Introduction
Leucine rich repeat LGI family member 3 (LGI3;
formerly known as leucine-rich glioma inactivated 3) is a secretory
protein member of the LGI family in vertebrates that has been
identified to be highly expressed in the brain (1). LGI3 expression in the brain has been
reported to be regulated at the transcription level by activating
enhancer-binding protein 2 (AP-2) and neuronal restrictive silencer
(1). Our previous studies
demonstrated that LGI3 regulates exocytosis and the differentiation
of neuronal cells (2,3). LGI3 is expressed in the epidermal layer
of the skin where it may act as a cutaneous cytokine (4). Our previous study found that LGI3 is
secreted in response to ultraviolet B irradiation and protects
keratinocytes (4). Additionally, we
previously reported that LGI3 increases migration and
differentiation of keratinocytes (5,6) and
melanocyte pigmentation (7).
Our previous studies revealed that LGI3 is expressed
in adipose tissues and its expression is downregulated during
adipogenesis and upregulated in adipose tissues in obesity
(8,9). Furthermore, one of our previous studies
demonstrated that LGI3 attenuates adipogenesis through a
disintegrin and metalloproteinase domain-containing protein
(ADAM)23, which is one of the receptors for LGI3 (ADAM22 and
ADAM23), and that LGI3 increases pro-inflammatory genes, including
tumor necrosis factor-α (TNF-α) in macrophage cells (9). LGI3 negatively regulates adiponectin
(8). LGI3 and TNF-α increase
expression mutually through NF-κB, suggesting their positive
cooperativity in promoting metabolic inflammation in obesity
(10). We postulated that LGI3 is a
pleiotropic cytokine and adipokine secreted by and acting at
multiple cell types, and that LGI3 may be a pro-inflammatory
cytokine that interacts with various cytokines, adipokines,
chemokines and signaling proteins (11).
Our recent studies proposed that LGI3 may be
involved in the cytokine network in cancer (11–13). Our
recent study reported that LGI3 expression is associated with the
prognosis of glioma (13).
Furthermore, the expression levels and genetic variations of LGI3
may have potential prognostic roles in various types of cancer
(12). The present study utilized
integrative analyses of gene expression microarrays, gene product
networks and patient cohorts, and presented evidence supporting the
potential prognostic role of LGI3 in non-small cell lung cancer
(NSCLC), which is the most common type of lung cancer affecting 85%
of patients (14).
Materials and methods
Gene expression microarray data
The mRNA expression microarray dataset was retrieved
from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). The dataset
GSE19804 is based on the GPL570 [HG-U133_Plus_2] Affymetrix Human
Genome U133 Plus 2.0 Array (Affymetrix; Thermo Fisher Scientific,
Inc.) platform (15). The dataset
contained 120 samples, including 60 samples of NSCLC tissues and 60
samples of paired adjacent normal lung tissues from nonsmoking
patients with NSCLC.
Data processing for identification of
differentially expressed genes (DEGs)
The microarray data were analyzed using the affy
package (version 1.62.0) in R 3.5.1 (http://www.r-project.org) (16). The dataset was subjected to
background correction, quantile normalization and probe
summarization of expression values. The log2 intensities of the
probeset were calculated by the Robust Multichip Average algorithm
of the affy package (16). Gene
expression data were averaged to provide the final expression
values for multiple probes for the same gene symbols. Probesets to
non-expressed mRNAs were excluded using the Affymetrix Microarray
Suite 5 calls algorithm (version 5.0; web.mit.edu/~r/current/arch/i386_linux26/lib/R/library/affy/html/mas5calls.html).
Differential expression analysis was conducted using the limma
package (version 3.40.2; bioconductor.org/packages/release/bioc/html/limma.html)
in R 3.5.1. Genes with P<0.05 and |log2 (fold
change)|≥1 were considered to be statistically significant
DEGs.
Comparative analysis, protein-protein
interaction network, and functional enrichment and gene
coexpression network (GCN) analyses of DEGs
Comparative analysis of categorized gene groups was
presented using a Venn diagram generated by Venny 2.1 (bioinfogp.cnb.csic.es/tools/venny). The
protein-protein interaction network was generated using data from
the Search Tool for the Retrieval of Interacting Genes (version
10.5; string-db.org) (17), and was visualized by Cytoscape 3.7.0
using an interaction degree-sorted circle layout (18). Functional enrichment analysis and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
were performed using the Database for Annotation, Visualization and
Integration Discovery (version 6.8; david.abcc.ncifcrf.gov) (19). Results were sorted by P-values and
the subsets of the entries with P<0.05 were presented. GCN
analysis was performed using the GCN of human lung (uuid:
0c14bd70-5cae-11e7-8f50-0ac135e8bacf) (20) from The Network Data Exchange database
(version 2.3.1; www.ndexbio.org) and visualized by Cytoscape 3.7.0
using the prefuse force directed layout. Gene Ontology (GO) terms
of the GCN were mapped by BiNGO (version 3.0.3; apps.cytoscape.org/apps/bingo) and
visualized by Cytoscape 3.7.0 using a hierarchical layout.
Transcriptional regulatory associations between transcription
factors and the groups of genes were analyzed by Transcription
Factor Affinity Prediction tools (trap.molgen.mpg.de) (21).
Meta-analysis of patient cohorts
The datasets of gene expression microarray analysis
for NSCLC cohorts were retrieved from the Prognoscan database
(http://www.prognoscan.org; Table III) (22). Datasets in the Prognoscan database
were previously processed through quality control tests,
normalization and batch effect adjustment and the exclusion of
low-quality samples. The association between gene expression values
and NSCLC prognosis was assessed by the minimum P-value method for
survival analysis of patient groups that calculates the cut-point
in a continuous gene expression measurement. Patients ranked by
gene expression values were divided at the cut-off point to
minimize the P-value, and the difference of survival between high
and low gene expression groups was calculated using a log-rank
test. The statistically significant (P<0.05) datasets were used
to generate Kaplan-Meier plots.
 | Table III.Dataset summary of expression
microarray analyses for non-small cell lung cancer studies. |
Table III.
Dataset summary of expression
microarray analyses for non-small cell lung cancer studies.
|
Groupa | Dataset | Array type | Number of patients,
n | Cut-off point | P-value |
|---|
| A | GSE31210 | HG-U133_Plus_2 | 204 | 0.72 | 0.0098 |
| B | GSE8894 | HG-U133_Plus_2 | 138 | 0.22 | 0.0341 |
| C | GSE11117 | Novachip human
34.5k | 41 | 0.83 | 0.0247 |
Somatic mutations in NSCLC
Somatic mutations of the LGI3 gene in NSCLC were
identified in Cbioportal (www.cbioportal.org), the Catalogue of Somatic
Mutations in Cancer (cancer.sanger.ac.uk/cosmic) and The Cancer Genome
Atlas (hportal.gdc.cancer.gov). Data of
conserved residues, phylogenetically coevolved residues and single
nucleotide polymorphisms (SNPs) of LGI3 have been previously
described (12). A Venn diagram of
the categorized genetic variations was generated using
InteractiVenn (www.interactivenn.net).
Results
Differential expression of LGI3 in
NSCLC
Our previous study reported that somatic mutations
and expression of LGI3 may have prognostic significance in various
types of cancer (12). Therefore, it
was hypothesized that LGI3 may be associated with the cytokine
network in lung cancer. In the present study, analysis of DEGs in
the lung tissues from patients with NSCLC revealed that LGI3
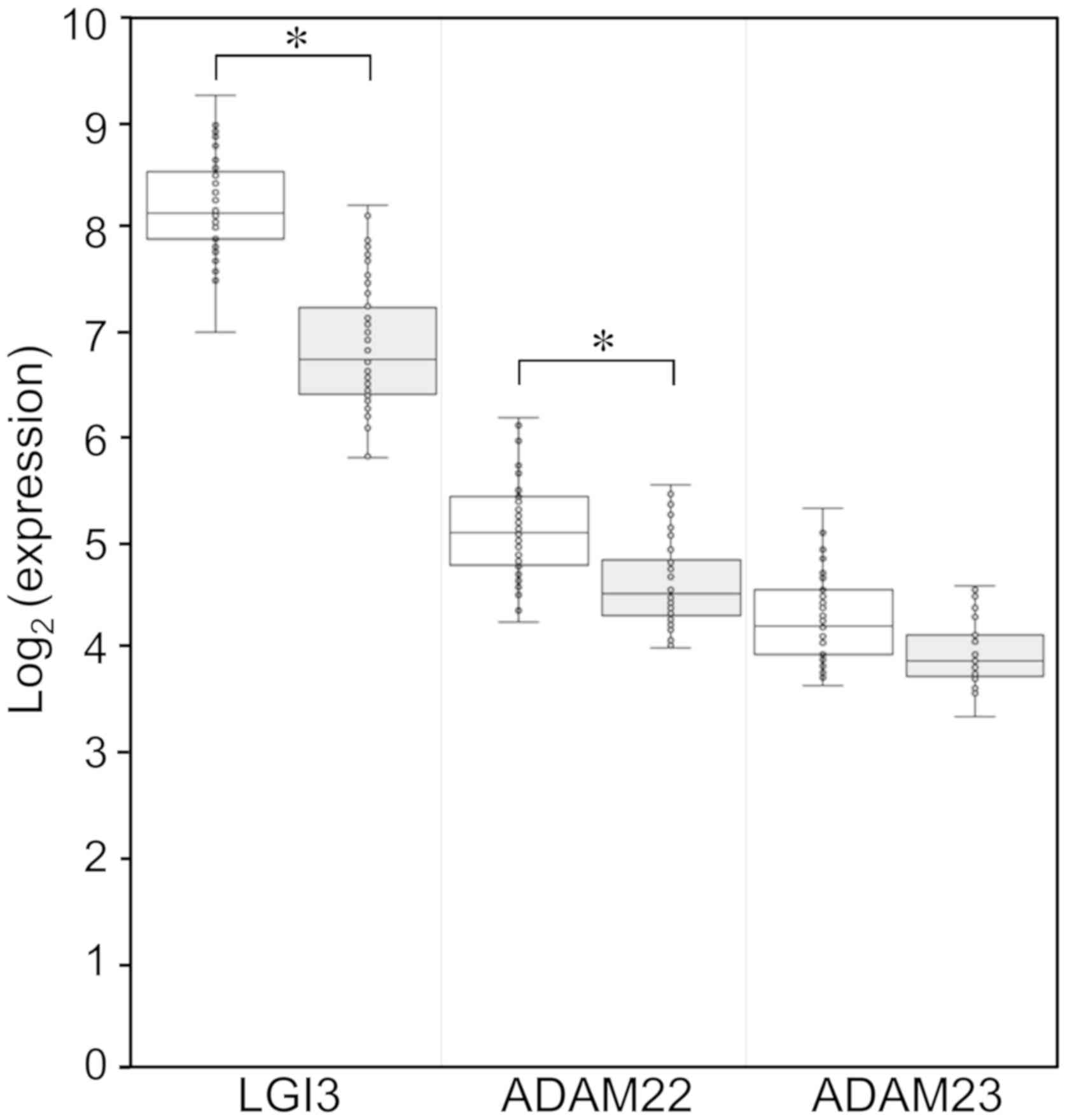
expression was significantly lower than in normal tissues (fold
change, 0.41; P=8.43×10−20; Fig. 1). The expression levels of the LGI3
receptor ADAM22 were significantly decreased in NSCLC tissues (fold
change, 0.72; P=7.01×10−8; Fig. 1). Expression of ADAM23 was not
significantly changed in NSCLC tissues (Fig. 1).
Identification of LGI3-regulated and
NSCLC-altered genes, and their protein-protein interaction
network
Analysis of DEGs in the expression microarray
dataset of NSCLC revealed that 1,158 genes were upregulated and
1,526 genes were downregulated in NSCLC tissues (|log2
(fold change)|≥1 and P<0.05). Our previous study identified 48
gene products that are regulated by LGI3 [adiponectin (ADIPOQ),
Akt1, BAD, complement C5 (C5), C-C motif chemokine ligand (CCL)2,
CCL12, CD68, CCAAT enhance binding protein α, C-reactive protein
(CRP), colony stimulating factor (CSF)1, CSF3, catenin β1, C-X-C
motif chemokine ligand (CXCL)2, CXCL13, cytochrome b-24 α chain,
cytochrome b-24 β chain, δ like non-canonical Notch ligand 1
(DLK1), eukaryotic translation initiation factor 4E binding protein
1, adhesion G protein-coupled receptor E1, endothelial cell
specific molecule 1 (ESM1), fatty acid binding protein 4 (FABP4),
glycogen synthase kinase 3 (GSK3)A, GSK3B, insulin like growth
factor 1 (IGF1), IGF binding protein (IGFBP)1, IGFBP5, interleukin
6 (IL6), integrin subunit αX, lipoprotein lipase (LPL),
mitogen-activated protein kinase (MAPK)1, MAPK3, MDM2
proto-oncogene (MDM2), microphthalmia-associated transcription
factor (MITF), neutrophil cytosolic factor (NCF)1, NCF2, NF-κB1,
nitric oxide synthase 2, PI3K catalytic subunit α, peroxisome
proliferator activated receptor γ (PPARG), protein kinase
AMP-activated catalytic subunit α1, PTEN,
prostaglandin-endoperoxide synthase 2 (PTGS2), protein tyrosine
kinase 2, serpin family E member 1 (SERPINE1), syntaxin 1A, TIMP
metallopeptidase inhibitor 1 (TIMP1), TNF and tumor protein p53]
(11). Venn diagram analysis of
NSCLC-altered and LGI3-regulated genes demonstrated that one
LGI3-upregulated gene and one LGI3-downregulated gene belonged to
the set of NSCLC-upregulated genes, and that seven LGI3-upregulated
genes and four LGI3-downregulated genes belonged to the set of
NSCLC-downregulated genes (Fig. 2A).
Overall, the expression levels of 27% (8/30) of the
LGI3-upregulated genes and 28% (5/18) of the LGI3-downregulated
genes were altered in NSCLC. Most of the LGI3-regulated gene
products (94%; 45/48 genes) have been demonstrated to form a
protein-protein interaction network cluster in our previous study
(11). Protein-protein interaction
network analysis of the 13 NSCLC-altered and LGI3-regulated genes
demonstrated that all gene products formed an interaction network
cluster (Fig. 2B). The proteins with
the highest interaction degrees (≥5) were CCL2 [also known as
monocyte chemoattractant protein (MCP-1)], IL6, PPARG, SERPINE1,
LPL and PTGS2 (also known as cyclooxygenase 2).
 | Figure 2.Comparative analysis of the up- and
downregulated genes in NSCLC and LGI3-regulated genes. (A) Venn
diagram showing the sets of the regulated gene categories. (B)
Protein-protein interaction network of NSCLC-altered and
LGI3-regulated products. The network includes nodes (gene products)
and lines (pairwise protein interactions) sorted by interaction
degrees. The symbols (*, +, # and
x) indicate the gene products in the common sets of the
regulated gene categories indicated in (A). CCL2, C-C motif
chemokine ligand 2; CSF3, colony stimulating factor 3; CXCL, C-X-C
motif chemokine ligand; DLK1, δ like non-canonical Notch ligand 1;
FABP4, fatty acid binding protein 4; IL6, interleukin 6; LGI3,
leucine rich repeat LGI family member 3; LPL, lipoprotein lipase;
NCF, neutrophil cytosolic factor; NSCLC, non-small cell lung
cancer; PPARG, peroxisome proliferator activated receptor γ; PTGS2,
prostaglandin-endoperoxide synthase 2; SERPINE1, serpin family E
member 1. |
Functional enrichment and KEGG pathway
analyses of LGI3-regulated and NSCLC-altered genes
The functional signature of LGI3-regulated genes
that are altered in NSCLC tissues was investigated by functional
enrichment analysis to obtain the GO terms of the gene groups
(Table I). The genes were
significantly associated with immune and inflammatory processes,
including lipopolysaccharide response, chemokines, vascular
endothelial growth factor receptor, cell redox homeostasis and cell
response to tumor necrosis factor. Additionally, KEGG pathway
analysis of LGI3-regulated and NSCLC-altered genes revealed that
the genes were associated with the signaling pathways of TNF,
chemokines, PPAR, and cytokines and infectious diseases (i.e.
malaria and leishmaniasis; Table
II). All associated KEGG pathways were closely associated with
immune and inflammatory systems.
 | Table I.Functional enrichment analysis of
leucine rich repeat LGI family member 3-regulated genes that are
altered in non-small cell lung cancer. |
Table I.
Functional enrichment analysis of
leucine rich repeat LGI family member 3-regulated genes that are
altered in non-small cell lung cancer.
| Category | Term | Count, n | P-value |
|---|
|
GOTERM_CC_DIRECT |
GO:0005615~extracellular space | 8 |
1.37×10−5 |
|
GOTERM_BP_DIRECT | GO:0050729~positive
regulation of inflammatory response | 4 |
2.19×10−5 |
|
GOTERM_CC_DIRECT |
GO:0005576~extracellular region | 8 |
4.41×10−5 |
|
GOTERM_BP_DIRECT | GO:0071222~cellular
response to lipopolysaccharide | 4 |
8.08×10−5 |
|
GOTERM_BP_DIRECT |
GO:0006954~inflammatory response | 5 |
1.55×10−4 |
|
GOTERM_BP_DIRECT | GO:0006955~immune
response | 5 |
2.33×10−4 |
|
GOTERM_BP_DIRECT | GO:0009409~response
to cold | 3 |
3.43×10−4 |
|
GOTERM_MF_DIRECT |
GO:0008009~chemokine activity | 3 |
6.31×10−4 |
|
GOTERM_BP_DIRECT |
GO:0070098~chemokine-mediated signaling
pathway | 3 |
1.33×10−3 |
|
GOTERM_BP_DIRECT | GO:0048010~vascular
endothelial growth factor receptor signaling pathway | 3 |
1.37×10−3 |
|
GOTERM_BP_DIRECT | GO:0042493~response
to drug | 4 |
1.47×10−3 |
|
GOTERM_BP_DIRECT | GO:0045454~cell
redox homeostasis | 3 |
1.57×10−3 |
|
GOTERM_BP_DIRECT | GO:0071356~cellular
response to tumor necrosis factor | 3 |
3.16×10−3 |
|
GOTERM_BP_DIRECT |
GO:0019221~cytokine-mediated signaling
pathway | 3 |
4.45×10−3 |
|
GOTERM_MF_DIRECT | GO:0008201~heparin
binding | 3 |
6.50×10−3 |
|
GOTERM_BP_DIRECT | GO:0032496~response
to lipopolysaccharide | 3 |
6.89×10−3 |
|
GOTERM_BP_DIRECT |
GO:0001525~angiogenesis | 3 |
1.24×10−2 |
|
GOTERM_MF_DIRECT | GO:0019899~enzyme
binding | 3 |
2.62×10−2 |
|
GOTERM_MF_DIRECT | GO:0005102~receptor
binding | 3 |
2.92×10−2 |
|
GOTERM_BP_DIRECT |
GO:0007186~G-protein coupled receptor
signaling pathway | 4 |
2.92×10−2 |
|
GOTERM_BP_DIRECT | GO:0045944~positive
regulation of transcription from RNA pol II promoter | 4 |
3.66×10−2 |
|
GOTERM_BP_DIRECT | GO:0045087~innate
immune response | 3 |
4.23×10−2 |
 | Table II.Kyoto Encyclopedia of Genes and
Genomes pathway analysis of leucine rich repeat LGI family member
3-regulated genes that are altered in non-small cell lung
cancer. |
Table II.
Kyoto Encyclopedia of Genes and
Genomes pathway analysis of leucine rich repeat LGI family member
3-regulated genes that are altered in non-small cell lung
cancer.
| Term | Count, n | P-value |
|---|
| hsa04668: TNF
signaling pathway | 4 | 0.00053 |
| hsa05144:
Malaria | 3 | 0.00260 |
| hsa04062: Chemokine
signaling pathway | 4 | 0.00270 |
| hsa03320: PPAR
signaling pathway | 3 | 0.00481 |
| hsa04060:
Cytokine-cytokine receptor interaction | 4 | 0.00493 |
| hsa05140:
Leishmaniasis | 3 | 0.00539 |
| hsa05142: Chagas
disease (American trypanosomiasis) | 3 | 0.01129 |
| hsa04380:
Osteoclast differentiation | 3 | 0.01754 |
GCN analysis of LGI3-regulated and
NSCLC-altered genes
To elucidate the roles of LGI3-regulated genes that
are altered in NSCLC, this gene set was queried against the GCN of
the lung. GCNs are a useful tool for exploring the roles of gene
sets, since coexpressed genes are controlled by common
transcriptional regulatory programs and are members of the same
protein complex or signaling pathway (23). A total of 10 gene products in the
gene set (Fig. 2B) were found in the
lung GCN (group a; Fig. 3A) and
these gene products were associated with 322 gene products in the
network (groups b and c; Fig. 3A).
The subnetwork of coexpression, consisting of 322 gene products
(Table SI), appeared to occupy a
domain with two adjacent clusters in the GCN (groups b and c;
Fig. 3A). A GO category map of the
subnetwork (groups b and c; Fig. 3B)
demonstrated that the gene products in the network were involved in
immune (groups b and c) and inflammatory responses, including
responses to wounding and stress (group c). Transcription factor
affinity prediction of the genes in groups b and c suggested that
these genes may be coexpressed under the common transcriptional
regulatory processes by various immune and inflammatory
transcription factors [Elf-1, ETS variant 4 (Pea3), Spi-1
proto-oncogene (Pu.1), C-ets-1, upstream transcription factor (Usf)
1/2, Stat6, NF-κB (Rela), cAMP response element-binding protein
(CREB) and AP-2; Table SII].
 | Figure 3.GCN analysis of NSCLC-altered and
LGI3-regulated gene products in the lung. (A) NSCLC-altered and
LGI3-regulated gene products found in the lung coexpression network
(group a), and the subnetwork of the gene group (groups b and c)
coexpressed with the genes in group a. (B) Gene Ontology category
map of the subnetwork consisting of group b and c genes. Letters
above the circles that represent leaf nodes of the hierarchical
tree indicated the groups of the subnetworks in (A). CCL2, C-C
motif chemokine ligand 2; CSF3, colony stimulating factor 3; CXCL2,
C-X-C motif chemokine ligand 2; FABP4, fatty acid binding protein
4; GCN, gene coexpression network; IL6, interleukin 6; LGI3,
leucine rich repeat LGI family member 3; NCF, neutrophil cytosolic
factor; NSCLC, non-small cell lung cancer; PPARG, peroxisome
proliferator activated receptor γ; PTGS2,
prostaglandin-endoperoxide synthase 2; SERPINE1, serpin family E
member 1. |
Association of LGI3 with prognosis of
NSCLC
Downregulation of LGI3 in NSCLC tissues (Fig. 1) suggested an association of LGI3
with the morbidity and mortality of NSCLC. To investigate the
prognostic significance of LGI3 expression in NSCLC, gene
expression microarray data of NSCLC patient cohorts were analyzed
(Table III). The results revealed
that the NSCLC studies demonstrated a significant association
(P<0.05) of low LGI3 expression with poor prognosis of NSCLC
(Table III, Fig. 4). Analysis of ADAM22 expression in
the NSCLC cohorts revealed no consistent association between its
expression and NSCLC prognosis (data not shown).
Somatic mutations of LGI3 in
NSCLC
The somatic mutations of the LGI3 gene in NSCLC with
amino acid alterations were identified in two major types of NSCLC,
lung adenocarcinoma and lung squamous cell carcinoma (Table IV). A total of seven mutations with
amino acid alterations were found in each NSCLC type, none of which
occurred in both types of cancer. Venn diagram analysis of the
amino acid variations in the five categories [conserved residues,
phylogenetically coevolved residues, SNPs (12) and somatic mutations in two types of
NSCLC] revealed that a subgroup of somatic mutation sites in NSCLC
belonged to phylogenetically coevolved residues (Y293H in lung
adenocarcinoma and A83V and L117F in lung squamous cell carcinoma)
or SNP sites (S171stop and R430G in lung squamous cell carcinoma;
Fig. 5). The amino acid alterations
(S171stop and R430G) at SNP sites were different from the residues
of the minor SNP alleles (Leu171, Cys430) and were not found in
somatic mutations in NSCLC (12). No
somatic mutation was identified at conserved residues.
 | Table IV.Somatic mutations of leucine rich
repeat LGI family member 3 in NSCLC tissues. |
Table IV.
Somatic mutations of leucine rich
repeat LGI family member 3 in NSCLC tissues.
| NSCLC type | Sample ID | Protein change | Mutation type | Chr 8 pos | Ref | Var | Allele freq
(T) |
|---|
| Adenocarcinoma | LUAD-5V8LT | S531C | Missense | 22005729 | T | A | NS |
| Adenocarcinoma | LUAD-5V8LT | P530H | Missense | 22005731 | G | T | NS |
| Adenocarcinoma |
TCGA-44-2656–01 | E507a | Nonsense | 22005801 | C | A | 0.21 |
| Adenocarcinoma |
TCGA-91-6829-01 | Q460L | Missense | 22005941 | T | A | 0.16 |
| Adenocarcinoma |
TCGA-55-A48X-01 | G82R | Missense | 22012939 | C | T | 0.09 |
| Adenocarcinoma |
TCGA-55-A490-01 | L185V | Missense | 22009455 | G | C | 0.40 |
| Adenocarcinoma |
TCGA-83-5908-01 | Y293H | Missense | 22006443 | A | G | 0.32 |
| Squamous cell
carcinoma |
TCGA-66-2795-01 | L117F | Missense | 22012074 | G | A | 0.47 |
| Squamous cell
carcinoma |
TCGA-52-7810-01 | S171a | Nonsense | 22009496 | G | C | 0.51 |
| Squamous cell
carcinoma |
TCGA-92-8065-01 | E78K | Missense | 22012951 | C | T | 0.24 |
| Squamous cell
carcinoma |
TCGA-NC-A5HF-01 | A83V | Missense | 22012935 | G | A | 0.38 |
| Squamous cell
carcinoma |
TCGA-85-8287-01 | A248V | Missense | 22009088 | G | A | 0.16 |
| Squamous cell
carcinoma |
TCGA-85-8287-01 | A248P | Missense | 22009089 | C | G | 0.17 |
| Squamous cell
carcinoma |
TCGA-66-2785-01 | R430G | Missense | 22006032 | G | C | 0.25 |
Discussion
LGI protein members (LGI1, −2, −3 and −4) are
differentially expressed in various tumor cells and LGI1 has been
proposed to be a tumor suppressor gene in brain tumors (24,25). Our
previous studies on LGI3 indicated that somatic mutations in
various types of cancer were found in the LGI3 gene and subsets of
the mutations affected SNP sites, phylogenetically coevolved amino
acids and a conserved amino acid (12). Additionally, our previous studies
demonstrated that the expression levels of LGI3 in tumor tissues
are associated with the prognosis of brain, colorectal and lung
cancers (12,13). It was postulated that LGI3 may
interact with the cytokine network in cancer through its genetic
variations and dysregulated expression (11,12).
The present study explored the potential prognostic
value and functional network of LGI3 in NSCLC using integrative
analysis of gene expression microarray data and the LGI3-regulated
cytokine network (11,12). Significant decreases in the
expression levels of LGI3 and one of its receptors, ADAM22, in
NSCLC tissues suggested that LGI3 may be involved in NSCLC
carcinogenesis and progression via its receptor-mediated signaling
pathway. Mediators of cellular signaling of LGI3 identified in our
previous studies were: Akt and focal adhesion kinase (FAK) in
LGI3-promoted neurite outgrowth (3),
p53 and MDM2 in LGI3-promoted cell protection in ultraviolet
B-irradiated keratinocytes (4),
β-catenin and GSK3B in LGI3-induced keratinocyte migration
(5), MITF in melanogenesis (7), and PPARG, CCAAT-enhancer binding
protein α and NF-κB in LGI3-regulated adipogenesis and metabolic
inflammation (9,10). Preadipocytes treated with LGI3
exhibit regulation of various signaling proteins (upregulated, Akt,
AMP-activated protein kinase, Erk and PTEN; downregulated,
eukaryotic translation initiation factor 4E binding protein 1, Bad
and GSK3A) (11). The mediators of
the LGI3-stimulated intracellular signaling pathway via ADAM22
should be addressed, and whether the signaling pathway is active
and perturbed in NSCLC cells requires further investigation.
Our cumulative studies found multiple gene products
that were regulated by LGI3 (1–5,7–11). A
majority of the LGI3-regulated gene products (45/48 gene products)
belong to a protein-protein interaction network that includes 16
cytokines, adipokines or chemokines, including ADIPOQ, CCL2/MCP-1,
CSF1, CRP, CXCL2, CXCL13, CSF3, C5/HC, ESM1, IGF1, IGFBP1, IGFBP5,
IL6, CCL12, TIMP1 and TNF-α (11).
Among the 13 LGI3-regulated and NSCLC-altered gene products that
formed a cluster of protein-protein interaction network, five gene
products belonged to the cytokines or chemokines (CCL2, CSF3,
CXCL2, CXCL13 and IL6). These results suggested that dysregulation
of LGI3 may account for perturbation of the cytokine network and
cell-cell communication in the microenvironment of NSCLC.
LGI3-regulated and NSCLC-altered genes were analyzed
by functional enrichment and KEGG pathway analyses, and were
determined to be significantly associated with the immune and
inflammatory responses, chemokines and cytokine activities. These
results were distinct from the same analyses of the LGI3-regulated
genes that were altered in glioma in that the gene products were
more significantly associated with angiogenesis, apoptosis,
hypoxia, proliferation, p53 and hypoxia-inducible factor-1
signaling pathways in glioma (13).
Therefore, LGI3 may be involved in the pathogenesis of NSCLC and
glioma through common and distinctive mechanisms.
All LGI3-regulated and NSCLC-altered genes were
found in the previous literature on NSCLC that reported an
association of genetic variations, expression and function of these
genes with NSCLC. Expression levels of PTGS2 (26), IL6 (27), CCL2 (28), NCF1/2 (29), CXCL2 (30), CSF3 (31), CXCL13 (32), LPL (33), SERPINE1 (34), PPARG (35) and FABP4 (36), and the promoter methylation of DLK1
(37) have been reported to be
associated with the prognosis and pathogenesis of NSCLC. A total of
seven genes (PTGS2, IL6, CCL2, NCF2, CXCL2, CSF3 and NCF1) that
have been reported to be increased by LGI3 may be downregulated in
NSCLC due to the suppression of LGI3 expression. DLK1, which has
been revealed to be decreased by LGI3 (11), may be increased in NSCLC due to
downregulation of LGI3. Thus, it may be postulated that the
perturbation of the expression of the eight genes (PTGS2, IL6,
CCL2, NCF2, CXCL2, CSF3, NCF1 and DLK1) was predominantly
influenced by LGI3 downregulation in NSCLC. LGI3 may functionally
interact with these gene products through protein-protein
interaction networks in the implicated mechanisms as determined by
the functional enrichment and KEGG pathway analyses.
LGI3 is abundantly expressed in the lung as well as
diverse tissues, including the brain, skin, adipose tissues, liver,
muscle, kidney and pancreas (1,4,9,38). LGI3
has been demonstrated to be expressed in a variety of cell types,
including neurons, keratinocytes, melanocytes, adipocytes and
macrophages (3,4,9,10). The RNA-sequencing data of the lung
from the Human Protein Atlas (https://www.proteinatlas.org) indicates that LGI3
transcripts are distributed in pneumocytes (20–40% of total
transcripts per million), bronchial epithelium (5–10%), endothelial
cells (20–35%) and macrophages (5–15%). LGI3-expressing cell types
in the mouse lung (mouse single cell transcriptome database;
http://tabula-muris.ds.czbiohub.org)
are notably similar to the human lung, supporting the validity of
mouse models in studying LGI3 in human lung cancer. GCN analysis of
LGI3-regulated and NSCLC-altered gene products in the present study
further supported the notion that LGI3 is involved in NSCLC
prognosis and pathogenesis as a regulatory cytokine in tumor
immunity and inflammation. The gene products in the subnetwork
(groups b and c) connected by gene coexpression with LGI3-regulated
and NSCLC-altered genes, were demonstrated to be associated with
common transcriptional regulatory processes of immune and
inflammatory transcription factors [Elf-1 (39), Pea3 (40), Pu.1 (41), C-ets-1 (42), Usf1/2 (43), Stat6 (44), NF-κB (45), CREB (46), and AP-2 (47)]. Our previous study reported that
NF-κB is a key transcription factor in mutual upregulation of LGI3
and TNF-α, implicated in adipose tissue inflammation in obesity
(10). Thus, downregulation of LGI3
in NSCLC may serve a role in the perturbation of the immune and
inflammatory GCN of the lung, and may account for prognostic
mechanisms of NSCLC.
The association of LGI3 expression with the
prognosis of NSCLC identified in the present study suggests that
LGI3 may serve antitumor roles in NSCLC progression. LGI3 has been
demonstrated to stimulate intracellular signaling proteins and
transcription factors involved in cancer, including p53, MDM2, Akt,
β-catenin, FAK, NF-κB and MITF (3,4,7,10,48–50).
Dysregulation of these proteins by decreased LGI3 may be
responsible for the prognostic role of LGI3 in NSCLC and glioma
(13). Our previous study
hypothesized that LGI3 may be a member of cytokine networks
involved in obesity-associated metabolic disorders and cancer
(11). Cytokine networks serve
critical roles in anticancer immunity in the NSCLC microenvironment
(51). By contrast, cytokine
networks may promote tumor growth and metastasis through chronic
inflammation (52). It was
postulated that upregulated LGI3 in adipose tissue and plasma in
obesity may promote chronic inflammation and cancer (8,9,11). Cytokine perturbation in obesity may
increase the risk of cancer in the digestive system, including the
liver, pancreas and gastrointestinal tract (53). Consistent association between obesity
and NSCLC has not been reported in previous studies; however,
previous reports have suggested that being overweight is a positive
prognostic factor (54,55). Thus, the LGI3-regulated adipokine
network in obesity may not account for the LGI3-regulated cytokine
network in NSCLC prognosis (8,10).
Our previous studies indicated that LGI3 increased
M1-polarized macrophage markers (TNF-α, inducible nitric oxide
synthase, CCL2/MCP-1, CD11c and IL-6) (9–11).
Macrophage polarization in the tumor microenvironment was
investigated in NSCLC prognosis, and predominant M1 macrophages
with inflammatory and antitumorigenic activities have been
associated with a positive NSCLC prognosis (56). Thus, LGI3 may contribute to antitumor
processes in the NSCLC microenvironment by promoting and
maintaining M1-polarization of tumor-associated macrophages.
The somatic mutations of LGI3 in two major types of
NSCLC suggested the potential prognostic value of the genetic
variations of LGI3 in NSCLC. The mutations were distributed
throughout the LGI3 protein domains (leucine-rich repeats and EPTP
domains) and the residue 248 was the only site with multiple
mutations (A248V and A248P). It was noted that two somatic
mutations in lung squamous cell carcinoma (S171stop and R430G) were
found at SNP sites with rare variants (global minor allele
frequency, 0.0002) (12). These
results warrant further studies on the prognostic and pathological
roles of the genetic variations of LGI3 in NSCLC. Studies with
LGI3-deficient or variant LGI3-expressing animal models and cell
lines may provide further insight into the prognostic and
pathological mechanisms of LGI3 in NSCLC.
In conclusion, the present study provided an
integrative insight into the prognostic value of LGI3 in NSCLC by
revealing the regulatory network of NSCLC-altered and
LGI3-regulated gene products, and the association of expression and
genetic variations of LGI3 with NSCLC. LGI3 may serve pathological
as well as prognostic roles in NSCLC through its pro-inflammatory
cytokine activity in immune and inflammatory processes of the tumor
microenvironment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education (grant no.
NRF-2018R1D1A1A09082440).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HYY conceived and designed the study, performed data
acquisition and analysis and wrote the manuscript. DSK and NSK
contributed to the analysis and interpretation of data. All authors
read and approved the final version.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lee SE, Lee AY, Park WJ, Jun DH, Kwon NS,
Baek KJ, Kim YG and Yun HY: Mouse LGI3 gene: Expression in brain
and promoter analysis. Gene. 372:8–17. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park WJ, Lee SE, Kwon NS, Baek KJ, Kim DS
and Yun HY: Leucine-rich glioma inactivated 3 associates with
syntaxin 1. Neurosci Lett. 444:240–244. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Park WJ, Lim YY, Kwon NS, Baek KJ, Kim DS
and Yun HY: Leucine-rich glioma inactivated 3 induces neurite
outgrowth through Akt and focal adhesion kinase. Neurochem Res.
35:789–796. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee SH, Jeong YM, Kim SY, Jeong HS, Park
KC, Baek KJ, Kwon NS, Yun HY and Kim DS: Ultraviolet B-induced LGI3
secretion protects human keratinocytes. Exp Dermatol. 21:716–718.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeong YM, Park WJ, Kim MK, Baek KJ, Kwon
NS, Yun HY and Kim DS: Leucine-rich glioma inactivated 3 promotes
HaCaT keratinocyte migration. Wound Repair Regen. 21:634–640. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim IW, Jeong HS, Kwon NS, Baek KJ, Yun HY
and Kim DS: LGI3 promotes human keratinocyte differentiation via
the Akt pathway. Exp Dermatol. 27:1224–1229. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jeong HS, Jeong YM, Kim J, Lee SH, Choi
HR, Park KC, Kim BJ, Baek KJ, Kwon NS, Yun HY and Kim DS:
Leucine-rich glioma inactivated 3 is a melanogenic cytokine in
human skin. Exp Dermatol. 23:600–602. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim HA, Kwon NS, Baek KJ, Kim DS and Yun
HY: Leucine-rich glioma inactivated 3 associates negatively with
adiponectin. Cytokine. 62:206–209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim HA, Park WJ, Jeong HS, Lee HE, Lee SH,
Kwon NS, Baek KJ, Kim DS and Yun HY: Leucine-rich glioma
inactivated 3 regulates adipogenesis through ADAM23. Biochim
Biophys Acta. 1821:914–922. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim HA, Kwon NS, Baek KJ, Kim DS and Yun
HY: Leucine-rich glioma inactivated 3 and tumor necrosis factor-α
regulate mutually through NF-κB. Cytokine. 72:220–223. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim HA, Kwon NS, Baek KJ, Kim DS and Yun
HY: Leucine-rich glioma inactivated 3: Integrative analyses support
its role in the cytokine network. Int J Mol Med. 40:251–259. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kwon NS, Baek KJ, Kim DS and Yun HY:
Leucine-rich glioma inactivated 3: Integrative analyses reveal its
potential prognostic role in cancer. Mol Med Rep. 17:3993–4002.
2018.PubMed/NCBI
|
|
13
|
Kwon NS, Kim DS and Yun HY: Leucine-rich
glioma inactivated 3: Integrative analyses support its prognostic
role in glioma. Onco Targets Ther. 10:2721–2728. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC,
Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC and Chuang EY:
Identification of a novel biomarker, SEMA5A, for non-small cell
lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers
Prev. 19:2590–2597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res 43
(Database Issue). D447–D452. 2015. View Article : Google Scholar
|
|
18
|
Lopes CT, Franz M, Kazi F, Donaldson SL,
Morris Q and Bader GD: Cytoscape Web: An interactive web-based
network browser. Bioinformatics. 26:2347–2348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee S, Zhang C, Liu Z, Klevstig M,
Mukhopadhyay B, Bergentall M, Cinar R, Ståhlman M, Sikanic N, Park
JK, et al: Network analyses identify liver-specific targets for
treating liver diseases. Mol Syst Biol. 13:9382017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thomas-Chollier M, Hufton A, Heinig M,
Heinig M, O'Keeffe S, Masri NE, Roider HG, Manke T and Vingron M:
Transcription factor binding predictions using TRAP for the
analysis of ChIP-seq data and regulatory SNPs. Nat Protoc.
6:1860–1869. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2:182009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li H, Sun Y and Zhan M: Exploring pathways
from gene co-expression to network dynamics. Methods Mol Biol.
541:249–267. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rossi MR, Huntoon K and Cowell JK:
Differential expression of the LGI and SLIT families of genes in
human cancer cells. Gene. 356:85–90. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chernova OB, Somerville RP and Cowell JK:
A novel gene, LGI1, from 10q24 is rearranged and downregulated in
malignant brain tumors. Oncogene. 17:2873–2881. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pan J, Yang Q, Shao J, Zhang L, Ma J, Wang
Y, Jiang BH, Leng J and Bai X: Cyclooxygenase-2 induced β1-integrin
expression in NSCLC and promoted cell invasion via the
EP1/MAPK/E2F-1/FoxC2 signal pathway. Sci Rep. 6:338232016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shintani Y, Fujiwara A, Kimura T, Kawamura
T, Funaki S, Minami M and Okumura M: IL-6 secreted from
cancer-associated fibroblasts mediates chemoresistance in NSCLC by
increasing epithelial-mesenchymal transition signaling. J Thorac
Oncol. 11:1482–1492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li L, Liu YD, Zhan YT, Zhu YH, Li Y, Xie D
and Guan XY: High levels of CCL2 or CCL4 in the tumor
microenvironment predict unfavorable survival in lung
adenocarcinoma. Thorac Cancer. 9:775–784. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song NY, Zhu F, Wang Z, Willette-Brown J,
Xi S, Sun Z, Su L, Wu X, Ma B, Nussinov R, et al: IKKα inactivation
promotes Kras-initiated lung adenocarcinoma development through
disrupting major redox regulatory pathways. Proc Natl Acad Sci USA.
115:E812–E821. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matsuo N, Azuma K, Hattori S, Ohtake J,
Kawahara A, Ishii H, Tokito T, Yamada K, Shibata Y, Shimokawaji T,
et al: Association between soluble immune mediators and tumor
responses in patients with non-small cell lung cancer treated with
anti-PD-1 inhibitor. Int J Cancer. 144:1170–1179. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tan X and Chen M: MYLK and MYL9 expression
in non-small cell lung cancer identified by bioinformatics analysis
of public expression data. Tumour Biol. 35:12189–12200. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Eide HA, Halvorsen AR, Sandhu V, Fåne A,
Berg J, Haakensen VD, Kure EH, Brustugun OT, Kiserud CE, Kyte JA
and Helland Å: Non-small cell lung cancer is characterised by a
distinct inflammatory signature in serum compared with chronic
obstructive pulmonary disease. Clin Transl Immunology. 5:e1092016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Podgornik H, Sok M, Kern I, Marc J and
Cerne D: Lipoprotein lipase in non-small cell lung cancer tissue is
highly expressed in a subpopulation of tumor-associated
macrophages. Pathol Res Pract. 209:516–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Eberlein C, Rooney C, Ross SJ, Farren M,
Weir HM and Barry ST: E-Cadherin and EpCAM expression by NSCLC
tumour cells associate with normal fibroblast activation through a
pathway initiated by integrin αvβ6 and maintained through TGFβ
signalling. Oncogene. 34:704–716. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bren-Mattison Y, Van Putten V, Chan D,
Winn R, Geraci MW and Nemenoff RA: Peroxisome
proliferator-activated receptor-gamma (PPAR(gamma)) inhibits
tumorigenesis by reversing the undifferentiated phenotype of
metastatic non-small-cell lung cancer cells (NSCLC). Oncogene.
24:1412–1422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang Z, Shen Q, Xie H, Zhou X, Li J, Feng
J, Liu H, Wang W, Zhang S and Ni S: Elevated expression of FABP3
and FABP4 cooperatively correlates with poor prognosis in non-small
cell lung cancer (NSCLC). Oncotarget. 7:46253–46262. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhong Z, Ye Y, Guo W, He Y and Hu W:
Relationship between DLK1 gene promoter region DNA methylation and
non-small cell lung cancer biological behavior. Oncol Lett.
13:4123–4126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gu W, Wevers A, Schröder H, Grzeschik KH,
Derst C, Brodtkorb E, de Vos R and Steinlein OK: The LGI1 gene
involved in lateral temporal lobe epilepsy belongs to a new
subfamily of leucine-rich repeat proteins. FEBS Lett. 519:71–76.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumazoe M, Yamashita M, Nakamura Y,
Takamatsu K, Bae J, Yamashita S, Yamada S, Onda H, Nojiri T,
Kangawa K and Tachibana H: Green tea polyphenol EGCG upregulates
Tollip expression by suppressing Elf-1 expression. J Immunol.
199:3261–3269. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kramer B, Wiegmann K and Kronke M:
Regulation of the human TNF promoter by the transcription factor
Ets. J Biol Chem. 270:6577–6583. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
DeKoter RP and Singh H: Regulation of B
lymphocyte and macrophage development by graded expression of PU.1.
Science. 288:1439–1441. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bevington SL, Cauchy P, Piper J, Bertrand
E, Lalli N, Jarvis RC, Gilding LN, Ott S, Bonifer C and Cockerill
PN: Inducible chromatin priming is associated with the
establishment of immunological memory in T cells. EMBO J.
35:515–535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Corre S and Galibert MD: USF as a key
regulatory element of gene expression. Med Sci (Paris). 22:62–67.
2006.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Swain SD, Meissner NN, Siemsen DW,
McInnerney K and Harmsen AG: Pneumocystis elicits a
STAT6-dependent, strain-specific innate immune response and airway
hyperresponsiveness. Am J Respir Cell Mol Biol. 46:290–298. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Q, Lenardo MJ and Baltimore D: 30
Years of NF-κB: A blossoming of relevance to human pathobiology.
Cell. 168:37–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wen AY, Sakamoto KM and Miller LS: The
role of the transcription factor CREB in immune function. J
Immunol. 185:6413–6419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Oyama N, Iwatsuki K, Homma Y and Kaneko F:
Induction of transcription factor AP-2 by inflammatory cytokines in
human keratinocytes. J Invest Dermatol. 113:600–606. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ji Z, Erin CY, Kumar R, Taylor M, Jenny
Njauw CN, Miao B, Frederick DT, Wargo JA, Flaherty KT, Jönsson G
and Tsao H: MITF modulates therapeutic resistance through EGFR
signaling. J Invest Dermatol. 135:1863–1872. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sulzmaier FJ, Jean C and Schlaepfer DD:
FAK in cancer: Mechanistic findings and clinical applications. Nat
Rev Cancer. 14:598–610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Uzdensky AB, Demyanenko SV and Bibov MY:
Signal transduction in human cutaneous melanoma and target drugs.
Curr Cancer Drug Targets. 13:843–866. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Domagala-Kulawik J: The role of the immune
system in non-small cell lung carcinoma and potential for
therapeutic intervention. Transl Lung Cancer Res. 4:177–190.
2015.PubMed/NCBI
|
|
52
|
West NR, McCuaig S, Franchini F and Powrie
F: Emerging cytokine networks in colorectal cancer. Nat Rev
Immunol. 15:615–629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Font-Burgada J, Sun B and Karin M: Obesity
and cancer: The oil that feeds the flame. Cell Metab. 23:48–62.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lam VK, Bentzen SM, Mohindra P, Nichols
EM, Bhooshan N, Vyfhuis M, Scilla KA, Feigenberg SJ, Edelman MJ and
Feliciano JL: Obesity is associated with long-term improved
survival in definitively treated locally advanced non-small cell
lung cancer (NSCLC). Lung Cancer. 104:52–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xie HJ, Zhang X, Wei ZQ, Long H, Rong TH
and Su XD: Effect of body mass index on survival of patients with
stage I non-small cell lung cancer. Chin J Cancer. 36:72017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jackute J, Zemaitis M, Pranys D,
Sitkauskiene B, Miliauskas S, Vaitkiene S and Sakalauskas R:
Distribution of M1 and M2 macrophages in tumor islets and stroma in
relation to prognosis of non-small cell lung cancer. BMC Immunol.
19:32018. View Article : Google Scholar : PubMed/NCBI
|