Introduction
Malignancies of the central nervous system represent
one of the most serious health burdens worldwide, accounting for
1.9% of newly diagnosed cancer cases and ~2.3% of cancer-associated
deaths worldwide (1). Glioma, the
most prevalent of the malignancies of the central nervous system,
has been extensively studied due to its poor outcome and the lack
of effective treatment (2). It is
clinicopathologically classified into four grades, including
pilocytic astrocytoma (grade I), diffuse astrocytoma (grade II),
anaplastic astrocytoma (grade III), and glioblastoma (GBM; grade
IV), according to the World Health Organization (WHO) grading
criteria (3). Among these grades,
GBM (WHO grade IV) is considered the most common type and is
characterized by high invasion rates (4,5). The
significant biological heterogeneity of glioma hinders the
understanding of the molecular mechanisms in tumor pathogenesis
(6,7). Currently, the 5-year survival rate of
patients with glioma has improved as a result of the advances in
therapeutic strategies, such as surgical procedures, radiotherapy
and chemotherapy. However, the prognosis of patients with advanced
stage glioma remains poor (8,9).
Therefore, the development of novel approaches is required to
improve early diagnosis, prognosis and treatment of patients with
glioma.
Accumulating evidence highlights the important roles
of microRNAs (miRs/miRNAs) in the pathogenesis of multiple types of
human cancer (10). Therefore,
miRNAs have attracted increasing attention due to their significant
diagnostic and prognostic values in patients with various types of
cancer (11,12). These small non-coding RNAs play
pivotal roles in biological processes by regulating the expression
of key genes at the post-transcriptional level (13). It is reported that miRNAs are stable
in serum and can be extracted from clinical specimens easily, thus
showing potential as diagnostic tools in the clinic (14). In addition, the abnormally expressed
miRNAs in tumors are involved in tumor progression, through
downregulation of oncogenes or tumor suppressors, and thus act as
therapeutic targets in cancer treatment (15,16).
Thus, the clinical significance of novel aberrant miRNAs in the
treatment of glioma was investigated. miR-193b has been
investigated in several types of human carcinoma, such as liver,
gastric and colorectal carcinoma (17–19).
This miR was demonstrated to enhance cell proliferation and
suppress cell apoptosis in liver cancer cells (17). In gastric cancer, overexpression of
miR-193b in tumor cells resulted in the inhibition of cell
proliferation, migration and invasion (18). In colorectal cancer, the aberrant
expression of miR-193b in tumor tissues was determined as a
prognostic biomarker for predicting overall survival in patients
(19). A study by Zhong et al
(20) demonstrated that miR-193b in
glioma cells led to increased cell proliferation. However, the
understanding of the clinical significance of miR-193b and its
functional role in the progression of glioma remains limited.
To investigate the role of miR-193b in glioma
further, the present study sought to assess the expression of
miR-193b in glioma serum, tissues and cells, evaluate its clinical
significance in diagnosis and prognosis, and explore its effects on
glioma cell function.
Materials and methods
Patients and specimens collection
A total of 122 patients who were histologically
diagnosed with glioma in Heze Municipal Hospital between June 2008
and May 2012 were recruited for the present study. The inclusion
criteria for the glioma patients were as follows: i) Pathologically
diagnosed as glioma; ii) had no preoperative therapy. The exclusion
criteria were as follows: i) systemic infection; ii) diagnosed with
other tumors. The present study also included 68 healthy
volunteers, who received routine physical examination and had no
history of malignancy. A volume of 5 ml venous blood was obtained
from the participants prior to surgical resection for the patients.
The blood samples were placed in EDTA tubes and were used for serum
isolation by centrifugation with 955.9 × g for 5 min at 4°C. Glioma
tissues and adjacent normal tissues, which were defined as 1 cm
away from the lesions, were collected from the 122 patients during
the surgery, and immediately frozen in liquid nitrogen for further
use. Demographic and clinicopathological characteristics, including
gender, age, tumor size, WHO grade (21) and Karnofsky Performance Scale (KPS)
(22) were recorded and are listed
in Table I. All patients were
enrolled on a 5-year follow-up survey, and their survival data were
obtained for the survival analysis. This study was approved by the
Ethics Committee of Heze Municipal Hospital, and each participant
provided written informed consent.
 | Table I.Association between miR-193b
expression level and clinicopathological features of patients with
glioma. |
Table I.
Association between miR-193b
expression level and clinicopathological features of patients with
glioma.
|
|
| Serum miR-193b
level |
| Tissue miR-193b
level |
|
|---|
|
|
|
|
|
|
|
|---|
| Features | Patients, n
(n=122) | Low (n=60) | High (n=62) | P-value | Low (n=58) | High (n=64) | P-value |
|---|
| Sex |
|
|
| 0.483 |
|
| 0.601 |
|
Female | 45 | 24 | 21 |
| 20 | 25 |
|
|
Male | 77 | 36 | 41 |
| 38 | 39 |
|
| Age, years |
|
|
| 0.910 |
|
| 0.458 |
|
≤50 | 34 | 17 | 17 |
| 18 | 16 |
|
|
>50 | 88 | 43 | 45 |
| 40 | 48 |
|
| Tumor size, cm |
|
|
| 0.570 |
|
| 0.088 |
| ≤3 | 68 | 35 | 33 |
| 37 | 31 |
|
|
>3 | 54 | 25 | 29 |
| 21 | 33 |
|
| WHO grade |
|
|
| 0.030 |
|
| 0.004 |
|
I–II | 59 | 35 | 24 |
| 36 | 23 |
|
|
III–IV | 63 | 25 | 38 |
| 22 | 41 |
|
| KPS |
|
|
| 0.016 |
|
| 0.001 |
|
≤90 | 83 | 47 | 36 |
| 48 | 35 |
|
|
>90 | 39 | 13 | 26 |
| 10 | 29 |
|
Cell culture and transfection
Glioma cell lines including T98G, A172 and LN229,
and a normal human astrocyte cell line UC2 were purchased from the
Type Culture Collection of the Chinese Academy of Sciences. Glioma
U87 cell line of undetermined origin was obtained from the American
Type Culture Collection (cat. no. ATCC® HTB-14™). All
cells were cultured in Dulbecco's modified Eagle's medium (DMEM),
supplemented with 10% FBS (both Gibco; Thermo Fisher Scientific,
Inc.), in a humidified atmosphere at 5% CO2 and
37°C.
The expression of miR-193b in glioma cells was
modulated by transfecting cells with 50 nM miR-193b mimic
(5′-AACUGGCCCUCAAAGUCCCGCU-3′), 100 nM miR-193b inhibitor
(5′-AGCGGGACUUUGAGGGCCAGUU-3′) and 50 nM corresponding negative
controls mimic NC (5′-ACUACUGAGUGACAGUAGA-3′) and 100 nM inhibitor
NC (5′-CAGUACUUUUGUGUAGUACAA-3′) (all from Shanghai GenePharma Co.,
Ltd.), using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). The subsequent cell experiments were
performed 48 h after the cell transfection.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from glioma serum samples,
tissues and cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). RT was conducted to synthesize cDNA from 1 µg
RNA, using a PrimeScript RT reagent kit (Takara Bio, Inc.)
following the manufacturer's instructions with a following reaction
condition: 42°C for 30 min, 85°C for 5 sec. The cDNA was
subsequently used as the template for qPCR, which was carried out
to evaluate the expression levels of miR-193b using a SYBR-Green I
Master Mix kit (Invitrogen; Thermo Fisher Scientific, Inc.) and the
7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The reactions in this analysis used U6 as an
internal control gene, and the thermocycling conditions were as
follows: 95°C for 10 min, 95°C for 30 sec, 60°C for 20 sec, 72°C
for 15 sec, for a total of 40 cycles. The oligonucleotide primer
sequences were as follows: miR-193b forward,
3′-GCGCAACTGGCCCTCAAAG-5′; miR-193b reverse,
3′-CAGTGCAGGGTCCGAGGT-5′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′; and U6 reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. The final relative miR-193b
expression was calculated using the 2−ΔΔCq method
(23) and normalized to U6.
Cell proliferation analysis
Following cell transfection, the effect of miR-193b
on the proliferation of glioma cell was examined using the Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.), as
per the manufacturer's protocols. The stable transfected cells were
seeded into 96-well plates with a cell density of 2×105
cells/well, and then maintained in a humidified incubator at 37°C
for 3 days. At the culture times of 0, 24, 48 and 72 h, CCK-8
reagent was added to the wells and incubated further for 2 h. The
cell viability was measured by reading the absorbance at a
wavelength of 450 nm, using a micro-plate analyzer (Bio-Rad
Laboratories, Inc.).
Cell migration and invasion
analysis
To explore the effect of miR-193b on cell migration
and invasion, Transwell chambers with 8-µm pore size (Corning,
Inc.) were used. The chambers were coated with Matrigel for the
invasion assay, and those without Matrigel were used for the
migration assay. Stably transfected cells (2×105) were
seeded in the upper chamber in serum-free DMEM. The lower chambers
contained DMEM, with 10% FBS as the chemoattractant. After 48 h of
incubation, the cells in the lower chamber were fixed with 4%
paraformaldehyde for 10 min at room temperature and stained with
0.1% crystal violet for 20 min at room temperature. The cells were
then counted using a light microscope (Olympus Corporation) at
magnification, ×200. All the experiments were performed in
triplicate.
Western blot analysis
Proteins were extracted from the cells using RIPA
lysis buffer (Thermo Fisher Scientific, Inc.), and the BCA method
was used to examine the concentration of proteins. Proteins (20 µg
per lane) were separated via SDS-PAGE (10% gel) and transferred
onto PVDF membranes (Merck KGaA). The membranes were incubated with
primary antibodies against matrix metalloproteinase (MMP)-2
(1:1,000; cat. no. 4022S; Cell Signaling Technology, Inc.) and
MMP-9 (1:1,000; cat. no. 2270S; Cell Signaling Technology, Inc.)
overnight at 4°C, after 4 h of blocking with 5% non-fat milk at
room temperature. Horseradish peroxidase-labeled secondary
antibodies (1:2,000; cat. no. A0192; Beyotime Institute of
Biotechnology) were incubated at room temperature for 4 h. β-actin
was used as an internal control with an anti-β-actin (1:1,000; cat.
no. AA128; Beyotime Institute of Biotechnology). The proteins were
detected using an enhanced chemiluminescence reagent (Seven Sea
Biotech), and were visualized using the Bio-Rad gel imaging system
(Beijing Thmorgan Biotechnology, Inc.). The quantitative analysis
for the proteins was performed using the IPP software (version 7.0;
Media Cybernetics, Inc.).
Statistical analysis
Data in this study are presented as the mean ± SD
and were analyzed using SPSS version 18.0 (IBM Corp.) and GraphPad
Prism version 5.0 software (GraphPad Software, Inc.). The
differences between groups were assessed using Student's t-test and
one-way ANOVA with Tukey's multiple comparison test. Association
analysis between miR-193b and clinicopathological data was
performed using the χ2 test. A receiver operating
characteristic (ROC) curve was plotted to evaluate the diagnostic
value of miR-193b, based on its serum expression levels.
Kaplan-Meier survival and Cox regression analyses were adopted to
examine the prognostic value of miR-193b. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of miR-193b in glioma
samples
To further understand the role of miR-193b in
glioma, its expression in glioma samples and cells lines was
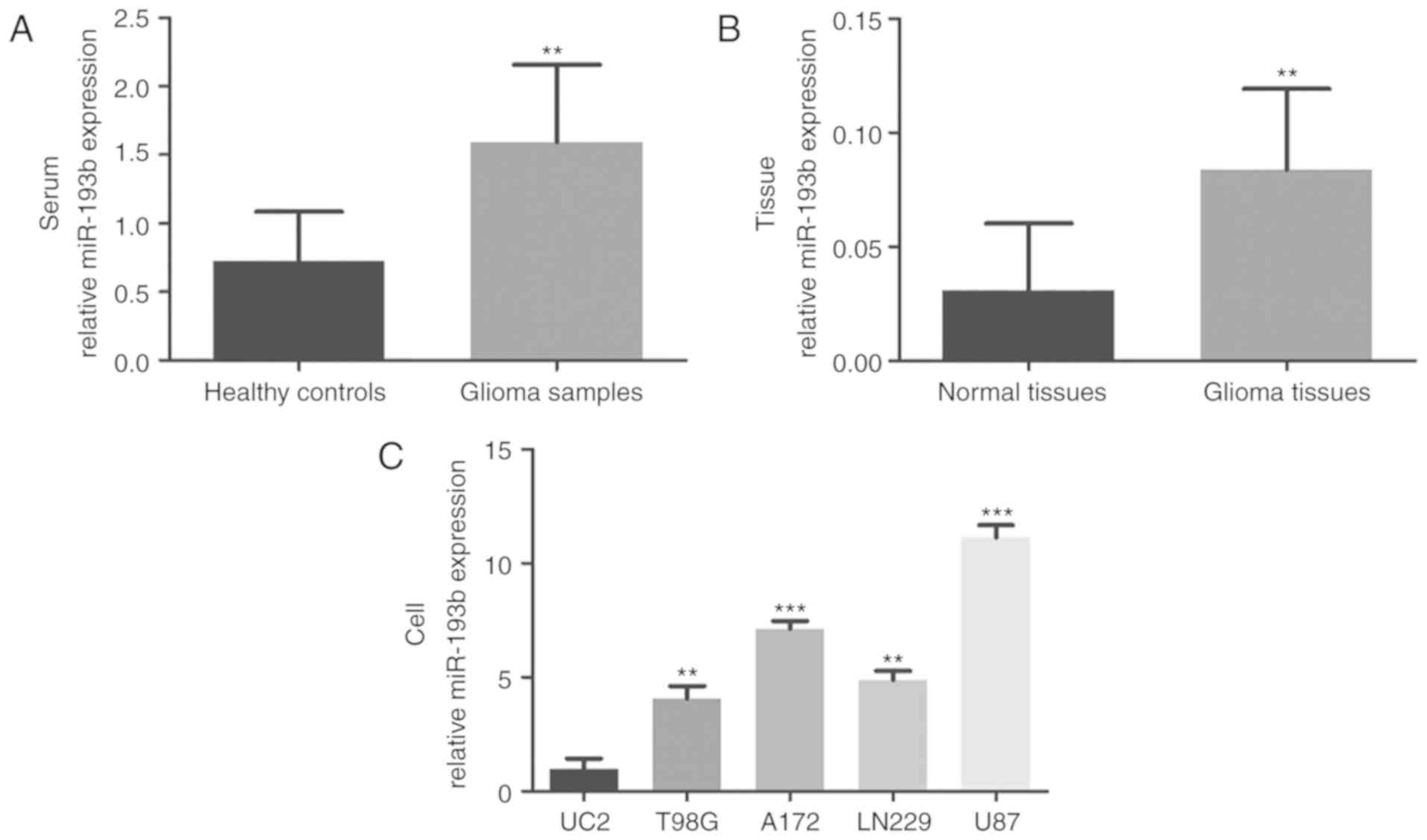
quantified by RT-qPCR. As shown in Fig.
1A, the serum expression of miR-193b was significantly
upregulated in patients with glioma compared with the healthy
controls (P<0.01). Consistently, increased miR-193b expression
was also observed in glioma tissues compared with the adjacent
normal tissues (P<0.01; Fig. 1B).
To confirm these findings, the levels of miR-193b in glioma cell
lines were also investigated, which demonstrated higher expression
of miR-193b in four glioma cell lines than in the normal human
astrocyte UC2 cells (all P<0.01; Fig.
1C).
Association between miR-193b
expression and clinicopathological features of patients with
glioma
The present study subsequently explored the role of
miR-193b in the development of glioma, by analyzing the association
between miR-193b and the clinical data of patients. Firstly, the
mean expression value of miR-193b (serum, 1.594; tissue, 0.084) was
used as the cutoff values to classify the patients into low and
high miR-193b expression groups. The findings from this analysis,
summarized in Table I, revealed that
the expression of miR-193b (in serum and tissues) was associated
with WHO grade (serum, P=0.030; tissue, P=0.004) and KPS (serum,
P=0.016; tissue, P=0.001) of patients. In contrast, no association
was found between miR-193b expression and other parameters, such as
gender, age and tumor size (all P>0.05).
Clinical significance of miR-193b in
the diagnosis and prognosis of glioma
The diagnostic and prognostic value of miR-193b
expression was evaluated in patients with glioma. The ROC curve was
generated, based on the serum levels of miR-193b in the patients
(Fig. 2A), and demonstrated that
miR-193b had high diagnostic value with an area under the curve of
0.903. The sensitivity was 79.5% and the specificity was 86.8% at
the cut-off value of 1.155, which represented an optimal relative
expression value of miR-193b to distinguish glioma patients from
healthy individuals.
Kaplan-Meier survival curves were plotted and Cox
regression analysis was performed to assess the prognostic value of
miR-193b. The expression of miR-193b in glioma tissues was used for
these analyses. As shown in Fig. 2B,
patients with high expression of miR-193b had low overall survival
compared with those with low miR-193b expression (P=0.002).
Furthermore, the univariate and multivariate Cox regression
analysis indicated that miR-193b was associated with the prognosis
of glioma and served as an independent prognostic factor of overall
survival in glioma (P=0.004; HR, 2.877; Table II).
 | Table II.Cox regression analysis of miR-193b
expression and clinicopathological features in patients with
glioma. |
Table II.
Cox regression analysis of miR-193b
expression and clinicopathological features in patients with
glioma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| miR-193b | 2.665 | 1.391–5.107 | 0.003 | 2.877 | 1.410–5.872 | 0.004 |
| Gender | 1.001 | 0.541–1.853 | 0.997 | 1.156 | 0.607–2.202 | 0.660 |
| Age | 1.502 | 0.721–3.126 | 0.277 | 1.298 | 0.610–2.762 | 0.498 |
| Tumor size | 1.203 | 0.666–2.173 | 0.541 | 1.140 | 0.620–2.096 | 0.672 |
| WHO grade | 2.284 | 1.211–4.138 | 0.008 | 1.798 | 0.951–3.399 | 0.071 |
| KPS | 2.166 | 1.199–3.915 | 0.010 | 1.687 | 0.898–3.171 | 0.104 |
Effects of miR-193b on the
proliferation of glioma cells
The current study conducted experiments on cells to
confirm the functional role of miR-193b in the progression of
glioma. Following stable transfection with a mimic or an inhibitor,
miR-193b expression was overexpressed or knocked down,
respectively, in A172 and U87 cells compared with the corresponding
negative controls (all P<0.01; Fig.
3A and B). The cell proliferation assay revealed that
overexpression of miR-193b contributed to increased glioma cell
proliferation, whereas the inhibition of miR-193b expression
suppressed the proliferation of the cells (all P<0.05, Fig. 3C and D).
Effects of miR-193b on the migration
and invasion of glioma cells
In addition to cell proliferation, the regulatory
effects of miR-193b on glioma cell migration and invasion were also
analyzed in A172 and U87 cells. Migration and invasion of glioma
A172 cells were promoted following miR-193b overexpression, whereas
they were suppressed following miR-193b inhibition (all P<0.05;
Fig. 4A-C). In addition, the
proteins involved in cell migration and invasion were examined, and
it was found that the protein levels of MMP-2 and MMP-9 were
increased following the overexpression of miR-193b and decreased
following its inhibition in A172 cells (all P<0.05; Fig. 4D and E). The migration and invasion
results were also obtained in another glioma U87 cell line, which
exhibited similar regulatory trends on cell migration and invasion
(all P<0.05; Fig. 5A-C) and the
associated protein levels (P<0.05; Fig. 5D and E) in U87 cells.
Discussion
Emerging studies have indicated that miRNAs are
involved in pivotal events during the pathogenesis of various human
diseases (24). Numerous oncogenes
and tumor suppressors are regulated by these miRNAs at the
post-transcriptional levels (25).
The biological function of miRNAs in cancer has received increased
attention as a growing number of miRNAs with abnormal expression
patterns in tumors have been observed (26). These deregulated miRNAs serve as
critical regulators in tumor progression, and can be used as
biomarkers for cancer diagnosis and prognosis due to their ectopic
expression patterns and stability in serum specimens (27,28). For
example, Long et al (29)
found that miR-124-3p was decreased in hepatocellular carcinoma
samples and predicted prognosis in patients with this malignancy.
Another study by Gao et al (14) indicated that serum miR-27a could act
as a diagnostic biomarker for the screening of patients with
prostate cancer, and demonstrated its effect of promoting tumor
cell proliferation. Hence, novel miRNAs, which can accurately
screen cancer cases and predict prognosis in patients are required
to improve the treatment of malignancies in humans.
Certain miRNAs have been investigated in glioma, and
their clinical significance and functional roles were explored in
previous studies. For instance, miR-424 expression was
downregulated in glioma tissues and cell lines, and participated in
the regulation of tumor cell migration, invasion and
epithelial-mesenchymal transition (EMT) by targeting the kinesin
family member 23 gene (30). The
dysregulation of miR-129-5p in glioma was demonstrated to suppress
glioma cell proliferation and cell cycle by regulating the DNA
methyltransferase 3 α gene (31).
Decreased expression of miR-218-5p was reported in glioma tissues
and cells, leading to enhanced cell proliferation, invasion and EMT
progression (32). Increased
expression of miR-423-3p in glioma predicted poor prognosis, and
promoted glioma progression (33).
The expression of miR-599 was decreased in glioma tissues compared
with normal controls, and was proved to be involved in the
progression of glioma (34). In the
present study, significantly increased expression of miR-193b was
observed in glioma serum, tissues and cell lines. Thus, it is
postulated that miR-193b may play a pivotal role in glioma.
Previous studies demonstrated that miR-193b
expression was altered in certain types of cancer (17–19). In
esophageal squamous cell carcinoma, serum miR-193b expression was
downregulated and served as a promising biomarker for the
prediction of chemoradiation sensitivity (35). In contrast, the increased expression
of miR-193b was detected in colon cancer tissues, and led to
increased cell proliferation and decreased cell apoptosis in cells
(36). Thus, the expression of
miR-193b varies depending on the type of cancer. In glioma, a study
reported elevated expression of miR-193b in both glioma tissues and
cells (20). Similarly, the present
study found upregulated expression of miR-193b in glioma serum
samples, tissues and cell lines compared with the corresponding
normal controls. Therefore, it is postulated that miR-193b may act
as an oncogene in glioma. Additionally, the association between
miR-193b expression and clinicopathological characteristics of
patients with glioma was further assessed to preliminarily analyze
the role of miR-193b in the development of glioma. As expected, the
expression of miR-193b was associated with WHO grade and KPS,
suggesting that miR-193b may be involved in tumor development.
Given the dysregulation of miR-193b in glioma
samples, its clinical significance in glioma diagnosis and
prognosis was analyzed. Serum miRNA levels are generally considered
as convenient diagnostic tools in cancer (37). Thus, a ROC analysis was performed,
based on the serum expression of miR-193b. The results revealed
miR-193b as a potential diagnostic biomarker with high sensitivity
and specificity. In addition, the prognostic value of miR-193b was
evaluated based on the 5-year survival information of the patients
with glioma. The Kaplan-Meier survival curves indicated that
patients with high miR-193b expression had poor overall survival
compared with those with low expression. Furthermore, miR-193b was
independently associated with the overall survival, which implied
that miR-193b may serve as an independent prognostic biomarker in
patients with glioma.
To discover the biological functions of miR-193b in
glioma progression, the effects of miR-193b on proliferation,
migration and invasion of glioma cells were investigated. The
expression of miR-193b was successfully upregulated by an miR-193b
mimic, and downregulated by an miR-193b inhibitor. The upregulation
of miR-193b was found to promote proliferation, whereas the
inhibition of miR-193b in glioma cells had the opposite effect. In
addition, similar regulatory effects of miR-193b were observed on
the glioma cell migration and invasion, evidenced by the increased
migratory and invading cells and the expression of key proteins
involved in these processes.
Overall, the findings of the present study suggest
that miR-193b may function as an oncogene in the tumor progression
of glioma. However, the molecular mechanisms underlying the role of
miR-193b in glioma remain unclear. A previous study by Zhong et
al (20) indicated that miR-193b
promoted glioma cell proliferation by regulating SMAD family member
3 (SMAD3). Thus, we suspected that the effects of miR-193b on cell
migration and invasion may also be achieved by targeting SMAD3.
Finally, no in vivo experiments were conducted, which is a
limitation of the present study. Therefore, further investigations
are needed to confirm the role of miR-193b in glioma in vivo
and to explore its precise mechanisms.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
MZ and JZ designed the present study and were
responsible for writing and revising the manuscript. WZ collected
the data and performed the clinical research. HZ performed the cell
experiments and data analysis.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Heze Municipal Hospital (approval no. 20070923), and
each participant provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pastuszak Z, Tomczykiewicz K,
Piusinska-Macoch R, Stępień A and Kordowska J: The occurrence of
tumors of the central nervous system in a clinical observation. Pol
Merkur Lekarski. 38:88–92. 2015.PubMed/NCBI
|
|
2
|
Ho VK, Reijneveld JC, Enting RH, Bienfait
HP, Robe P, Baumert BG and Visser O; Dutch Society for
Neuro-Oncology (LWNO), : Changing incidence and improved survival
of gliomas. Eur J Cancer. 50:2309–2318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Malzkorn B and Reifenberger G: Practical
implications of integrated glioma classification according to the
World Health Organization classification of tumors of the central
nervous system 2016. Curr Opin Oncol. 28:494–501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Olar A and Aldape KD: Using the molecular
classification of glioblastoma to inform personalized treatment. J
Pathol. 232:165–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang L, Liu J, Yu X, Shi L, Liu J, Xiao H
and Huang Y: Drug-drug interactions between moxifloxacin and
rifampicin based on pharmacokinetics in vivo in rats. Biomed
Chromatogr. 30:1591–1598. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Panditharatna E, Yaeger K, Kilburn LB,
Packer R and Nazarian J: Clinicopathology of diffuse intrinsic
pontine glioma and its redefined genomic and epigenomic landscape.
Cancer Genet. 208:367–373. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stoltz K, Sinyuk M, Hale JS, Wu Q, Otvos
B, Walker K, Vasanji A, Rich JN, Hjelmeland AB and Lathia JD:
Development of a Sox2 reporter system modeling cellular
heterogeneity in glioma. Neuro Oncol. 17:361–371. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang H, Song X, Huang Q, Xu T, Yun D, Wang
Y, Hu L, Yan Y, Chen H, Lu D and Chen J: LGALS3 promotes treatment
resistance in glioblastoma and is associated with tumor risk and
prognosis. Cancer Epidemiol Biomarkers Prev. 28:760–769. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Zhu Y, Cheng H, Zhang J, Zhu Y,
Chen H, Chen L, Qi H, Ren G, Tanget J, et al: The increased
expression of estrogen related receptor α correlates with wnt5a and
poor prognosis in patients with glioma. Mol Cancer Ther.
152018.(18): 173–184, 2019.
|
|
10
|
Wang WT and Chen YQ: Circulating miRNAs in
cancer: From detection to therapy. J Hematol Oncol. 7:862014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Harrandah AM, Mora RA and Chan EKL:
Emerging microRNAs in cancer diagnosis, progression, and immune
surveillance. Cancer Lett. 438:126–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Hu J, Pan L, Yin X, Wang Q and
Chen H: Diagnostic and prognostic value of serum miR-99a expression
in oral squamous cell carcinoma. Cancer Biomark. 23:333–339. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deng YH, Deng ZH, Hao H, Wu XL, Gao H,
Tang SH and Tang H: MicroRNA-23a promotes colorectal cancer cell
survival by targeting PDK4. Exp Cell Res. 373:171–179. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao W, Hong Z, Huang H, Zhu A, Lin S,
Cheng C, Zhang X, Zou G and Shi Z: miR-27a in serum acts as
biomarker for prostate cancer detection and promotes cell
proliferation by targeting Sprouty2. Oncol Lett. 16:5291–5298.
2018.PubMed/NCBI
|
|
15
|
Xiang J, Wu Y, Li DS, Wang ZY, Shen Q, Sun
TQ, Guan Q and Wang YJ: miR-584 suppresses invasion and cell
migration of thyroid carcinoma by regulating the target oncogene
ROCK1. Oncol Res Treat. 38:436–440. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kong Q, Han J, Deng H, Wu F, Guo S and Ye
Z: miR-431-5p alters the epithelial-to-mesenchymal transition
markers by targeting UROC28 in hepatoma cells. Onco Targets Ther.
11:6489–6503. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yin W, Nie Y, Chen L, Wang Q, Liu S, He X
and Wang W: Deregulation of microRNA-193b affects the proliferation
of liver cancer via myeloid cell leukemia-1. Oncol Lett.
15:2781–2788. 2018.PubMed/NCBI
|
|
18
|
Wang L, Zhang Y, Zhao L, Liu S, Yu S, Ma Y
and Sun G: MicroRNA-193b inhibits the proliferation, migration and
invasion of gastric cancer cells via targeting cyclin D1. Acta
Histochem. 118:323–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo F, Luo Y, Mu YF, Qin SL, Qi Y, Qiu YE
and Zhong M: miR-193b directly targets STMN1 and inhibits the
malignant phenotype in colorectal cancer. Am J Cancer Res.
6:2463–2475. 2016.PubMed/NCBI
|
|
20
|
Zhong Q, Wang T, Lu P, Zhang R, Zou J and
Yuan S: miR-193b promotes cell proliferation by targeting Smad3 in
human glioma. J Neurosci Res. 92:619–626. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chambless LB, Kistka HM, Parker SL,
Hassam-Malani L, McGirt MJ and Thompson RC: The relative value of
postoperative versus preoperative Karnofsky Performance Scale
scores as a predictor of survival after surgical resection of
glioblastoma multiforme. J Neurooncol. 121:359–364. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fan Z, Cui H, Xu X, Lin Z, Zhang X, Kang
L, Han B, Meng J, Yan Z, Yan X and Jiao S: MiR-125a suppresses
tumor growth, invasion and metastasis in cervical cancer by
targeting STAT3. Oncotarget. 6:25266–25280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dassow H and Aigner A: MicroRNAs (miRNAs)
in colorectal cancer: From aberrant expression towards therapy.
Curr Pharm Des. 19:1242–1252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bertoli G, Cava C and Castiglioni I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo SJ, Zeng HX, Huang P, Wang S, Xie CH
and Li SJ: MiR-508-3p inhibits cell invasion and
epithelial-mesenchymal transition by targeting ZEB1 in
triple-negative breast cancer. Eur Rev Med Pharmacol Sci.
22:6379–6385. 2018.PubMed/NCBI
|
|
29
|
Long HD, Ma YS, Yang HQ, Xue SB, Liu JB,
Yu F, Lv ZW, Li JY, Xie RT, Chang ZY, et al: Reduced hsa-miR-124-3p
levels are associated with the poor survival of patients with
hepatocellular carcinoma. Mol Biol Rep. 45:2615–2623. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao C, Wang XB, Zhang YH, Zhou YM, Yin Q
and Yao WC: MicroRNA-424 inhibits cell migration, invasion and
epithelial-mesenchymal transition in human glioma by targeting
KIF23 and functions as a novel prognostic predictor. Eur Rev Med
Pharmacol Sci. 22:6369–6378. 2018.PubMed/NCBI
|
|
31
|
Gu X, Gong H, Shen L and Gu Q:
MicroRNA-129-5p inhibits human glioma cell proliferation and
induces cell cycle arrest by directly targeting DNMT3A. Am J Transl
Res. 10:2834–2847. 2018.PubMed/NCBI
|
|
32
|
Li Z, Qian R, Zhang J and Shi X:
MiR-218-5p targets LHFPL3 to regulate proliferation, migration and
epithelial-mesenchymal transitions of human glioma cells. Biosci
Rep. 39(pii): BSR201808792019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu J, He J, Huang H, Peng R and Xi J:
MicroRNA-423-3p promotes glioma growth by targeting PANX2. Oncol
Lett. 16:179–188. 2018.PubMed/NCBI
|
|
34
|
Jiang Y, Wang X, Zhang J and Lai R:
MicroRNA-599 suppresses glioma progression by targeting RAB27B.
Oncol Lett. 16:1243–1252. 2018.PubMed/NCBI
|
|
35
|
Chan CM, Lai KKY, Ng EKO, Kiang MN, Kwok
TWH, Wang HK, Chan KW, Law TT, Tong DK, Chan KT, et al: Serum
microRNA-193b as a promising biomarker for prediction of
chemoradiation sensitivity in esophageal squamous cell carcinoma
patients. Oncol Lett. 15:3273–3280. 2018.PubMed/NCBI
|
|
36
|
Wu K, Zhao Z, Ma J, Chen J, Peng J, Yang S
and He Y: Deregulation of miR-193b affects the growth of colon
cancer cells via transforming growth factor β and regulation of the
SMAD3 pathway. Oncol Lett. 13:2557–2562. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu X and Lu J: The significance of
detection of serum miR-423-5p and miR-484 for diagnosis of
colorectal cancer. Clin Lab. 61:187–190. 2015. View Article : Google Scholar : PubMed/NCBI
|