Introduction
Epidermal growth factor receptor (EGFR) is a
receptor tyrosine kinase found on the cell surface that is often
upregulated in tumor cells (1). EGFR
plays a pivotal role in cell proliferation by activating downstream
signaling pathways (2). Cetuximab, a
chimeric immunoglobulin G1 monoclonal antibody against EGFR, blocks
the function of EGFR by competitively antagonizing and/or
internalizing the receptor (3,4).
Clinical studies have demonstrated its efficacy in the treatment of
a number of different types of cancer, including advanced and
metastatic colorectal cancer (mCRC) (1,4).
Signaling from EGFR is relayed by a GTPase
transducer protein named RAS, and RAS-associated mutations in tumor
cells are associated with resistance to cetuximab treatment
(5,6). Therefore, the clinical use of cetuximab
is limited to patients with RAS wild-type mCRC. In addition, RAS is
not a sufficient biomarker for predicting tumor response, and
disease control is observed in only half of patients with KRAS
wild-type mCRC subjected to monotherapy as first- or later-line
treatment (7,8). Therefore, additional predictors of
tumor response to cetuximab are required in order to avoid poor
treatment efficacy with unnecessary adverse reactions.
Antibody-dependent, cell-mediated cytotoxicity
(ADCC) is proposed as a distinct mechanism of antitumor activity by
cetuximab, and thus has gathered attention as a potential predictor
of treatment efficacy and/or safety (9,10).
Cetuximab has an antigen-binding and crystalline fragment (Fc
fragment) in its structure (9,11)
allowing it to bind to both the tumor antigen (EGFR) and fragment C
γ receptor (FcγR) located on immune cells, and to trigger ADCC
(11). A histidine (H)/arginine (R)
polymorphism at position 131 on FcγR2A and a valine
(V)/phenylalanine (F) polymorphism at position 158 on FcγR3A are
associated with different affinities for human IgG (12). According to the accumulating
evidence, patients harboring FcγR2A-131H/H and FcγR3A-158V/V
mutations are expected to have stronger ADCC during monoclonal
antibody therapies (13–15).
Furthermore, higher EGFR expression levels due to
lower numbers of CA repeats in EGFR intron 1 may increase the
response to cetuximab (16,17). In addition, a substitution from R to
lysine (K) in codon 521 of the extracellular domain of EGFR could
result in lower ligand binding affinity, downregulation of the
target gene, and consequent favorable response to cetuximab
treatment (18,19). Despite these promising findings,
clinical studies remain scarce, and interpretations of the results
are conflicting due to several limiting factors. The present study
thus investigated the association between gene polymorphisms in
FcγR2A, FcγR3A and EGFR and the efficacy of first-line cetuximab
and oxaliplatin treatment in patients with extended RAS/BRAF
wild-type mCRC.
Materials and methods
Patients
The present study reviewed the clinical data of
patients participating in one of two trials evaluating the efficacy
of combination therapy with cetuximab and oxaliplatin-based
chemotherapy as a first-line treatment (UMIN 000003253 and
UMIN000007195) (20,21). The patients were recruited, and the
specimen was collected from 31 institutes in Japan between April
2010 and May 2011, and between February, 2012 and February, 2013.
These institutes included Chiba Cancer Center (Chiba, Japan);
Fukui-Ken Saiseikai Hospital (Fukui, Japan); Gifu University
Hospital (Gifu, Japan); Hokkaido Cancer Center (Sapporo, Japan);
Ishikawa Prefectural Central Hospital (Kanazawa, Japan); Japan
Community Health Care Organization (JCHO) Osaka Hospital (Osaka,
Japan); Kagawa University Hospital (Kita, Japan); Kanagawa Cancer
Center (Yokohama, Japan); Kanazawa Medical University Hospital
(Kahoku, Japan); Kansai Medical University Hospital (Hirakata,
Japan); Kitakyushu General Hospital (Kitakyushu, Japan); Kobe
Ekisaikai Hospital (Kobe, Japan); Kochi Medical School Hospital
(Nankoku, Japan); Matsunami General Hospital (Hashima, Japan);
Nakadori General Hospital (Akita, Japan); National Hospital
Organization Nagoya Medical Center (Nagoya, Japan); National
Hospital Organization Osaka National Hospital (Osaka, Japan); Osaka
City University Graduate School and Faculty of Medicine (Osaka,
Japan); Osaka General Medical Center (Osaka, Japan); Osaka Rosai
Hospital (Sakai, Japan); Osakakita Teishin Hospital (Osaka, Japan);
Rinku General Medical Center (Izumisano, Japan); Sakai City Medical
Center (Sakai, Japan); Sano Hospital (Kobe, Japan); Showa
University Fujigaoka Hospital (Yokohama, Japan); Teikyo University
Chiba Medical Center (Ichihara, Japan); Toyama Prefectural Central
Hospital (Toyama, Japan); University of Occupational and
Environmental Health (Kitakyushu, Japan); Toyonaka Municipal
Hospital (Toyonaka, Japan); Yamaguchi University Hospital (Ube,
Japan); and Yokoyama Hospital for Gastroenterological Diseases
(Nagoya, Japan). The primary endpoint of these two trials was
response rates (RRs) with confirmation, as evaluated by computed
tomography at 4- to 8-weekly intervals. The RR in the present study
was regarded as the clinically important primary endpoint as per
the previous two clinical trials (20,21). In
total, 90 patients, with RAS/BRAF wild-type mCRC were identified,
and the polymorphisms of these patients were analyzed. The mean age
of the patients was 66.3±9.9 years (standard deviation). The
present study was approved by the Institutional Review Board of
Yamaguchi University School of Medicine (approval number, H28-171)
and performed in accordance with the Declaration of Helsinki. The
requirement for informed consent was waived as the present study
was a retrospective analysis of previously collected samples and
data. The patients were given the opportunity to refuse the use of
their samples in the present study, according to the ethics
guidelines of the Institutional Review Board. Formalin-fixed,
paraffin-embedded (FFPE) samples collected as part of the previous
studies were used to analyze polymorphisms in FcγR2A, FcγR3A and
EGFR for the present study. The data used in the present study was
collected by Case Report Form for each clinical trial. No
additional data/samples were collected for the present study.
Evaluation of polymorphisms in FcγR2A,
FcγR3A and EGFR
DNA was extracted from the FFPE samples. Applying
micro-dissection on 10 µm sections, non-tumor tissues were
dissected from the FFPE samples. DNA extraction was performed using
a QIAamp DNA FFPE Tissue kit (Qiagen GmbH) according to the
manufacturer's protocol. The TaqMan technique was then used to
determine FcγR2A-H131R rs1801274, FcγR3A-V158F rs396991 and
EGFR-R521K rs2227983 polymorphisms using established primers
(22), TaqMan SNP Genotyping Assays
C—9077561_20, C—25815666_10 and C—16170352_20 (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and the TaqMan Genotyping Master
Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). In brief,
a 5-µl reaction solution, containing TaqMan Genotyping Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.), Assay Mix,
and 20–40 ng of genomic DNA diluted in dH2O, was
incubated in 384-well microtiter plates at 50°C for 2 min to
degrade dU-containing DNA, followed by incubation at 95°C for 10
min (denaturation), followed by 40 cycles of 15 sec at 95°C and 1
min of annealing and extension at 60°C. The ABI Prism 7900HT
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for
end-point reading of the fluorescence generated during PCR
amplification. EGFR CA Repeats in Intron 1 Genotyping was
determined via direct sequencing, as previously described (23,24).
Statistical analysis
The primary endpoint was RR and the secondary
endpoint was the maximum change in tumor diameter from baseline,
calculated using the formula (tumor diameter at evaluation-tumor
diameter at baseline)/tumor diameter at baseline ×100, whereby
negative numbers indicate tumor shrinkage during the treatment and
positive numbers indicate tumor enlargement.
The Eastern Cooperative Oncology Group Performance
Status (ECOG-PS) (25), combined
chemotherapy, patient sex and primary tumor sites were used as
variables that may potentially affect treatment efficacy. The
detailed information regarding tumor location in the colon (i.e.
left- or right-side colon) was not collected in the previous
clinical trials and was therefore unavailable in the present study.
In order to analyze the association between the tumor response and
variables, χ2 test was performed, followed by logistic
regression analysis. For the association between tumor shrinkage
and variables, Welch's t-test was performed, followed by multiple
linear regression analysis. P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses and
graph depiction were performed using Microsoft Excel (version 2013;
Microsoft Corporation) and KaleidaGraph 4.5 (version 4.5; Synergy
Software). The final figures were created using Photoshop CS2
(Adobe Systems).
Results
Frequency of polymorphisms and
mutation status
H/H in FcγR2A was the most frequent genetic
polymorphism observed in the present study (61.1%), while V/V in
FcγR3A was observed in only 12 patients (13.3%) (Table I). K/K in EGFR was observed in 33
patients (36.7%), while 27.8% of the patients had <36 CA repeats
in EGFR. In one patient, FcγR2A polymorphisms could not be
determined due to DNA fragmentation, and their data concerning
FcγR2A were excluded from further analysis.
 | Table I.Clinical characteristics and
frequencies of polymorphisms. |
Table I.
Clinical characteristics and
frequencies of polymorphisms.
| Variables | Number of patients
(%) |
|---|
| Sex |
|
|
Female | 34 (37.8) |
|
Male | 56 (62.2) |
| ECOG-PS |
|
| 0 | 79 (87.8) |
| 1 | 11 (12.2) |
| Treatment |
|
|
FOLFOX | 37 (41.1) |
|
CapeOX | 53 (58.9) |
| Primary tumor
site |
|
|
Colon | 51 (56.7) |
|
Rectum | 39 (43.3) |
| FcγR2A (H131R) |
|
|
H/H | 55 (61.1) |
|
H/R | 31 (34.4) |
|
R/R | 3 (3.3) |
| Not
determined | 1 (1.1) |
| FcγR3A (V158F) |
|
|
V/V | 12 (13.3) |
|
V/F | 35 (38.9) |
|
F/F | 43 (47.8) |
| EGFR (R521K) |
|
|
K/K | 33 (36.7) |
|
K/R | 39 (43.3) |
|
R/R | 18 (20.0) |
| EGFR (CA
repeat) |
|
|
<36 | 25 (27.8) |
|
≥36 | 65 (72.2) |
In terms of the mutation status, one of the two
clinical trials only recruited patients with KRAS wild-type CRC.
Therefore, the total rate of RAS/BRAF mutations could not be
assessed in this retrospective study.
RR
The univariate analysis demonstrated no significant
difference in RR between patients with and without the tested
polymorphisms (Table II).
Therefore, the odds ratio for tumor response was estimated using
all listed variables (Fig. 1). The
patients with an H/H polymorphism in FcγR2A (vs. non-H/H
polymorphism) had an odds ratio of 2.25, although this was not
statistically significant (P=0.09).
 | Table II.Univariate analysis for response
rate. |
Table II.
Univariate analysis for response
rate.
| Variables | CR or PR, n | RR, % | χ2 | P-value |
|---|
| Sex |
|
| 0.48 | 0.49 |
|
Female | 20 | 58.8 |
|
|
|
Male | 37 | 66.1 |
|
|
| ECOG-PS |
|
| 0.42 | 0.52 |
| 0 | 51 | 64.6 |
|
|
| 1 | 6 | 54.5 |
|
|
| Treatment |
|
| 0.06 | 0.80 |
|
FOLFOX | 24 | 64.9 |
|
|
|
CapeOX | 33 | 62.3 |
|
|
| Primary tumor
site |
|
| 0.33 | 0.57 |
|
Colon | 31 | 60.8 |
|
|
|
Rectum | 26 | 66.7 |
|
|
| FcγR2A (H131R) |
|
| 2.95 | 0.09 |
|
H/H | 39 | 70.9 |
|
|
|
Non-H/H | 18 | 52.9 |
|
|
| FcγR3A (V158F) |
|
| 0.07 | 0.80 |
|
V/V | 8 | 66.7 |
|
|
|
Non-V/V | 49 | 62.8 |
|
|
| EGFR (R521K) |
|
| 0.25 | 0.62 |
|
K/K | 22 | 66.7 |
|
|
|
Non-K/K | 35 | 61.4 |
|
|
| EGFR (CA
repeat) |
|
| 0.32 | 0.57 |
|
<36 | 17 | 68.0 |
|
|
|
≥36 | 40 | 61.5 |
|
|
Maximum change in tumor diameter from
baseline
The maximum change in tumor diameter from baseline
was used as a secondary endpoint in the present study to
investigate the influence of gene polymorphisms. As the present
study included patients who had received cetuximab in addition to
conventional cytotoxic chemotherapy, the maximum change in tumor
diameter from baseline (continuous scale) was considered to be a
more sensitive endpoint for measuring the association between
polymorphisms and treatment efficacy. As the tumor diameter
information was unavailable for one patient, the analyses were
performed using the data of 89 patients.
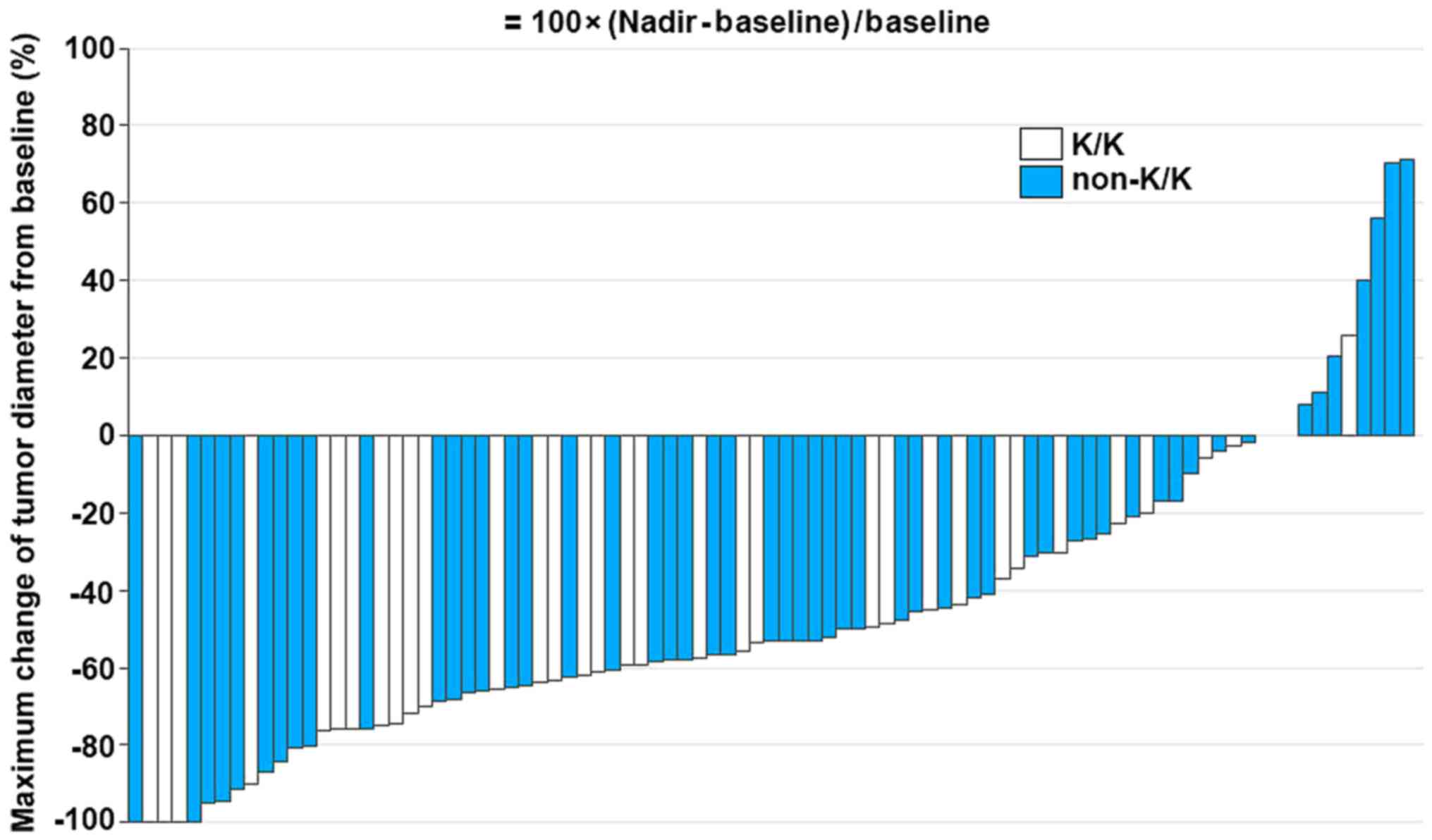
Notably, patients with a K/K polymorphism in the
EGFR gene exhibited greater tumor shrinkage compared with the
patients with K/R or R/R in the EGFR gene (P=0.04; Fig. 2). The multivariate analysis
demonstrated a significant association between the K/K polymorphism
in EGFR and tumor shrinkage [multiple linear regression analysis
estimate, −19.3; 95% confidence interval (CI), −35.5 to 3.0;
P=0.02] (Table III). Patients with
<36 CA repeats in the EGFR gene exhibited a tendency toward a
better tumor response (estimate, −16.9; 95% CI, −34.4 to 0.6;
P=0.06). Tumor size change at individual level was also compared
between patients with and without the K/K EGFR polymorphism
(Fig. 3), and was consistent with
the univariate analysis in that those with the polymorphism
exhibited greater tumor shrinkage than those without the gene
change. However, it should be noted that there were certain
patients that exhibited sufficient tumor shrinkage among patients
with K/R or R/R in the EGFR gene.
 | Figure 2.Degree of tumor size change and
univariate analysis. Maximum changes in tumor diameter from
baseline is presented according to the variables. Negative numbers
indicate tumor shrinkage, while positive numbers indicate tumor
enlargement. Enhanced tumor shrinkage was observed among the
patients with K/K polymorphisms in EGFR (R521K) compared with the
patients with no K/K polymorphism. EGFR, epidermal growth factor
receptor; CI, confidence interval; FcγR, fragment C γ receptor; H,
histidine; V, valine; K, lysine; R, arginine; F, phenylalanine; PS,
performance status. |
 | Table III.Multiple linear regression analysis
for maximum tumor change from baseline. |
Table III.
Multiple linear regression analysis
for maximum tumor change from baseline.
| Variables | Groups | Estimate (SE) | 95% CI | P-value |
|---|
| Sex | Male vs.
female | 1.6 (8.7) | −15.4, 18.6 | 0.85 |
| ECOG-PS | 1 vs. 0 | −0.8 (12.6) | −25.7, 24.2 | 0.95 |
| Treatment | FOLFOX vs.
CapeOX | −10 (8.2) | −26.4, 6.3 | 0.23 |
| Primary tumor
site | Colon vs.
rectum | 6.5 (8.1) | −9.7, 22.6 | 0.43 |
| FcγR2A (H131R) | H/H vs.
non-H/H | −6.2 (8) | −22.1, 9.7 | 0.44 |
| FcγR3A (V158F) | V/V vs.
non-V/V | −6.1 (11.6) | −29.2, 17 | 0.60 |
| EGFR (R521K) | K/K vs.
non-K/K | −19.3 (8.2) | −35.5, −3.0 | 0.02 |
| EGFR (CA
repeat) | <36 vs. ≥36 | −16.9 (8.8) | −34.4, 0.6 | 0.06 |
Discussion
Previous studies investigating the association
between polymorphisms of FcγR and cetuximab treatment had several
limitations. First, only mCRC harboring KRAS exon2 mutations were
excluded (10,26,27), and
thus the influence of other mutations, such as extended RAS and
BRAF, could not be ruled out. Secondly, a number of studies
analyzed the combined data of patients with substantially divergent
backgrounds, including line of the treatment, backbone of the
chemotherapy (oxaliplatin, irinotecan or monotherapy) and even
monotherapy (9,28). As discussed by Inoue et al
(22), the deteriorated systemic and
local immune systems in heavily treated patients could possibly
exert only limited antitumor activity mediated by ADCC; analyzing
these data without considering these factors may have led to
conflicting results. In contrast, the uniquely valuable
characteristic features of the present study are the exclusion of
patients with mCRC that exhibited BRAF or extended RAS mutations,
the inclusion of only first-line treatment regimens, and limiting
the backbone treatment to oxaliplatin and fluoropyrimidines.
Under these conditions, two results were revealed:
i) A clear association between the K/K polymorphism of EGFR and
maximum tumor shrinkage from baseline; and ii) a tendency toward
greater efficacy in tumors carrying the H/H polymorphism of FcγR2A.
The former result is partly consistent with previous suggestions of
an improved prognosis in patients with the K/K polymorphism
(18,19), including the observation that tumors
harboring K/K or K/R exhibited favorable tumor characteristics and
a higher RR to cetuximab combined chemotherapy in 112 patients with
KRAS wild-type colorectal carcinoma (18). Such a result could reflect attenuated
EGFR signaling and the higher sensitivity to signaling blockade by
cetuximab in patients with the R521K polymorphism (18). Unlike colorectal cancer, expression
of the K-allele in head and neck cancers has been associated with
shorter progression free survival (PFS) and resistance to
cetuximab, with stronger treatment required to induce K-alleles in
ADCC cells in vitro, due to lower affinity (29). Although no clear explanation has yet
emerged for these inconsistent observations, differing dependencies
on EGFR signaling among different tumor types and different degrees
of required antibody affinity for signal inhibition by cetuximab
are both possible underlying mechanisms (29). Further studies are required in order
to elucidate these aspects.
In contrast to the tumor shrinkage effects,
polymorphisms of FcγR2A, FcγR3A and EGFR had no statistically
significant association with tumor response.. Nevertheless, the
multivariate analysis demonstrated a tendency for an improved tumor
response in H/H tumors compared with non-H/H tumors. Specifically,
a H/R polymorphism at position 131 on FcγR2A was associated with
enhanced affinities for human IgG, and patients harboring
FcγR2A-131H/H mutations were predicted to have stronger ADCC
(13–15). A study using the data and samples
from patients receiving cetuximab monotherapy for colorectal cancer
demonstrated a significant association between efficacy of
late-line cetuximab monotherapy and an H/H polymorphism in FcγR2A
(10). The present study therefore
investigated whether combination oxaliplatin-based chemotherapy
could obscure the association between tested polymorphisms and RR,
as cytotoxic-doublet treatment is generally effective in 50% of
patients with mCRC. In addition, a number of the patients recruited
in the clinical trials assessed during the present study received a
hepatectomy with curative intent, which would significantly
influence PFS and overall survival.
The incidence of H/H polymorphisms in FcγR2A and V/V
in FcγR3A in the present study was 61 and 13%, respectively. A
previous study demonstrated that the incidence of H/H in FcγR2A was
higher among Japanese patients than patients from Europe and the
USA (9), and 61% in the present
study is consistent with this and other previous study (9,22). In
contrast, a 4–9% incidence rate of the V/V polymorphism in FcγR3A
has been demonstrated (9,21), suggesting a slightly higher incidence
of V/V in FcγR3A in the present study; however, the frequency of
polymorphisms of certain gene differs within Japan (30), and such variability could account for
the small differences in incidence between previous studies and the
present study. Although external validation was not performed in
the present study, the similarities in polymorphism frequencies and
the use of an established primers (22) and methods described above ensure
reliability in the laboratory evaluations.
A limitation of the present study was the potential
effect of polymorphisms on the treatment efficacy of cytotoxic
agents, such as oxaliplatin and fluoropyrimidines. Although this is
an unlikely outcome, the issue may be overcome by setting
FOLFOX/CapeOX alone as a control arm and demonstrating the lack of
polymorphism effects in this control group. Analyzing the data of
patients receiving cetuximab monotherapy would be an alternative
resolution, although cetuximab monotherapy is rarely utilized as a
first- or second-line treatment. Instead, monotherapy is often used
only in patients with deteriorated general status (31) or as a later treatment option, and in
both these cases the immune system and ADCC may not function as
expected. Another limitation was the lack of information regarding
the sidedness of the primary tumors. As sidedness of the primary
tumor is recognized as being significantly associated with the
efficacy of anti-EGFR therapy (32),
adding sidedness to the other clinical data may further clarify the
impact of polymorphisms.
In conclusion, the present study provides
preliminary evidence suggesting an association between treatment
efficacy and polymorphisms in the EGFR gene in patients with
RAS/BRAF wild-type mCRC. Individuals harboring the K/K polymorphism
in EGFR demonstrated significantly greater tumor shrinkage during
treatment than those with the non-K/K polymorphism. Further studies
with an appropriate control arm and endpoints of clinical
importance are necessary.
Acknowledgements
Not applicable.
Funding
The present study was supported, in part, by the
non-profit organization Epidemiological & Clinical Research
Organization (ECRIN).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
HMa, SH, SI, KO, JS, HMi and NN conceptualized the
study, curated the data and wrote the original draft of the
manuscript. KO, RT, NO, YS, TY, YN, NS, HN, JS, HMa and NN
performed the analyses, investigations and all methodology. SH, KO,
RT, NO, YS, TY, YN, JS, HMi and NN performed the project
administration. HMa, SH, JS and HMi wrote, reviewed and edited the
manuscript. HMi and NN supervised the investigations. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Yamaguchi University School of Medicine. The
requirement for informed consent was waived as the samples had been
collected as part of previous clinical trials. The patients were
given the opportunity to refuse that their samples be used in the
present study.
Patient consent for publication
Not applicable.
Competing interests
JS has received honoraria from Tsumura, Chugai
Pharmaceutical and consulting fees from Takeda Pharmaceutical. KO
has received honoraria (lecture and/or manuscript fees) from Takeda
Pharmaceutical Company Ltd., Bristol-Myers Squibb Company Ltd., Ono
Pharmaceutical Co. Ltd. and Chugai Pharmaceutical Co. Ltd. SH and
HN received research funding from NEC Corporation, Toyo Kohan
Corporation and Merck Serono Co., Ltd. HMis has received honoraria
from Chugai Pharmaceutical, Takeda Pharmaceutical and Taiho
Pharmaceutical. NN has received honoraria from Takeda
Pharmaceutical Company Ltd. The other co-authors declare that they
have no competing interests.
References
|
1
|
Kim ES, Khuri FR and Herbst RS: Epidermal
growth factor receptor biology (IMC-C225). Curr Opin Oncol.
13:506–513. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Woodburn JR: The epidermal growth factor
receptor and its inhibition in cancer therapy. Pharmacol Ther.
82:241–250. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Galizia G, Lieto E, De Vita F, Orditura M,
Castellano P, Troiani T, Imperatore V and Ciardiello F: Cetuximab,
a chimeric human mouse anti-epidermal growth factor receptor
monoclonal antibody, in the treatment of human colorectal cancer.
Oncogene. 26:3654–3660. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harris M: Monoclonal antibodies as
therapeutic agents for cancer. Lancet Oncol. 5:292–302. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chetty R and Govender D: Gene of the
month: KRAS. J Clin Pathol. 66:548–550. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao B, Wang L, Qiu H, Zhang M, Sun L,
Peng P, Yu Q and Yuan X: Mechanisms of resistance to anti-EGFR
therapy in colorectal cancer. Oncotarget. 8:3980–4000.
2017.PubMed/NCBI
|
|
7
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD,
Robitaille S, et al: K-ras mutations and benefit from cetuximab in
advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pessino A, Artale S, Sciallero S,
Guglielmi A, Fornarini G, Andreotti IC, Mammoliti S, Comandini D,
Caprioni F, Bennicelli E, et al: First-line single-agent cetuximab
in patients with advanced colorectal cancer. Ann Oncol. 19:711–716.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Geva R, Vecchione L, Kalogeras KT, Jensen
BV, Lenz HJ, Yoshino T, Paez D, Montagut C, Souglakos J, Cappuzzo
F, et al: FCGR polymorphisms and cetuximab efficacy in
chemorefractory metastatic colorectal cancer: An international
consortium study. Gut. 64:921–928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu G, Tu D, Lewis M, Cheng D, Sullivan
LA, Chen Z, Morgen E, Simes J, Price TJ, Tebbutt NC, et al: Fc-γ
receptor polymorphisms, cetuximab therapy, and survival in the NCIC
CTG CO.17 trial of colorectal cancer. Clin Cancer Res.
22:2435–2444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weiner LM, Surana R and Wang S: Monoclonal
antibodies: Versatile platforms for cancer immunotherapy. Nat Rev
Immunol. 10:317–327. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
van Sorge NM, van der Pol WL and van de
Winkel JG: FcgammaR polymorphisms: Implications for function,
disease susceptibility and immunotherapy. Tissue Antigens.
61:189–202. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Koene HR, Kleijer M, Algra J, Roos D, von
dem Borne AE and de Haas M: Fc gammaRIIIa-158V/F polymorphism
influences the binding of IgG by natural killer cell Fc gammaRIIIa,
independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood.
90:1109–1114. 1997.PubMed/NCBI
|
|
14
|
Dall'Ozzo S, Tartas S, Paintaud G, Cartron
G, Colombat P, Bardos P, Watier H and Thibault G:
Rituximab-dependent cytotoxicity by natural killer cells: Influence
of FCGR3A polymorphism on the concentration-effect relationship.
Cancer Res. 64:4664–4669. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Binstadt BA, Geha RS and Bonilla FA: IgG
Fc receptor polymorphisms in human disease: Implications for
intravenous immunoglobulin therapy. J Allergy Clin Immunol.
111:697–703. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Amador ML, Oppenheimer D, Perea S, Maitra
A, Cusatis G, Iacobuzio-Donahue C, Baker SD, Ashfaq R, Takimoto C,
Forastiere A and Hidalgo M: An epidermal growth factor receptor
intron 1 polymorphism mediates response to epidermal growth factor
receptor inhibitors. Cancer Res. 64:9139–9143. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Buerger H, Gebhardt F, Schmidt H, Beckmann
A, Hutmacher K, Simon R, Lelle R, Boecker W and Brandt B: Length
and loss of heterozygosity of an intron 1 polymorphic sequence of
egfr is related to cytogenetic alterations and epithelial growth
factor receptor expression. Cancer Res. 60:854–857. 2000.PubMed/NCBI
|
|
18
|
Hsieh YY, Tzeng CH, Chen MH, Chen PM and
Wang WS: Epidermal growth factor receptor R521K polymorphism shows
favorable outcomes in KRAS wild-type colorectal cancer patients
treated with cetuximab-based chemotherapy. Cancer Sci. 103:791–796.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gonçalves A, Esteyries S, Taylor-Smedra B,
Lagarde A, Ayadi M, Monges G, Bertucci F, Esterni B, Delpero JR,
Turrini O, et al: A polymorphism of EGFR extracellular domain is
associated with progression free-survival in metastatic colorectal
cancer patients receiving cetuximab-based treatment. BMC Cancer.
8:1692008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Soda H, Maeda H, Hasegawa J, Takahashi T,
Hazama S, Fukunaga M, Kono E, Kotaka M, Sakamoto J, Nagata N, et
al: Multicenter phase II study of FOLFOX or biweekly XELOX and
Erbitux (cetuximab) as first-line therapy in patients with
wild-type KRAS/BRAF metastatic colorectal cancer: The FLEET study.
BMC Cancer. 15:6952015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hazama S, Maeda H, Iwamoto S, Kim HM,
Takemoto H, Kobayashi K, Sakamoto J, Nagata N, Oba K and Mishima H:
A phase II study of XELOX and cetuximab as first-line therapy in
patients with KRAS wild type metastatic colorectal cancer (FLEET2
Study). Clin Colorectal Cancer. 15:329–336. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Inoue Y, Hazama S, Iwamoto S, Miyake Y,
Matsuda C, Tsunedomi R, Okayama N, Hinoda Y, Yamasaki T, Suehiro Y,
et al: FcγR and EGFR polymorphisms as predictive markers of
cetuximab efficacy in metastatic colorectal cancer. Mol Diagn Ther.
18:541–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Okayama N, Nishioka M, Hazama S, Sakai K,
Suehiro Y, Maekawa M, Sakamoto J, Iwamoto S, Kato T, Mishima H, et
al: The importance of evaluation of DNA amplificability in KRAS
mutation testing with dideoxy sequencing using formalin-fixed and
paraffin-embedded colorectal cancer tissues. Jpn J Clin Oncol.
41:165–171. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maruta Y, Okayama N, Hiura M, Suehiro Y,
Hirai H and Hinoda Y: Determination of ancestral allele for
possible human cancerassociated polymorphisms. Cancer Genet
Cytogenet. 180:24–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the Eastern Cooperative Oncology Group. Am J Clin
Oncol. 5:649–655. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pander J, Gelderblom H, Antonini NF, Tol
J, van Krieken JH, van der Straaten T, Punt CJ and Guchelaar HJ:
Correlation of FCGR3A and EGFR germline polymorphisms with the
efficacy of cetuximab in KRAS wild-type metastatic colorectal
cancer. Eur J Cancer. 46:1829–1834. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kjersem JB, Skovlund E, Ikdahl T, Guren T,
Kersten C, Dalsgaard AM, Yilmaz MK, Fokstuen T, Tveit KM and Kure
EH: FCGR2A and FCGR3A polymorphisms and clinical outcome in
metastatic colorectal cancer patients treated with first-line
5-fluorouracil/folinic acid and oxaliplatin +/- cetuximab. BMC
Cancer. 14:3402014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park SJ, Hong YS, Lee JL, Ryu MH, Chang
HM, Kim KP, Ahn YC, Na YS, Jin DH, Yu CS, et al: Genetic
polymorphisms of FcγRIIa and FcγRIIIa are not predictive of
clinical outcomes after cetuximab plus irinotecan chemotherapy in
patients with metastatic colorectal cancer. Oncology. 82:83–89.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Braig F, Kriegs M, Voigtlaender M, Habel
B, Grob T, Biskup K, Blanchard V, Sack M, Thalhammer A, Ben Batalla
I, et al: Cetuximab Resistance in Head and Neck Cancer Is Mediated
by EGFR-K521 Polymorphism. Cancer Res. 77:1188–1199. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kobayashi M, Hazama S, Takahashi K, Oba K,
Okayama N, Nishioka M, Hinoda Y, Oka M, Okamoto K, Maeda H, et al:
Is there diversity among UGT1A1 polymorphism in Japan? World J
Gastrointest Oncol. 4:170–175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Van Cutsem E, Cervantes A, Adam R, Sobrero
A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson
A, Bodoky G, et al: ESMO consensus guidelines for the management of
patients with metastatic colorectal cancer. Ann Oncol.
27:1386–1422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tejpar S, Stintzing S, Ciardiello F,
Tabernero J, Van Cutsem E, Beier F, Esser R, Lenz HJ and Heinemann
V: Prognostic and predictive relevance of primary tumor location in
patients with RAS wild-type metastatic colorectal cancer:
Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA
Oncol. 3:194–201. 2017. View Article : Google Scholar : PubMed/NCBI
|