithelial GES-1 cells and 293T cells were obtained from
the Cell Bank of the Chinese Academy of Sciences (Shanghai, China).
All cells were grown in DMEM (Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific,
Inc.) at 37°C under a humidified atmosphere containing 5%
CO2.
Human tissues
GC tumor tissues and adjacent normal tissues were
obtained for the current study from 20 patients (age range, 49–71
years old; 12 males and 8 females) who underwent surgery for
stomach cancer at the Affiliated Hospital of Weifang Medical
University (Weifang, China) between June 2015 and June 2016. Prior
written informed consent was obtained from each patient. Patients
who underwent chemotherapy or radiotherapy prior to surgery were
excluded from the current study. The study methodologies and the
experiments were approved by the ethics committee of the Affiliated
Hospital of Weifang Medical University (approval no.
WYFY20150109002). Following removal, all tissues were stored at
−80°C immediately, until use.
Cell transfection
The miR-761 mimic
(5′-UCAGCAGGCAGGCUGGUGCAGCGGTATTCGCACTGGATACGACGAACTTT-3′), miRNA
negative control (miR-NC; 5′-ACUACUGAGUGACAGUAGA-3′), miR-761
inhibitor (5′-GCUCCUGGAGCGUGUUAAUCCUGA-3′) and inhibitor negative
control (anti-NC; 5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from
Guangzhou RiboBio Co., Ltd. The open reading frame of RIN1 was
inserted into pcDNA3.1 vector (Invitrogen; Thermo Fisher
Scientific, Inc.) to generate the pcDNA3.1/RIN1 vector. The empty
pcDNA3.1 vector (empty vector) was used as the control. A total of
5×105 SGC-7901 cells were transfected with miR-NC (2.5
µg) or miR-761 mimic (2.5 µg) and/or pcDNA3.1/RIN1 vector (100 nM)
or empty vector (100 nM) using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. A total of 5×105 BGC-823
cells were transfected with anti-NC (2.5 µg) or miR-761 inhibitor
(2.5 µg) using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Reverse transcription-quantitative PCR (RT-qPCR) was
carried out to analyze the transfection efficiency after 48 h.
Western blot analysis was carried out 72 h after transfection.
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay (Beyotime Institute of Biotechnology)
was carried out to detect cell proliferation. GC cells (3,000
cells/well) were seeded into 96-well plates. Then, 10 µl CCK-8
solution was added to the wells. The absorbance of each well at 0,
24, 48 and 72 h was detected at 450 nm.
Colony-formation assay
Cells were transfected with miR-761 mimic, miR-NC,
miR-761 inhibitor or anti-NC, as described above. The transfected
cells were trypsinized, counted, and re-plated in a 6-well plate at
a density of 400 cells/well. After 10 days, colonies resulting from
the surviving cells were fixed with 4% paraformaldehyde for 10 min
at room temperature, stained with 0.1% crystal violet for 5 min at
room temperature and counted. Colonies containing ≥50 cells were
scored. Each assay was performed in triplicate.
Transwell assay
The migration and invasion assay was carried out
using Transwell chambers (Corning Inc.). GC cells
(5×104) were plated on the top chamber in DMEM (Thermo
Fisher Scientific, Inc.) without serum. Medium containing 10% FBS
was added to the bottom chamber. Following incubation at 37°C in an
atmosphere containing 5% CO2 for 24 h, cells on the
lower filter were fixed with 4% paraformaldehyde (Beyotime
Institute of Biotechnology) at room temperature for 20 min, and
stained with 0.5% crystal violet at room temperature for 10 min.
Cells migrating through the membrane were counted using a light
microscope. For detecting cell invasion potential, 50 µl
pre-diluted Matrigel (BD Biosciences) was firstly added into the
chambers and incubated at 37°C for 2 h. The cells were later
processed similar to that of cell migration assay.
Bioinformatics prediction
To investigate the possible target genes of miR-761,
the online prediction system, TargetScan 7.1 software (http://www.targetscan.org), was used.
Dual-luciferase reporter assay
The wild-type (WT) or mutant (MUT) RIN1-3′UTR which
contained the miR-761 binding sites was inserted into the psiCHECK2
vector (Promega Corporation). 293T cells (2×104) were
co-transfected with 0.1 mg psiCHECK2-WT RIN1-3′-UTR or 0.1 mg
psiCHECK2-MUT RIN1-3′-UTR and 10 nM miR-761 mimic or 10 nM miR-761
inhibitor using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Cells were cultured 37°C for 24 h and luciferase
activities were analyzed using the Dual-Luciferase Reporter Assay
system (GeneCopoeia, Inc.) according to the manufacturer's
protocol. The activity of firefly luciferase was normalized to the
corresponding Renilla luciferase activity.
Western blot assay
Total proteins were extracted from cells using
radioimmunoprecipitation lysis buffer (Thermo Fisher Scientific,
Inc.) and the protein content was determined using the
Bicinchoninic Acid Protein assay kit (Beyotime Institute of
Biotechnology). Proteins (30 µg) were separated by SDS-PAGE (10%
gels) and transferred to polyvinylidene fluoride membranes (EMD
Millipore). Membranes were blocked at room temperature with 5%
non-fat milk for 1 h and incubated at 4°C overnight with the
following antibodies: GAPDH (cat. no. ab9485; 1:500; Abcam), RIN1
(cat. no. ab251835; 1:200; Abcam), and subsequently with goat
anti-rabbit secondary antibodies (cat. no. ab150077; 1:1,000;
Abcam) at room temperature for 2 h. The blots were detected by an
enhanced chemiluminescence (ECL) system (Thermo Fisher Scientific,
Inc.), and developed using X-ray film.
RT-qPCR
Total RNA was isolated from GC tumor tissues,
adjacent normal tissues, GC cells and GES-1 cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was
synthesized at 70°C for 5 min using a Maxima First Strand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The gene expression levels of RIN1 and
miR-761 were determined using a SYBR Green qPCR kit (Thermo Fisher
Scientific, Inc.), and calculated using the 2−ΔΔCq
method (15). The PCR thermocycling
conditions were: 94°C for 2 min followed by 30 cycles of 94°C for
30 sec, 61°C for 30 sec, and 72°C for 30 sec. To determine RIN1
mRNA expression levels, GAPDH served as an internal control. To
examine the expression levels of miR-761, U6 acted as the internal
control. The primers used for amplification were as follows:
miR-761 forward, 5′-ACAGCAGGCACAGAC-3′; miR-761 reverse,
5′-GAGCAGGCTGGAGAA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′; U6
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; RIN1 forward,
5′-GCACCTGGCGAGAGAAAAG-3′; RIN1 reverse
5′-TAGATTTCCGCACGAGGAACG-3′; GAPDH forward, 5′-GGTGATGCTGAGTA-3′;
reverse 5′-GGATGCAGGGATGATGTTCT-3′.
Statistical analysis
Each experiment was repeated ≥three times.
Statistical analysis was carried out using SPSS software (version
13.0; SPSS, Inc.). Unless otherwise indicated, the data were
evaluated as the mean ± standard deviation. Pearson's correlation
analysis was used to analyze the correlation between miR-761 and
RIN1 mRNA expression. Data of >two groups were analyzed using
one-way analysis of variance with Tukey's post hoc test. Student's
t-test was used to analyze the results between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-761 expression is downregulated in
GC tissues and cell lines
RT-qPCR was carried out to analyze the expression
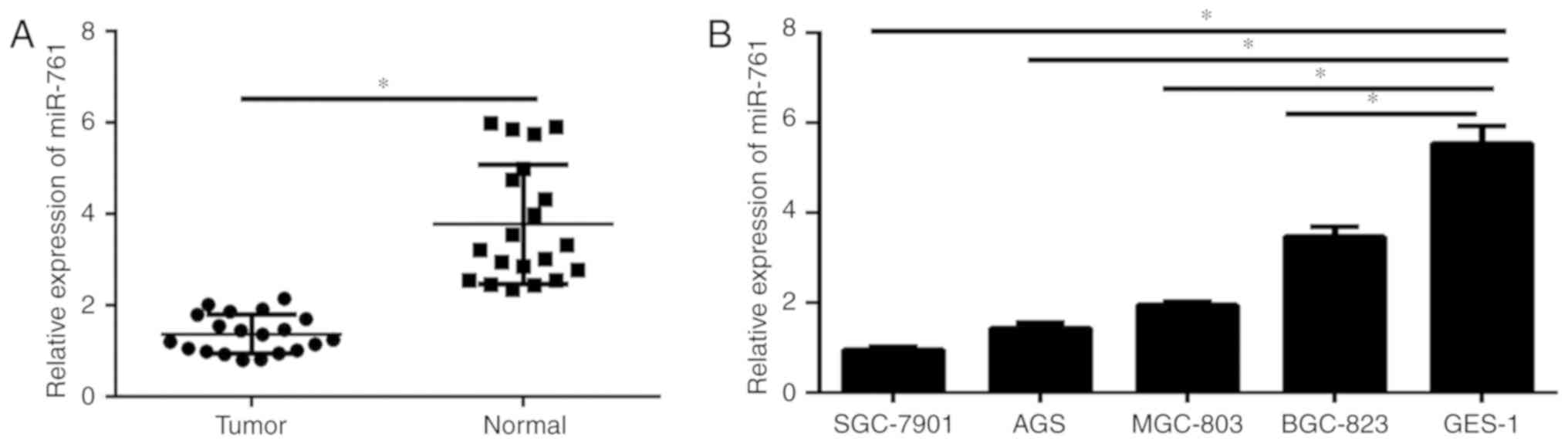
level of miR-761 in GC tissues and cell lines. As demonstrated in
Fig. 1A, the expression of miR-761
was significantly decreased in GC tissues compared with that in the
corresponding normal adjacent tissues. In addition, miR-761
expression in GC cells and normal gastric epithelium GES-1 cells
was analyzed. The results illustrated that miR-761 was
significantly decreased in GC cell lines compared with that in the
GES-1 cells (P<0.05; Fig. 1B).
These data indicate that miR-761 may play a critical role in the
progression of GC.
miR-761 inhibits the proliferation of
GC cells
To study the functional role of miR-761 in the
progression of GC, SGC-7901 and BGC-823 cells were selected for
biological function experiments, based on the miR-761 expression
results in the GC cells analyzed via RT-qPCR (relatively lower in
the SGC-7901 cells and relatively higher in BGC-823). miR-761 mimic
oligonucleotides were transfected into SGC-7901 cells and mir-761
inhibitor into BGC-823 cells, and its expression was detected via
RT-qPCR. The results confirmed the transfection efficiency
(P<0.05; Fig. 2A). The results of
the CCK-8 assay showed that miR-761 overexpression decreased the
proliferation of SGC-7901 cells transfected with miR-761 mimic
compared with that of the cells transfected with miR-NC (P<0.05;
Fig. 2B), and the opposite effect
was observed in BGC-823 cells subjected to miR-761 silencing
(P<0.05; Fig. 2C). A
colony-formation assay was performed to further examine the
proliferation effects of miR-761. Consistent with the CCK-8 assay,
the results of the colony-formation assay showed that miR-761
overexpression significantly decreased the colony-formation ability
of SGC-7901 cells, whereas miR-761 inhibition exhibited the
opposite effects in the BGC-823 cells (P<0.05; Fig. 2D). These data suggest that miR-761
may act as a tumor suppressor in GC.
miR-761 inhibits the migration and
invasion of GC cells
To further illustrate the function of miR-761 in the
progression of GC, a Transwell migration and invasion assay was
carried out. The results showed that the number of migrating and
invading cells was significantly decreased in the SGC-7901 cells
transfected with miR-761 mimic, and the opposite results were
observed when miR-761 was silenced in the BGC-823 cells (P<0.05;
Fig. 3). These data further indicate
that miR-761 may serve as tumor suppressor in GC.
RIN1 is a direct downstream target of
miR-761 in GC cells
The molecular mechanisms underlying the
aforementioned functions were further explored. Among the potential
candidates, detected via bioinformatic analysis (data not shown),
RIN1 was selected as the focus of this study. The TargetScan target
gene prediction analysis identified position 44–51 at the
3′untranslated region (3′-UTR) of RIN1 mRNA as a possible site of
action of miR-761 (Fig. 4A). A
luciferase reporter assay was performed to evaluate whether RIN1 is
a direct target gene of miR-761. The results demonstrated that
miR-761 markedly decreased the luciferase activity in the WT group
compared with the miR-NC (Fig. 4B),
whereas no significant differences were observed among the groups
of cells with the Mut expression (Fig.
4B). Next, the results of the RT-qPCR and western blot assays
showed decreased RIN1 expression in miR-761-overexpression cells,
and enhanced RIN1 expression in miR-761-silenced cells (P<0.05;
Fig. 4C, D). In addition, the mRNA
expression of RIN1 in tumor tissues was significantly elevated
compared with that in normal tissues (P<0.05; Fig. 4E), and a negative correlation between
RIN1 mRNA and miR-761 expression was revealed in GC tissues
(P<0.05; Fig. 4F). These data
suggest that miR-761 directly targets and regulates RIN1 expression
in GC.
RIN1 is a functional downstream target
of miR-761 in GC cells
This study investigated whether RIN1 overexpression
was able to rescue the antitumor activity of miR-761 mimic in GC
cells. SGC-7901 cells were transfected with miR-761 mimic and RIN1
overexpression plasmid. The western blotting results illustrated
that the expression of RIN1 was enhanced in the RIN1 + miR-761
mimic group compared with that in the empty vector + miR-761 mimic
group (Fig. 5A). The results of the
CCK-8 and Transwell assays showed that RIN1 overexpression reversed
the suppressive effects induced by the miR-761 mimic on GC cell
proliferation (P<0.05; Fig. 5B),
migration and invasion (P<0.05; Fig.
5C and D). These data indicate that miR-761 may inhibit GC
progression partially through the downregulation of RIN1.
Discussion
Although metastasis is the predominant cause of
cancer-associated mortality, a comprehensive review of the modular
and cellular determinants governing the related processes has not
been conducted. Accumulating evidence has shown that miRNAs are
aberrant in various types of cancer, and closely associated with
progression and metastasis. Recent studies revealed that miR-761
presents important functions in human cancer types: It functions as
an oncogene in breast cancer, hepatocellular carcinoma and
non-small cell lung cancer (11,12,14), and
serves as a tumor suppressor in glioma, colorectal cancer and
ovarian cancer (9,10,13).
However, the expression, function and underlying molecular
mechanism of miR-761 in GC have not been evaluated.
In the present study, miR-761 was found to be
downregulated in GC tissues and cell lines, consistent with data
from cancer types in which miR-761 acts as a tumor suppressor
(9,10,13).
However, the association between miR-761 and the differentiation of
GC cells was not investigated in this study. The functional assays
demonstrated that miR-761 suppressed cell proliferation, migration
and invasion. These data suggest that miR-761 serves as a tumor
suppressor in GC.
Each miRNA targets certain genes, and the
identification of these targets may help illustrate the
pathogenesis of cancer (7). In the
present study, the potential target of miR-761 was predicted using
bioinformatics analysis, and RIN1 (a Ras effector protein) was
identified as a molecule of interest. Previous studies have shown
that RIN1 promotes tumor progression in non-small cell lung cancer,
bladder urothelial carcinoma, renal cell carcinoma, melanoma,
gastric adenocarcinoma and hepatocellular cancer (16–22).
However, the mechanism of RIN1 in promoting tumor progression had
not been determined. In this study, the relative expression of RIN1
in GC tissues was revealed, and RIN1 was significantly
overexpressed in GC samples compared with normal tissues. It was
also demonstrated that RIN1 is a direct target gene of miR-761,
using a luciferase assay, RT-qPCR and western blotting.
Furthermore, a negative correlation was revealed between RIN1 mRNA
and miR-761 expression in GC tissues. Finally, the restoration of
RIN1 was found to rescue the inhibition of proliferation, migration
and invasion induced by the miR-761 mimic. These data therefore
suggest that miR-761 suppresses GC progression by directly
targeting RIN1. However, the signaling pathway regulated by the
miR-761/RIN1 axis has not been explored.
A previous study reported that RIN1 regulated the
epidermal growth factor receptor (EGFR) signaling pathway and
promoted renal cell carcinoma malignancy in renal carcinogenesis
(15). The association between
miR-761 and the EGFR signaling pathway has not been illustrated,
and this topic may be a novel research direction. In addition, the
present study described the function and mechanism of miR-761 only
in vitro. In future studies, in vivo experiments will
be performed to support the present results. Thirdly, it is worth
mentioning that each miRNA can regulate numerous genes, and
multiple miRNAs may regulate the same gene. Therefore, the
possibility of other target genes that may also be involved in the
suppressive effects of miR-761 cannot be excluded, highlighting a
further novel research direction.
In summary, this study revealed for the first time
that miR-761 was significantly decreased in GC tissues and cell
lines compared with normal tissues. miR-761 may serve as a tumor
suppressor and decrease cell proliferation, migration and invasion
by repressing RIN1 in GC cells. Therefore, miR-761 may be a
potential biomarker for the prognosis of GC and a novel therapeutic
target for treating advanced stages of this disease.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ and XS conceived and designed the experiments.
QZ, YS and XS conducted all of the experiments. YS and XS wrote and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of the Affiliated Hospital of Weifang Medical University
(approval no. WYFY20150109002). Prior written informed consent was
obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Choi IJ, Lee JH, Kim YI, Kim CG, Cho SJ,
Lee JY, Ryu KW, Nam BH, Kook MC and Kim YW: Long-term outcome
comparison of endoscopic resection and surgery in early gastric
cancer meeting the absolute indication for endoscopic resection.
Gastrointest Endosc. 81:333–341 e1. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fukagawa T, Katai H, Mizusawa J, Nakamura
K, Sano T, Terashima M, Ito S, Yoshikawa T, Fukushima N, Kawachi Y,
et al: A prospective multi-institutional validity study to evaluate
the accuracy of clinical diagnosis of pathological stage III
gastric cancer (JCOG1302A). Gastric Cancer. 21:68–73. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smyth EC, Verheij M, Allum W, Cunningham
D, Cervantes A and Arnold D; ESMO Guidelines Committee, : Gastric
cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 27 (Suppl 5):v38–v49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Valinezhad Orang A, Safaralizadeh R and
Kazemzadeh-Bavili M: Mechanisms of miRNA-mediated gene regulation
from common downregulation to mRNA-specific upregulation. Int J
Genomics. 2014:9706072014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan HL, Wang T and Zhang KH: MicroRNAs as
potential biomarkers for diagnosis, therapy and prognosis of
gastric cancer. Onco Targets Ther. 11:3891–3900. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dehghanzadeh R, Jadidi-Niaragh F, Gharibi
T and Yousefi M: MicroRNA-induced drug resistance in gastric
cancer. Biomed Pharmacother. 74:191–199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li GF, Li L, Yao ZQ and Zhuang SJ:
Hsa_circ_ 0007534/miR-761/ZIC5 regulatory loop modulates the
proliferation and migration of glioma cells. Biochem Biophys Res
Commun. 499:765–771. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cao S, Lin L, Xia X and Wu H: MicroRNA-761
promotes the sensitivity of colorectal cancer cells to
5-Fluorouracil through targeting FOXM1. Oncotarget. 9:321–331.
2017.PubMed/NCBI
|
|
11
|
Guo GC, Wang JX, Han ML, Zhang LP and Li
L: microRNA-761 induces aggressive phenotypes in triple-negative
breast cancer cells by repressing TRIM29 expression. Cell Oncol
(Dordr). 40:157–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou X, Zhang L, Zheng B, Yan Y, Zhang Y,
Xie H, Zhou L, Zheng S and Wang W: MicroRNA-761 is upregulated in
hepatocellular carcinoma and regulates tumorigenesis by targeting
Mitofusin-2. Cancer Sci. 107:424–432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi C and Zhang Z: miR-761 inhibits tumor
progression by targeting MSI1 in ovarian carcinoma. Tumour Biol.
37:5437–5443. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan A, Yang C, Chen Z, Li C and Cai L:
MiR-761 promotes progression and metastasis of non-small cell lung
cancer by targeting ING4 and TIMP2. Cell Physiol Biochem. 37:55–66.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feng ZH, Fang Y, Zhao LY, Lu J, Wang YQ,
Chen ZH, Huang Y, Wei JH, Liang YP, Cen JJ, et al: RIN1 promotes
renal cell carcinoma malignancy by activating EGFR signaling
through Rab25. Cancer Sci. 108:1620–1627. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li F, Fang Z, Zhang J, Li C, Liu H, Xia J,
Zhu H, Guo C, Qin Z, Li F, et al: Identification of TRA2B-DNAH5
fusion as a novel oncogenic driver in human lung squamous cell
carcinoma. Cell Res. 26:1149–1164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu HY, Wu G, He H, Wang YW, Cheng Y and
Liu YF: RIN1 expression in hepatocellular cancer and the affection
on prognosis and tumor invasion ability. Zhonghua Wai Ke Za Zhi.
51:1025–1029. 2013.(In Chinese). PubMed/NCBI
|
|
19
|
Fang P, Zhao Z, Tian H and Zhang X: RIN1
exhibits oncogenic property to suppress apoptosis and its aberrant
accumulation associates with poor prognosis in melanoma. Tumour
Biol. 33:1511–1518. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu HF, Zhao G, Ge ZJ, Wang DR, Chen J,
Zhang Y, Zha TZ, Zhang K, Zhang M, Tan YF, et al: High RIN1
expression is associated with poor prognosis in patients with
gastric adenocarcinoma. Tumour Biol. 33:1557–1563. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shan GY, Zhang Z, Chen QG, Yu XY, Liu GB
and Kong CZ: Overexpression of RIN1 associates with tumor grade and
progression in patients of bladder urothelial carcinoma. Tumour
Biol. 33:847–855. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Q, Gao Y, Tang Y, Ma L, Zhao M and
Wang X: Prognostic significance of RIN1 gene expression in human
non-small cell lung cancer. Acta Histochem. 114:463–468. 2012.
View Article : Google Scholar : PubMed/NCBI
|