Introduction
Cancer has placed a great burden on human health and
the economy worldwide. Lung cancer has the highest rate of
mortality and morbidity among both sexes, making it one of the most
significant types of cancer, according to a major global
epidemiological survey in 2018 (1).
The financial burden of cancer not only takes a toll on an
individual's life, but it also leads to a further loss of
productivity. When adolescents with cancer face unbearable economic
costs, their drug treatment and subsequent treatment will be
affected, forming a vicious circle and causing further losses to
society (2). Small cell lung cancer
(SCLC) accounts for approximately 15% of the total cases of lung
cancer (1). Compared with other
types of lung cancer, SCLC is usually more sensitive to
radiotherapy and chemotherapy, but its diagnosis and treatment are
more difficult (3). Although SCLC is
sensitive to chemotherapy, it is more likely to develop drug
resistance and is highly prone to systemic metastasis (4). Patients with late-stage SCLC who are
treated with standard chemotherapy have a median survival time of
only 9–10 months from diagnosis (5).
Unlike non-small cell lung cancer (NSCLC), there are no driver
genes yet identified that can be used for clinically targeted
therapy, so the treatment of SCLC primarily relies on radiotherapy
and chemotherapy.
The etiology of malignant tumors is not yet fully
understood. In addition to smoking, environmental exposure
(6,7)
and genetics (8) have been linked to
the risk of developing cancer. Genetic factors are currently
recognized as one of the endogenous causes of cancer. In addition,
genetic mutations in patients with cancer can lead to resistance to
cancer drugs. As a result, it is very important to investigate the
molecular mechanisms underlying the occurrence and development of
SCLC and to identify effective biomarkers.
With the continuous development of microarray and
high-throughput sequencing technologies, molecular biomarkers
associated with the occurrence, development, diagnosis and
treatment of tumors can be identified (9–12).
Compared with research on NSCLC, fewer bioinformatics analyses have
been conducted in SCLC. However, certain studies have revealed
potential biomarkers associated with SCLC (13,14). To
further investigate the crucial biomarkers associated with SCLC and
to overcome the limitations regarding the results that are due to
different technology platforms or small sample sizes in various
studies, integrated bioinformatics methods were adopted in the
present study in order to conduct investigations across multiple
platforms with larger sample sizes.
Materials and methods
Gene expression profile data
The GEO database was used to obtain 4 microarray
expression profiles: GSE60052 (15),
GSE43346 (16), GSE15240 (17) and GSE6044 (18).
The inclusion criteria for the gene expression
profiles were as follows: i) The tumor tissue samples were from
patients with SCLC, and the control group was normal tissue; ii)
the number of samples was >10; iii) all included studies were
published in the English language; iv) the included studies
provided enough data or raw data to enable further bioinformatics
analysis; and v) the research subjects were human. The
characteristics of all the gene expression profiles are presented
in Table I.
 | Table I.Basic characteristics of the gene
expression profile data. |
Table I.
Basic characteristics of the gene
expression profile data.
| Record | Tissue | Platform | Normal | Tumor | Country | Race | (Refs.) |
|---|
| GSE60052 | SCLC | GPL11154 Illumina
HiSeq 2000 (Homo sapiens) | 7 | 79 | USA | Asian | (15) |
| GSE43346 | SCLC | GPL570
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 42 | 23 | Japan | Asian | (16) |
| GSE15240 | SCLC | GPL570
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 3 | 42 | USA | Caucasian | (17) |
| GSE6044 | SCLC | GPL201 [HG-Focus]
Affymetrix Human HG-Focus Target Array | 5 | 9 | Germany | Caucasian | (18) |
Integrated analysis of microarray
datasets
R software (version 3.5.1; 64-bit; http://www.r-project.org/) (19) was used to identify relevant DEGs. The
expression data for each gene expression profile were downloaded
from the GEO database. For values not reported in logarithmic form,
log2 conversion was performed. If standardized data were
not available, the raw data were downloaded. For CEL files, the
affy package (19) was used to read
the expression data. If a gene had multiple probes in the same
chip, the average value of all probes was taken as the expression
value of the gene. The limma package (20) was used to standardize the data for
each experiment. Finally, the RobustRankAggreg package (21) was used to integrate the DEGs
identified in the four gene expression profiles. The included DEGs
conformed to the following criteria: |log2FC| >1.5
(where FC means fold change) and adjusted P<0.05.
Functional and pathway enrichment
analysis
FunRich software (version 3.1.3; 64-bit) (22) was used to perform the functional and
pathway enrichment analyses. The FunRich tool was designed to
process a variety of gene and protein datasets and to perform
powerful functional enrichment analyses (22). The GO database (geneontology.org) was chosen for the functional
pathway enrichment analysis of downregulated and upregulated DEGs
in the present study. This database contains information associated
with biological processes (BPs), cellular components (CCs) and
molecular functions. The DOSE package (23) for R software was selected for the
KEGG pathway enrichment analysis. P<0.05 was considered to
indicate a statistically significant result.
PPI network construction and analysis
of modules
The STRING database (version 10.5; string-db.org/) is an online database that facilitates
searches for interactions between proteins (24). The DEGs with interaction scores
>0.9 were mapped into PPIs. Then, Cytoscape (version 3.6.1;
64-bit; www.cytoscape.org/) (25) was used to identify hub genes.
Cytoscape is an open-source software platform for visualizing
complex networks and integrating those networks with any type of
attributed data (25). The modules
with degree cut-off=2, node score cut-off=0.2 and K-core value =2
were screened by Molecular Complex Detection (MCODE) (26). The modules with MCODE scores >10
and >15 nodes were selected as notable modules. FunRich software
and R software were used to perform functional and KEGG pathway
enrichment analyses, respectively, of the DEGs in these different
modules. The genes with degrees >20 were considered hub
genes.
Expression level analysis of hub
genes
Oncomine (oncomine.org)
is currently the largest oncogene chip database and integrated data
mining platform in the world; it is intended to be used to mine
cancer gene information. To date, the database has collected data
from 729 gene expression datasets and >90,000 cancer and normal
tissue samples. It can be used to analyze the differential
expression of individual genes in samples of major cancers and
their associated normal tissues and to visualize the results
(27). The expression value data for
normal and SCLC tissues were downloaded from Oncomine
(pre-processed expression levels were Log2 normalized and median
centered), and GraphPad Prism software (version 7.0; 64-bit;
GraphPad Software, Inc.) was used to draw box diagrams and identify
significant differences using unpaired Student's t-test. P<0.05
was considered to indicate a statistically significant result.
Results
DEG identification
In all four microarray expression profiles
(GSE60052, GSE43346, GSE15240 and GSE6044), 412 DEGs were
identified, which contained 57 normal tissue samples and 153 SCLC
tissue samples. Compared with normal tissues, SCLC tissues had 146
upregulated genes and 266 downregulated genes. To visualize the
differential expression levels in each microarray expression
profile following standardization, volcano maps were used for the 4
datasets (Fig. 1). Red points
represent upregulated genes (log2FC >1.5 and adjusted
P<0.05), whereas green points represent downregulation genes
(log2FC <-1.5 and adjusted P<0.05). The black
points represent genes with no significant difference (adjusted
P≥0.05). Then, a cluster heat map was used to present the
differences in the expression levels of the top 20 DEGs according
to adjusted P-values integrated from the four different microarray
expression profiles (Fig. 2). The
top 20 integrated DEGs presented the smallest adjusted P-values,
and were S1PR1, MAD2L1, CDKN2A, STIL, NDC80, NCAPG, PAD51AP1, TTK,
PRM2, EZH2, PRC1, UBE2C, RFC4, CENPF, TCP2A, HMGB3, TYMS, SOX4,
MCM2 and SMC4.
Enrichment analyses
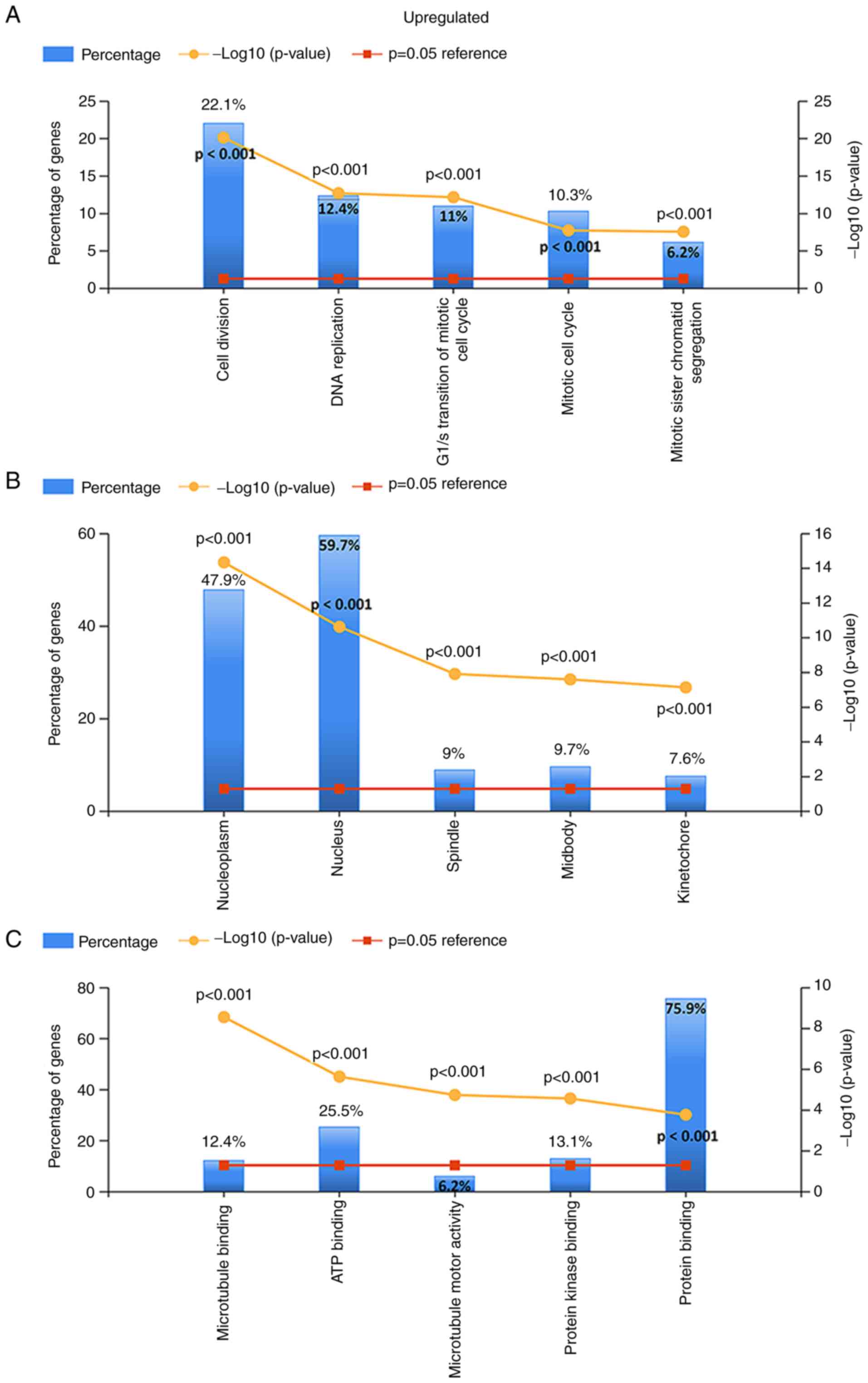
For the upregulated and downregulated DEGs, GO and
KEGG pathway enrichment analyses were performed. The top 5 enriched
results are presented in Figs.
3–5. For upregulated DEGs, the
enriched BPs were cell division, DNA replication, G1/S
transition of the mitotic cell cycle, mitotic cell cycle and
mitotic sister chromatid segregation. The enriched BPs among the
downregulated genes included the cytokine-mediated signaling
pathway, positive regulation of angiogenesis, the inflammatory
response, cell adhesion and vasculogenesis. Regarding the enriched
CCs, the upregulated DEGs were enriched in the nucleoplasm,
nucleus, spindle, midbody and kinetochore, while the downregulated
DEGs were enriched in extracellular vesicular exosomes, the
extracellular space, the cell surface, focal adhesion and the
extracellular region. In addition, with regard to molecular
function, the upregulated DEGs were associated with microtubule
binding, ATP binding, microtubule motor activity and protein kinase
binding, while the downregulated DEGs were associated with protein
binding, protein homodimerization activity, integrin binding,
β-amyloid binding and tumor necrosis factor binding (all adjusted
P-values <0.05).
There were 5 KEGG pathways that were significantly
associated with the upregulated genes (all adjusted P-values
<0.05; Fig. 5), including the
cell cycle, DNA replication, mismatch repair, homologous
recombination and oocyte meiosis (Fig.
5A). Complement and coagulation cascades, fluid shear stress
and atherosclerosis, Staphylococcus aureus infection,
cytokine-cytokine receptor interaction and viral myocarditis were
primarily associated with downregulated DEGs (Fig. 5B).
Construction of PPI network and module
identification
To predict the interactions between recognized DEGs
at the protein level, a PPI network was constructed using the
STRING database. In total, 410 nodes and 852 edges were presented
in this PPI network (Fig. 6).
According to the inclusion criteria used within the present study,
one significant module was identified (Fig. 7). In this module, 8 hub genes with
degrees >20 were identified: BIRC5, NDC80, BUB1B, CENPE, KIF2C,
PLK1, CDC20 and MAD2L1 (Table II).
The GO pathway enrichment analysis revealed that microtubule
binding, tubulin binding, microtubule motor activity, motor
activity, protein C-terminus binding and ATPase activity were the
primarily enriched pathways. Via the KEGG analysis, the following
four crucial pathways were identified: The cell cycle, oocyte
meiosis, human T-cell leukemia virus 1 infection and
progesterone-mediated oocyte maturation (Fig. 8).
 | Table II.Hub genes with a high degree of
connectivity. |
Table II.
Hub genes with a high degree of
connectivity.
| Gene | Degree | Type | MCODE Cluster |
|---|
| MAD2L1 | 36 | Upregulated | Cluster 1 |
| CDC20 | 33 | Upregulated | Cluster 1 |
| PLK1 | 29 | Upregulated | Cluster 1 |
| CENPE | 25 | Upregulated | Cluster 1 |
| KIF2C | 25 | Upregulated | Cluster 1 |
| BUB1B | 23 | Upregulated | Cluster 1 |
| NDC80 | 22 | Upregulated | Cluster 1 |
| BIRC5 | 20 | Upregulated | Cluster 1 |
Expression level analysis of hub
genes
Using the Ocomine database, two important results
were obtained from the Garber et al (28) and Bhattacharjee et al
(29) lung cancer gene expression
data. From the analysis of their data, it can be concluded that
when compared with normal tissues, SCLC tissues had five highly
expressed hub genes. These genes were the same as those identified
in the bioinformatics investigation of the present study, except
CENPE, PLK1 and BIRC5. Three of the identified genes were
associated with cell cycle pathways and were highly expressed
(CDC20, BUB1B and MAD2L1) (all P<0.05; Fig. 9).
Discussion
SCLC has been termed a recalcitrant cancer. Despite
being sensitive to radiotherapy and chemotherapy, the recurrence
rate is very high. Although there have been breakthroughs in the
treatment of SCLC, the pathogenesis remains unclear (30–32).
Therefore, the discovery of potential molecular mechanisms
underlying SCLC may be very important for the treatment and
diagnosis of SCLC. In particular, the development of
high-throughput sequencing facilitates the search for potentially
involved genes and tumor mechanisms. Cancer is a polygenic genetic
disease; with the development of tumors, cancer cells are still
constantly dividing, multiplying and accumulating mutations, and
their genomes are unstable. Bioinformatics, on the other hand, uses
genetic information for molecular diagnoses and drug therapies. To
the best of our knowledge, the present study is currently the first
to use four gene chips for cross-platform analysis. The sample size
was increased, the effect of batch difference was eliminated using
the RobustRankAggreg package, and the results were more accurate.
The more accurate evaluation of the previously discovered genes and
pathways will provide a theoretical basis for subsequent studies,
and also guide future studies.
In the present study, 412 DEGs were identified,
including 146 upregulated genes and 266 downregulated genes. The
upregulated genes were primarily enriched in the cell cycle, DNA
replication, mismatch repair, homologous recombination and oocyte
meiosis. The downregulated genes were primarily enriched in
cytokine-cytokine receptor interactions. Among these DEGs, a
crucial module containing 8 hub genes was selected from the PPI
network. Furthermore, the important enriched module pathways
containing the 8 hub genes revealed that the cell cycle was the
most significant pathway. A previous study (32) that performed whole genome sequencing
on a large sample of 152 freshly frozen samples from patients
diagnosed with stage I–IV SCLC revealed that a loss of the tumor
suppressors TP53 and retinoblastoma protein 1 promoted the
development of SCLC. The present study obtained 5 new promising hub
genes, which were highly expressed in SCLC tissues compared with
normal tissue, and three of those genes were strongly associated
with the cell cycle pathway (TP53, TP73 and RB1; all P-values
<0.05).
An abnormal cell cycle may lead to the occurrence of
a variety of different types of tumor, such as hepatocellular
carcinoma (33,34), breast cancer (35), pancreatic cancer (36), ovarian cancer (37) and NSCLC in non-smoking females
(38). Wang et al (39) used different microarray expression
profiles in their study of primitive neuro-ectodermal tumors, and
revealed the same four pathways that were identified in the present
study, and several of the same genes in the modules that they
identified as being important. These similarities may be the result
of both tumors being neuroendocrine tumors. These results support
the view that disordered cell cycle regulation may contribute to
the development of SCLC. Previous studies (14,40) have
also indicated that the cell cycle pathway is an important pathway
involved in the development and progression of SCLC. It has also
been demonstrated that certain associated drugs can restore the
abnormal cell cycle in SCLC, thus playing anti-tumor roles
(41).
Following the discovery of cell division cycle
protein 20 (CDC20) ~40 years ago, the original researchers
demonstrated that CDC20 mutations can cause abnormal cessation of
mitosis, resulting in abnormal chromosome separation in anaphase
(42). Wan et al (43) revealed that the anaphase-promoting
complex (APC) CDC20 plays an important role in the regulation of
the cell cycle and apoptosis, and it can also inhibit the
apoptosis-induced resistance of cancer cells to chemotherapy and
radiotherapy. Furthermore, CDC20 is highly expressed in a number of
different types of tumor, and is associated with the poor prognosis
of patients, such as those with colorectal cancer (44), hepatocellular carcinoma (45), gastric cancer (46), bladder cancer (47) and cervical cancer (48). Taniguchi et al (49) discovered that downregulation of CDC20
can increase the sensitivity of pancreatic cancer cells to
radiotherapy and paclitaxel. In addition, Wan et al
(43) demonstrated that hyperactive
CDC20 plays an important role in chemoresistance, and is associated
with the human T-cell leukemia virus 1 infection pathway. The
downregulation of CDC20 expression slows the growth of both NSCLC
and SCLC and the formation rate of lung cancer cell colonies
(50). In conclusion, CDC20 is
expected to be a new target for research on SCLC.
The BUB1B gene encodes a kinase involved in the
spindle assembly checkpoint and chromosome separation (51). During the G2 phase of
mitosis, BUB1B binds to CDC20 and inhibits APC/cyclostome (APC/C)
activity, allowing cyclin B to accumulate before mitosis begins and
slows the cell cycle (52). This
gene also contributes to the progression of certain types of tumor
and is associated with the prognosis of patients with
hepatocellular carcinoma (53) and
glioblastoma (54). Ma et al
(55) revealed that high expression
levels of BUB1B may promote the tolerance of glioblastoma to
radiotherapy. Chen et al (56) used gene expression profiling of the
GSE40791 (57) database to
demonstrate that BUB1B is a candidate gene associated with the
pathogenesis of lung adenocarcinoma. Therefore, this gene is a
promising candidate, and should be the focus of future research on
SCLC.
Another gene enriched in cell cycle pathways is
mitotic arrest deficient 2 like 1 (MAD2L1), which is a component of
the mitotic spindle assembly checkpoint that ensures that all
chromosomes are properly aligned at the metaphase plate (58). The destruction of the function of
MAD2L1 in mammalian cells can affect the function of the spindle,
leading to cell aneuploidy or the occurrence of tumors. Partial
deletion of MAD2L1 (MAD2L1+/− cells) accompanied by
chromosomal instability can cause lung cancer in mice (59). MAD2L1 is highly expressed in patients
with breast cancer (60) and gastric
cancer (61), and its expression is
associated with the tumor stage. A recent study (62) also demonstrated that patients with
lung adenocarcinoma with high expression levels of MAD2L1 have
worse prognoses than those with low expression levels of
MAD2L1.
The kinesin family member 2C (KIF2C) gene encodes a
protein that is part of the kinase-like protein family. This
protein plays a role in regulating microtubule dynamics in cells,
and is important for chromosome separation during anaphase
(63). The GO analysis in the
present study suggests that it has functional enrichment in two
aspects: Microtubule binding and tubulin binding. Therefore, its
abnormal expression affects mitosis and can lead to cancer. In
addition, KIF2C has been highly expressed not only in patients with
esophageal squamous cell carcinoma (64) and glioma (65) with poor prognoses, but also in those
with adenocarcinoma of the lung, as indicated by two different
bioinformatics analyses (66,67).
Nuclear division cycle 80 (Ndc80) is an
isotetrameric protein complex that plays an important role in cell
mitosis (68). A liver
cancer-associated study (69)
revealed that liver cancer tumor tissue samples from 47 patients
had higher Ndc80 mRNA expression levels than the adjacent normal
tissue samples. The virus carrying Ndc80-siRNA silences the Ndc80
gene, inhibits the in vitro proliferation of the cultured
liver cancer SMMC-7721 cell line and induces cell apoptosis,
demonstrating that Ndc80 primarily acts through its effect on
overcoming cell cycle arrest and antiapoptosis mechanism-induced
liver cancer. In a study that focused on osteosarcoma, Ndc80 mRNA
expression was higher in 84.6% of tumor tissues compared with in
adjacent normal tissues, and the expression level was associated
with the Tumor-Node-Metastasis stage (70) and distant metastasis of the tumor. In
addition, the level of Ndc80 was indicated to have value as an
independent prognostic evaluation index (71). Yuan et al (72) screened 13 GSE datasets and revealed
that NDC80 and 7 other hub genes can interact with ZW10-interacting
protein and participate in the formation of both NSCLC and
SCLC.
There were limitations to the present study. First,
the conclusions were drawn based on data from public databases
rather than actual experiments, meaning that the quality of the
data cannot be guaranteed, and that the results may be inaccurate.
Secondly, the inability to include samples from all ethnic groups,
such as African-Americans, may have affected the levels of gene
expression observed. Finally, due to the lack of public data on the
prognosis of patients with SCLC, the impact of the identified genes
on survival was not further investigated, and factors such as the
sex, age and tumor stage of patients were not included. All of
these factors could affect the conclusions of the present
study.
In conclusion, the present study has revealed five
genes that may be associated with the pathogenesis of SCLC: NDC80,
BUB1B, KIF2C, CDC20 and MAD2L1. The cell cycle may play an
important role in the development of SCLC; however, more
well-designed experiments with larger sample sizes need to be
performed in order to confirm these conclusions. The results from
the present study will provide new ideas for the treatment of
SCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL and GY conceived and designed the study, acquired
and analyzed data and wrote the manuscript. XW and PZ contributed
to data analysis and manuscript drafting. XF and CH designed the
study and revised the manuscript. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of
interest.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Landwehr MS, Watson SE, Macpherson CF,
Novak KA and Johnson RH: The cost of cancer: A retrospective
analysis of the financial impact of cancer on young adults. Cancer
Med. 5:863–870. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gao H, Niu Y, Li M, Fang S and Guo L:
Identification of DJ-1 as a contributor to multidrug resistance in
human small-cell lung cancer using proteomic analysis. Int J Exp
Pathol. 98:67–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang C, Huang M, Chen W, Zhu W, Meng H,
Guo L, Wei T and Zhang J: N-acetylglucosaminyltransferase V
modulates radiosensitivity and migration of small cell lung cancer
through epithelial-mesenchymal transition. FEBS J. 282:4295–4306.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Demedts IK, Vermaelen KY and van Meerbeeck
JP: Treatment of extensive-stage small cell lung carcinoma: Current
status and future prospects. Eur Respir J. 35:202–215. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lam WK: Lung cancer in Asian women-the
environment and genes. Respirology. 10:408–417. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hosgood HD III, Boffetta P, Greenland S,
Lee YC, McLaughlin J, Seow A, Duell EJ, Andrew AS, Zaridze D,
Szeszenia-Dabrowska N, et al: In-home coal and wood use and lung
cancer risk: A pooled analysis of the international lung cancer
consortium. Environ Health Perspect. 118:1743–1747. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu PF, Lee CH, Wang MJ, Goggins WB, Chiang
TA, Huang MS and Ko YC: Cancer aggregation and complex segregation
analysis of families with female non-smoking lung cancer probands
in Taiwan. Eur J Cancer. 40:260–266. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matamala N, Vargas MT, González-Cámpora R,
Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G,
Yanowsky K, Calvete-Candenas J, et al: Tumor microRNA expression
profiling identifies circulating microRNAs for early breast cancer
detection. Clin Chem. 61:1098–1106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lusito E, Felice B, D'Ario G, Ogier A,
Montani F, Di Fiore PP and Bianchi F: Unraveling the role of
low-frequency mutated genes in breast cancer. Bioinformatics.
35:36–46. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang L, Yang Y, Cheng L, Cheng Y, Zhou HH
and Tan ZR: Identification of common genes refers to colorectal
carcinogenesis with paired cancer and noncancer samples. Dis
Markers. 2018:34527392018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu H, Wei S, Zhang L, Yuan C, Duan Y and
Wang Q: Secreted phosphoprotein 1 promotes the development of small
cell lung cancer cells by inhibiting autophagy and apoptosis.
Pathol Oncol Res. 2018. View Article : Google Scholar
|
|
14
|
Ni Z, Wang X, Zhang T, Li L and Li J:
Comprehensive analysis of differential expression profiles reveals
potential biomarkers associated with the cell cycle and regulated
by p53 in human small cell lung cancer. Exp Ther Med. 15:3273–3282.
2018.PubMed/NCBI
|
|
15
|
Jiang L, Huang J, Higgs BW, Hu Z, Xiao Z,
Yao X, Conley S, Zhong H, Liu Z, Brohawn P, et al: Genomic
landscape survey identifies SRSF1 as a key oncodriver in small cell
lung cancer. PLoS Genet. 12:e10058952016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sato T, Kaneda A, Tsuji S, Isagawa T,
Yamamoto S, Fujita T, Yamanaka R, Tanaka Y, Nukiwa T, Marquez VE,
et al: PRC2 overexpression and PRC2-target gene repression relating
to poorer prognosis in small cell lung cancer. Sci Rep. 3:19112013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Daniel VC, Marchionni L, Hierman JS,
Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M,
Peacock CD and Watkins DN: A primary xenograft model of small-cell
lung cancer reveals irreversible changes in gene expression imposed
by culture in vitro. Cancer Res. 69:3364–3373. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rohrbeck A, Neukirchen J, Rosskopf M,
Pardillos GG, Geddert H, Schwalen A, Gabbert HE, von Haeseler A,
Pitschke G, Schott M, et al: Gene expression profiling for
molecular distinction and characterization of laser captured
primary lung cancers. J Transl Med. 6:692008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kolde R, Laur S, Adler P and Vilo J:
Robust rank aggregation for gene list integration and
meta-analysis. Bioinformatics. 28:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pathan M, Keerthikumar S, Chisanga D,
Alessandro R, Ang CS, Askenase P, Batagov AO, Benito-Martin A,
Camussi G, Clayton A, et al: A novel community driven software for
functional enrichment analysis of extracellular vesicles data. J
Extracell Vesicles. 6:13214552017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu G, Wang LG, Yan GR and He QY: DOSE: An
R/Bioconductor package for disease ontology semantic and enrichment
analysis. Bioinformatics. 31:608–609. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saito R, Smoot ME, Ono K, Ruscheinski J,
Wang PL, Lotia S, Pico AR, Bader GD and Ideker T: A travel guide to
cytoscape plugins. Nat Methods. 9:1069–1076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bhattacharjee A, Richards WG, Staunton J,
Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et
al: Classification of human lung carcinomas by mRNA expression
profiling reveals distinct adenocarcinoma subclasses. Proc Natl
Acad Sci USA. 98:13790–13795. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pietanza MC, Byers LA, Minna JD and Rudin
CM: Small cell lung cancer: Will recent progress lead to improved
outcomes? Clin Cancer Res. 21:2244–2255. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Karachaliou N, Pilotto S, Lazzari C, Bria
E, de Marinis F and Rosell R: Cellular and molecular biology of
small cell lung cancer: An overview. Transl Lung Cancer Res.
5:2–15. 2016.PubMed/NCBI
|
|
32
|
George J, Lim JS, Jang SJ, Cun Y, Ozretić
L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al:
Comprehensive genomic profiles of small cell lung cancer. Nature.
524:47–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Z, Zhong Y, Chen YJ and Chen H: SOX11
regulates apoptosis and cell cycle in hepatocellular carcinoma via
Wnt/β-catenin signaling pathway. Biotechnol Appl Biochem.
66:240–246. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
An MJ, Kim DH, Kim CH, Kim M, Rhee S, Seo
SB and Kim JW: Histone demethylase KDM3B regulates the
transcriptional network of cell-cycle genes in hepatocarcinoma
HepG2 cells. Biochem Biophys Res Commun. 508:576–582. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun X, Hu Y, Wu J, Shi L, Zhu L, Xi PW,
Wei JF and Ding Q: RBMS2 inhibits the proliferation by stabilizing
P21 mRNA in breast cancer. J Exp Clin Cancer Res. 37:2982018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Matsushita Y, Furutani Y, Matsuoka R and
Furukawa T: Hot water extract of Agaricus blazei Murrill
specifically inhibits growth and induces apoptosis in human
pancreatic cancer cells. BMC Complement Altern Med. 18:3192018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee KS, Kim SW and Lee HS: Orostachys
japonicus induce p53-dependent cell cycle arrest through the MAPK
signaling pathway in OVCAR-3 human ovarian cancer cells. Food Sci
Nutr. 6:2395–2401. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang G, Chen Q, Xiao J, Zhang H, Wang Z
and Lin X: Identification of genes and analysis of prognostic
values in nonsmoking females with non-small cell lung carcinoma by
bioinformatics analyses. Cancer Manag Res. 10:4287–4295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang GY, Li L, Liu B, Han X, Wang CH and
Wang JW: Integrated bioinformatic analysis unveils significant
genes and pathways in the pathogenesis of supratentorial primitive
neuroectodermal tumor. Onco Targets Ther. 11:1849–1859. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hubaux R, Thu KL, Coe BP, MacAulay C, Lam
S and Lam WL: EZH2 promotes E2F-driven SCLC tumorigenesis through
modulation of apoptosis and cell-cycle regulation. J Thorac Oncol.
8:1102–1106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin YC, Su JH, Lin SC, Chang CC, Hsia TC,
Tung YT and Lin CC: A soft coral-derived compound,
11-dehydrosinulariolide, induces G2/M cell cycle arrest and
apoptosis in small cell lung cancer. Mar Drugs. 16:2018. View Article : Google Scholar
|
|
42
|
Hartwell LH, Culotti J and Reid B: Genetic
control of the cell-division cycle in yeast. I. Detection of
mutants. Proc Natl Acad Sci USA. 66:352–359. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wan L, Tan M, Yang J, Inuzuka H, Dai X, Wu
T, Liu J, Shaik S, Chen G, Deng J, et al: APC(Cdc20) suppresses
apoptosis through targeting Bim for ubiquitination and destruction.
Dev Cell. 29:377–391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu WJ, Hu KS, Wang DS, Zeng ZL, Zhang DS,
Chen DL, Bai L and Xu RH: CDC20 overexpression predicts a poor
prognosis for patients with colorectal cancer. J Transl Med.
11:1422013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li J, Gao JZ, Du JL, Huang ZX and Wei LX:
Increased CDC20 expression is associated with development and
progression of hepatocellular carcinoma. Int J Oncol. 45:1547–1555.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ding ZY, Wu HR, Zhang JM, Huang GR and Ji
DD: Expression characteristics of CDC20 in gastric cancer and its
correlation with poor prognosis. Int J Clin Exp Pathol. 7:722–727.
2014.PubMed/NCBI
|
|
47
|
Choi JW, Kim Y, Lee JH and Kim YS: High
expression of spindle assembly checkpoint proteins CDC20 and MAD2
is associated with poor prognosis in urothelial bladder cancer.
Virchows Arch. 463:681–687. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim Y, Choi JW, Lee JH and Kim YS: MAD2
and CDC20 are upregulated in high-grade squamous intraepithelial
lesions and squamous cell carcinomas of the uterine cervix. Int J
Gynecol Pathol. 33:517–523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Taniguchi K, Momiyama N, Ueda M, Matsuyama
R, Mori R, Fujii Y, Ichikawa Y, Endo I, Togo S and Shimada H:
Targeting of CDC20 via small interfering RNA causes enhancement of
the cytotoxicity of chemoradiation. Anticancer Res. 28:1559–1563.
2008.PubMed/NCBI
|
|
50
|
Kidokoro T, Tanikawa C, Furukawa Y,
Katagiri T, Nakamura Y and Matsuda K: CDC20, a potential cancer
therapeutic target, is negatively regulated by p53. Oncogene.
27:1562–1571. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Guo Y, Kim C, Ahmad S, Zhang J and Mao Y:
CENP-E-dependent BubR1 autophosphorylation enhances chromosome
alignment and the mitotic checkpoint. J Cell Biol. 198:205–217.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Malureanu LA, Jeganathan KB, Hamada M,
Wasilewski L, Davenport J and van Deursen JM: BubR1 N terminus acts
as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in
interphase. Dev Cell. 16:118–131. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhuang L, Yang Z and Meng Z: Upregulation
of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted
worse overall survival and disease-free survival in hepatocellular
carcinoma patients. Biomed Res Int. 2018:78973462018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee E, Pain M, Wang H, Herman JA, Toledo
CM, DeLuca JG, Yong RL, Paddison P and Zhu J: Sensitivity to BUB1B
inhibition defines an alternative classification of glioblastoma.
Cancer Res. 77:5518–5529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ma Q, Liu Y, Shang L, Yu J and Qu Q: The
FOXM1/BUB1B signaling pathway is essential for the tumorigenicity
and radioresistance of glioblastoma. Oncol Rep. 38:3367–3375.
2017.PubMed/NCBI
|
|
56
|
Chen L, Zhuo D, Chen J and Yuan H:
Screening feature genes of lung carcinoma with DNA microarray
analysis. Int J Clin Exp Med. 8:12161–12171. 2015.PubMed/NCBI
|
|
57
|
Zhang Y, Foreman O, Wigle DA, Kosari F,
Vasmatzis G, Salisbury JL, Van Deursen J and Galardy PJ: USP44
regulates centrosome positioning to prevent aneuploidy and suppress
tumorigenesis. J Clin Invest. 122:4362–4374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cheng Y, Li K, Diao D, Zhu K, Shi L, Zhang
H, Yuan D, Guo Q, Wu X, Liu D and Dang C: Expression of KIAA0101
protein is associated with poor survival of esophageal cancer
patients and resistance to cisplatin treatment in vitro. Lab
Invest. 93:1276–1287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Michel L, Diaz-Rodriguez E, Narayan G,
Hernando E, Murty VV and Benezra R: Complete loss of the tumor
suppressor MAD2 causes premature cyclin B degradation and mitotic
failure in human somatic cells. Proc Natl Acad Sci USA.
101:4459–4464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon
DS, Wersto RP, Tully E, Wilsbach K and Gabrielson E: Increased
expression of mitotic checkpoint genes in breast cancer cells with
chromosomal instability. Clin Cancer Res. 12:405–410. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kim HS, Park KH, Kim SA, Wen J, Park SW,
Park B, Gham CW, Hyung WJ, Noh SH, Kim HK and Song SY: Frequent
mutations of human Mad2, but not Bub1, in gastric cancers cause
defective mitotic spindle checkpoint. Mutat Res. 578:187–201. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shi YX, Zhu T, Zou T, Zhuo W, Chen YX,
Huang MS, Zheng W, Wang CJ, Li X, Mao XY, et al: Prognostic and
predictive values of CDK1 and MAD2L1 in lung adenocarcinoma.
Oncotarget. 7:85235–85243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wordeman L, Wagenbach M and von Dassow G:
MCAK facilitates chromosome movement by promoting kinetochore
microtubule turnover. J Cell Biol. 179:869–879. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Duan H, Zhang X, Wang FX, Cai MY, Ma GW,
Yang H, Fu JH, Tan ZH, Fu XY, Ma QL, et al: KIF-2C expression is
correlated with poor prognosis of operable esophageal squamous cell
carcinoma male patients. Oncotarget. 7:80493–80507. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bie L, Zhao G, Wang YP and Zhang B:
Kinesin family member 2C (KIF2C/MCAK) is a novel marker for
prognosis in human gliomas. Clin Neurol Neurosurg. 114:356–360.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bidkhori G, Narimani Z, Hosseini Ashtiani
S, Moeini A, Nowzari-Dalini A and Masoudi-Nejad A: Reconstruction
of an integrated genome-scale co-expression network reveals key
modules involved in lung adenocarcinoma. PLoS One. 8:e675522013.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Song YJ, Tan J, Gao XH and Wang LX:
Integrated analysis reveals key genes with prognostic value in lung
adenocarcinoma. Cancer Manag Res. 10:6097–6108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
D'Archivio S and Wickstead B: Trypanosome
outer kinetochore proteins suggest conservation of chromosome
segregation machinery across eukaryotes. J Cell Biol. 216:379–391.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ju LL, Chen L, Li JH, Wang YF, Lu RJ, Bian
ZL and Shao JG: Effect of NDC80 in human hepatocellular carcinoma.
World J Gastroenterol. 23:3675–3683. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang L, Dou X, Liu T, Lu W, Ma Y and Yang
Y: Tumor size and lymph node metastasis are prognostic markers of
small cell lung cancer in a Chinese population. Medicine
(Baltimore). 97:e117122018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xu B, Wu DP, Xie RT, Liu LG and Yan XB:
Elevated NDC80 expression is associated with poor prognosis in
osteosarcoma patients. Eur Rev Med Pharmacol Sci. 21:2045–2053.
2017.PubMed/NCBI
|
|
72
|
Yuan W, Xie S, Wang M, Pan S, Huang X,
Xiong M, Xiao RJ, Xiong J, Zhang QP and Shao L: Bioinformatic
analysis of prognostic value of ZW10 interacting protein in lung
cancer. Onco Targets Ther. 11:1683–1695. 2018. View Article : Google Scholar : PubMed/NCBI
|