Introduction
Cancer of the thyroid gland is the most common
endocrine malignancy, comprising ~1% of all cancer cases in the
United States of America (1,2). According to the pathological type,
thyroid cancers are divided into papillary, follicular, medullary
and anaplastic thyroid cancer (3),
the first two of which are typically indolent tumors with a 5-year
survival rate of >95% (4).
However, the metastasis of thyroid cancer to lung or other tissue
is often observed at an early stage in certain cases (5–7). Primary
thyroid tumors are usually well differentiated, but highly
aggressive and respond poorly to standard therapies, which usually
results in a poor prognosis and overall survival rate (8). Therefore, it is important to find a
molecular biomarker that can predict and inhibit the malignant
biological behavior of tumors.
Coiled-coil domain containing 67 (CCDC67) is a tumor
suppressor gene that exhibits significant inhibitory effects on
gastric, prostate and hepatocellular cancer (9–11). Our
previous study demonstrated that the upregulation of CCDC67 gene
has a significant inhibitory effect on thyroid cancer in
vitro, which promotes the apoptosis of thyroid cancer cells and
inhibits their proliferation, invasion and migration (12). However, due to the lack of a suitable
cell tool, particularly cells with a fluorescent label that can be
detected in living animals, these results were not tested in
vivo.
Luciferase reporter gene, one of the most common
gene reporter systems in animal experiments, achieves real-time
monitoring of micrometastasis in non-invasive conditions (13). Luciferase is safe and sensitive, and
exhibits high specificity and signal-to-noise ratio. Therefore, it
can be used as a molecular label to observe the proliferation and
metastasis of tumor cells indirectly through in vivo imaging
systems (IVIS) (14).
In order to generate a reliable and convenient cell
tool for further research on CCDC67 in vivo, a thyroid
cancer cell line expressing CCDC67, luciferase and puromycin
acetyltransferase (PAC) genes was generated. In addition, the
tumorigenic ability of the generated cell line
TPC-1-Luc-Puromycin-CCDC67 was validated in vivo. The newly
generated cell line may provide a useful tracer cell for future
studies.
Materials and methods
Cell lines and cell culture
The human papillary thyroid cancer cell line (TPC-1)
and human embryonic kidney cell line (293T) were kindly provided by
Dr Ye Lei of Shanghai Rui Jin Hospital (Shanghai, China). TPC-1
cells were cultured in RPMI-1640 (Beijing Solarbio Science &
Technology Co., Ltd.) medium supplemented with 10% FBS (Gemini Bio
Products), and 293T cells were cultured in DMEM (Beijing Solarbio
Science & Technology Co., Ltd.) supplemented with 10% FBS. All
cells were cultured in an atmosphere containing 5% CO2
at 37°C and digested by 0.25% trypsin with 0.01% EDTA.
Construction and identification of a
positive recombinant plasmid of CCDC67
To construct the lentiviral vector carrying the
target gene, a positive recombinant plasmid was first constructed.
The primer of the CCDC67 gene was designed and synthesized by
Shanghai GeneChem Co., Ltd. Primer sequences were as follows:
CCDC67 forward,
5′-GAGGATCCCCGGGTACCGGTCGCCACCATGGAGAACCAAGCCCATAATAC-3′ and
reverse, 5′-TCACCATGGTGGCGACCGGTATGTGTCTATTTTGTTTTAGC-3′. The
reverse transcription procedure was performed using a PrimeScript™
II kit (Takara Bio, Inc.) according to the manufacturer's protocol,
and the cDNA fragment following amplification was cloned into a
modified pCV146-Luc-Puromycin vector (Shanghai GeneChem Co., Ltd.).
pCV146-Luc-Puromycin was linearized by BamHI/AgeI
(Shanghai GeneChem Co., Ltd.) prior to mixing with purified cDNA
fragment in a 1:5 ratio. T4 DNA ligase (Shanghai GeneChem Co.,
Ltd.) was used for the ligation reaction, and the ligation reaction
system was as follows: 1 µl linearized pCV146 vector (100 ng/µl), 1
µl double-stranded DNA oligo, 2 µl 10X T4 DNA ligase buffer, 1 µl
T4 DNA ligase, and 15 µl ddH2O. The subsequent
conversion reaction and sequencing of positive clones were
performed by Majorbio Technology Co., Ltd. Sequencing results were
analyzed and compared with the sequence of CCDC67 gene in GenBank
(https://www.ncbi.nlm.nih.gov/nuccore/KJ906442.1?report=fasta)
using Chromas software (version 2.0; Miaolingbio) The positive
recombinant plasmid was termed pCV146-Luc-Puromycin-CCDC67.
Packaging and concentration of
lentiviral vectors
293T cells were seeded in a 15-cm culture dish at a
density of 6×106 cells/ml, and the serum-free DMEM was
replaced when the cell density reached ~80%. Transfection complex
solution comprised 20 µg pCV146-Luc-Puromycin-CCDC67 vectors, 15 µg
pHelper l.0 vectors (Shanghai GeneChem Co., Ltd.), 10 µg pHelper
2.0 vectors (Shanghai GeneChem Co., Ltd.), 100 µl
Lipofectamine® 2000 (Shanghai GeneChem Co., Ltd.) and
4.9 ml Opti-MEM medium (Shanghai GeneChem Co., Ltd.). Following
configuration according to the manufacturer's protocol, the
transfection complex solution was added to the culture dish with
293T cells. At 8 h, the culture medium was replaced with DMEM
containing 10% FBS. The supernatant of 293T cells was collected at
48 h, and the lentivirus was concentrated by ultracentrifugation
(4.472×104 × g at 4°C for 3 h).
Determination of the lentivirus
titer
HIV-1 p24 Antigen ELISA 2.0 kit (ZeptoMetrix
Corporation) was used to determine the titer of the lentivirus.
HIV-1 p24 Antigen Standard was diluted to 125.0, 62.5, 31.3, 15.6,
7.8 and 3.9 pg/ml in PBS. The lentivirus solution was diluted with
PBS and the dilution ratios of 1:1×106 and
1:1×107 were selected for testing. A total of 200 µl
Antigen Standard in different concentrations and diluted lentivirus
samples were added into a microwell plate separately. Subsequently,
the plate was sealed by Parafilm® and placed in an oven
at 37°C for 1.5 h. The samples were removed and 100 µl HIV-1 p24
Detector Antibody was added to each well, with the exception of the
control wells. The plate was maintained at room temperature for 30
min in the dark. After the sample wells with p24 turned blue, 100
µl Stop Solution was added to stop the reaction. Optical density at
450 nm was detected within 15 min by an automatic enzyme-linked
immunosorbent assay plate reader (Bio-Rad Laboratories, Inc.).
Cell transfection and screening
TPC-1 cells were seeded at 2×105
cells/well in 6-well plates. After the cells attached to the wall,
10 µl lentivirus with a titer of 2×108 TU/ml (MOI=10)
and 40 µl HitransG P infection enhancer (Shanghai GeneChem Co.,
Ltd.) were added into each well. At 8 h, the culture medium was
replaced with RPMI-1640 medium containing 10% FBS. The cells were
screened by culture medium containing 2.5 µg/ml puromycin
(PerkinElmer, Inc.). After 48 h, culture medium containing 1.5
µg/ml puromycin was used to screen for ≥2 weeks to obtain a stable
transfected cell line. The generated thyroid cancer cell line was
termed TPC-1-Luc-Puromycin-CCDC67. An empty lentiviral vector was
used for negative control.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The expression of CCDC67 gene was detected by
RT-qPCR. The primers of CCDC67 and GAPDH genes were synthesized by
Shanghai GeneChem Co., Ltd., and the sequences were as follows:
CCDC67 forward,
5′-GAGGATCCCCGGGTACCGGTCGCCACCATGGAGAACCAAGCCCATAATAC-3′ and
reverse, 5′-TCACCATGGTGGCGACCGGTATGTGTCTATTTTGTTTTAGC-3′; GAPDH
forward, 5′-TGAAGGTCGGAGTCAACGG-3′ and reverse,
5′-CTGGAAGATGGTGATGGGATT-3′. Total RNA was extracted from TPC-1 and
TPC-1-Luc-Puromycin-CCDC67 cells (continuously cultured in
puromycin-free medium for ≥4 weeks) using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and 1 µl
total RNA was reverse-transcribed to cDNA using PrimeScript™ II kit
(Takara Bio, Inc.) according to the manufacturer's protocol. The
resulting cDNA was quantified using a RT-qPCR mRNA SYBR Green
Detection kit (Takara Bio, Inc.). The resulting cDNA (2 µl) was
used as the template for PCR in a 20-µl reaction volume containing
10 µl 2X SYBR Premix Ex Taq II, 0.8 µl each of 10 µmol/l forward
and reverse primers and 6.4 µl ddH2O. The thermocycling
conditions were as follows: 5 sec at 95°C, followed by 50 cycles of
95°C for 5 sec, 60°C for 50 sec. The mRNA expression level of GAPDH
was used for normalization. The threshold cycle (Cq) value was
recorded, and the data were analyzed by the comparative
2−ΔΔCq method (15).
Luciferase activity assay in
vitro
TPC-1 and TPC-1-Luc- Puromycin-CCDC67 cells
(continuously cultured in puromycin-free medium for ≥4 weeks) were
diluted to 5.0×105, 2.5×105,
1.3×105, 0.6×105 and 0.3×105
cells/ml and inoculated into a 24-well plate. This was done to
demonstrate that the fluorescence intensity increased with
increasing cell number. After the cells attached to the wall,
D-luciferin (PerkinElmer, Inc.) was added to a final concentration
of 100 mg/ml. At 3 min, fluorescence intensity was measured using
IVIS.
Tumorigenesis assay in vivo
A total of 5 female SCID Beige mice (4–5 weeks old;
initial body weight: 15–18 g) were used to establish an animal
model of pulmonary metastasis (16).
The mice were maintained in specific pathogen-free housing at Henan
Key Laboratory for Pharmacology of Liver Diseases (temperature,
27°C; humidity: 50%; light cycle, 10 h light, 14 h darkness; food
and water, ad libitum). A preliminary experiment revealed
that a total of 200 µl TPC-1-Luc-Puromycin-CCDC67 cells
(1×107 cells/ml) achieved a higher tumor formation rate
and a longer cell survival time. Cells were administered to SCID
Beige mice by tail vein injection. At 2, 3 and 4 weeks after
injection, mice (n=5) were intraperitoneally injected with 200 µl
D-luciferin solution (150 mg/kg body weight; PerkinElmer, Inc.) and
anesthetized with 2% isoflurane. After 10 min, the mice were placed
into IVIS for bioluminescence imaging. Body weights were recorded
daily. Considering requirements of experimental animal ethics, mice
were sacrificed by cervical dislocation after CO2
euthanasia (flow rate of CO2: 5 l/min; size of chamber:
40×30×25 cm) when the mice lost >2 g body weight (17). Lungs and other organs containing
metastatic foci were excised for ex vivo bioluminescence
imaging. A control experiment was not performed as the main purpose
of the tumorigenesis assay was to examine the tumorigenic ability
of the generated cell line.
Histopathological assay
Lungs and other organs in which metastatic foci were
detected were fixed in 4% paraformaldehyde at room temperature for
24 h, embedded in paraffin and cut into 4-µm sections. Following
staining with hematoxylin for 3 min and eosin for 2 min at room
temperature, the sections were observed and imaged using an upright
fluorescence microscope (cat. no. TS100-F; magnification, ×40 or
×200; Nikon Corporation) with a digital image capturing system
using NIS-Elements D software (version 2.30; Nikon
Corporation).
Statistical analysis
The results of at least three independent
experiments are presented as mean ± standard deviation. Simple
linear regression was used to detect the titer of lentivirus.
Student's t-test was performed to evaluate the differences in the
expression levels of CCDC67 gene. Comparison between multiple
groups was performed using one-way analysis of variance followed by
the Student-Newman-Keuls post hoc test. P<0.05 was considered to
indicate a statistically significant difference. Statistical
analyses were performed using SPSS 21.0 software for Windows (IBM
Corp.). GraphPad Prism 5 (GraphPad Software, Inc.) was used to plot
the data.
Results
Construction and identification of a
positive recombinant plasmid of CCDC67
The full length of CCDC67 gene was selected as the
template for PCR amplification. The PCR products were tested by
agarose gel electrophoresis. The results demonstrated that a
specific bright band was amplified at 1,862 bp (Fig. 1A). The purified CCDC67 gene was
digested, ligated and transformed with the vector
pCV146-luc-puromycin. A total of eight positive clones were
selected to be mixed with Escherichia coli, and the
identification results of the bacterial liquid revealed that a
specific bright band was amplified at 1,392 bp in three of the
eight positive clones (Fig. 1B). The
sequencing results of the three positive clones were compared with
the original sequence of CCDC67 gene in GenBank (NM-181645), and
only one of them fully matched the sequence in GenBank NM-181645
(Fig. 2). The positive recombinant
plasmid pCV146-luc-puromycin-CCDC67 was constructed by introducing
the selected positive clone to the vector pCV146-luc-puromycin.
Determination of the virus titer
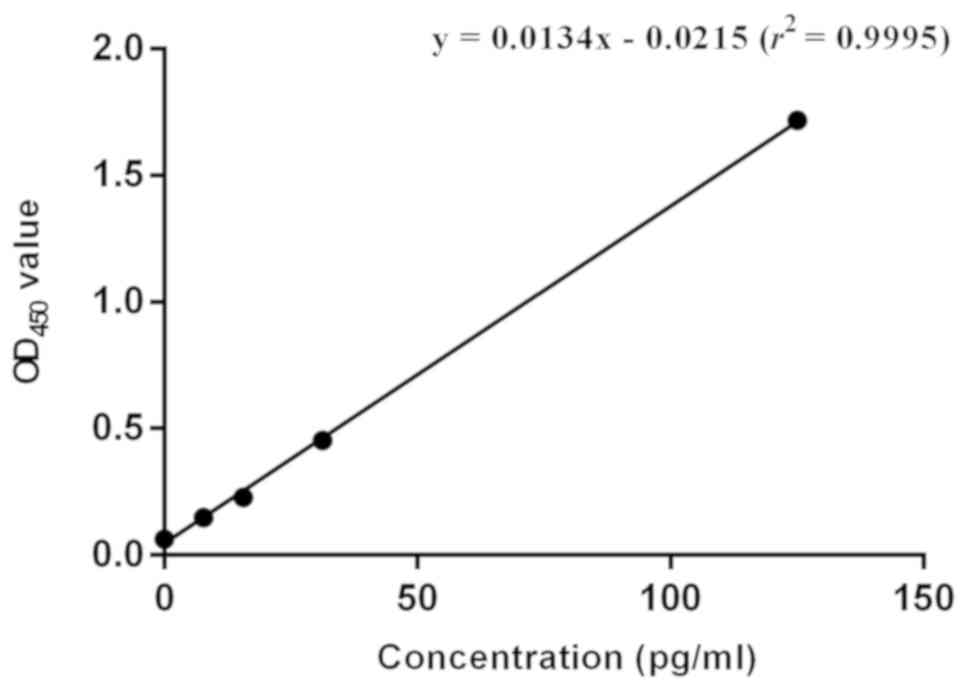
A standard curve was drawn using the corresponding
OD450 values of different concentrations of HIV-1 p24
Antigen Standard (Fig. 3). The
linear equation of the obtained standard curve was: y=0.0134× -
0.0215 (r2=0.9995). Based on the OD450 values
of the two samples (dilution ratios of 1:1×106 and
1:1×107), the average concentration of the original
lentivirus solution was 5.0×107 pg/ml (Table I). According to the conversion
relation between concentration and titer of virus provided by ELISA
kit (10 TU=1 pg), the titer of original lentivirus was
5.0×108 TU/ml.
 | Table I.The OD450 value of virus
solution in two dilution ratios. |
Table I.
The OD450 value of virus
solution in two dilution ratios.
| Sample | Dilution ratio |
OD450 | Concentration,
pg/ml | Raw virus solution,
pg/ml | Average, pg/ml |
|---|
| 1 |
1×106 | 0.413 | 32.4 |
3.2×107 |
5.0×107 |
| 2 |
1×107 | 0.071 |
6.9 |
6.9×107 |
|
Detection of the relative mRNA level
of CCDC67 gene
TPC-1 cells transfected with empty vectors (negative
control group) and TPC-1-Luc-Puromycin-CCDC67 cells were screened
with puromycin and continuously cultured in puromycin-free medium
for ≥4 weeks. The relative mRNA expression level of CCDC67 in the
two groups was assessed by RT-qPCR. The results demonstrated that
the expression level of CCDC67 gene in TPC-1-Luc-Puromycin-CCDC67
cells was 19,446.782-fold higher compared with that in the negative
control group (P<0.01; Fig.
4).
Detection of luciferase activity
All cells used for luciferase activity assay were
screened with puromycin and continuously cultured in puromycin-free
medium for ≥4 weeks. The results of the luciferase activity assay
demonstrated that high luciferase activity was detected in
TPC-1-Luc-Puromycin-CCDC67 cells, whereas no fluorescence was
detected in untransfected TPC-1 cells (Fig. 5). In addition, the fluorescence
intensity increased with the number of cells (Fig. 5).
Identification of tumorigenic ability
in vivo
The tumorigenic ability of generated cell line were
verified using an animal model of pulmonary metastasis. The growth
of tumors was monitored by bioluminescence imaging (Fig. 6A). At week 2, fluorescent foci were
only detected in the lungs of one mouse. At week 3, fluorescent
foci in one or both lungs were detected in all mice, resulting in a
tumor formation rate of 100% (5/5). Quantification analysis of
bioluminescence intensity demonstrated time-dependent tumor growth
(Fig. 6B). In addition, the body
weight of the mice exhibited a slight drop in the duration of tumor
bearing (Fig. 6C), which suggested
that the involvement of thyroid tumors affected the normal growth
of mice.
Bioluminescence imaging and
histopathological analysis
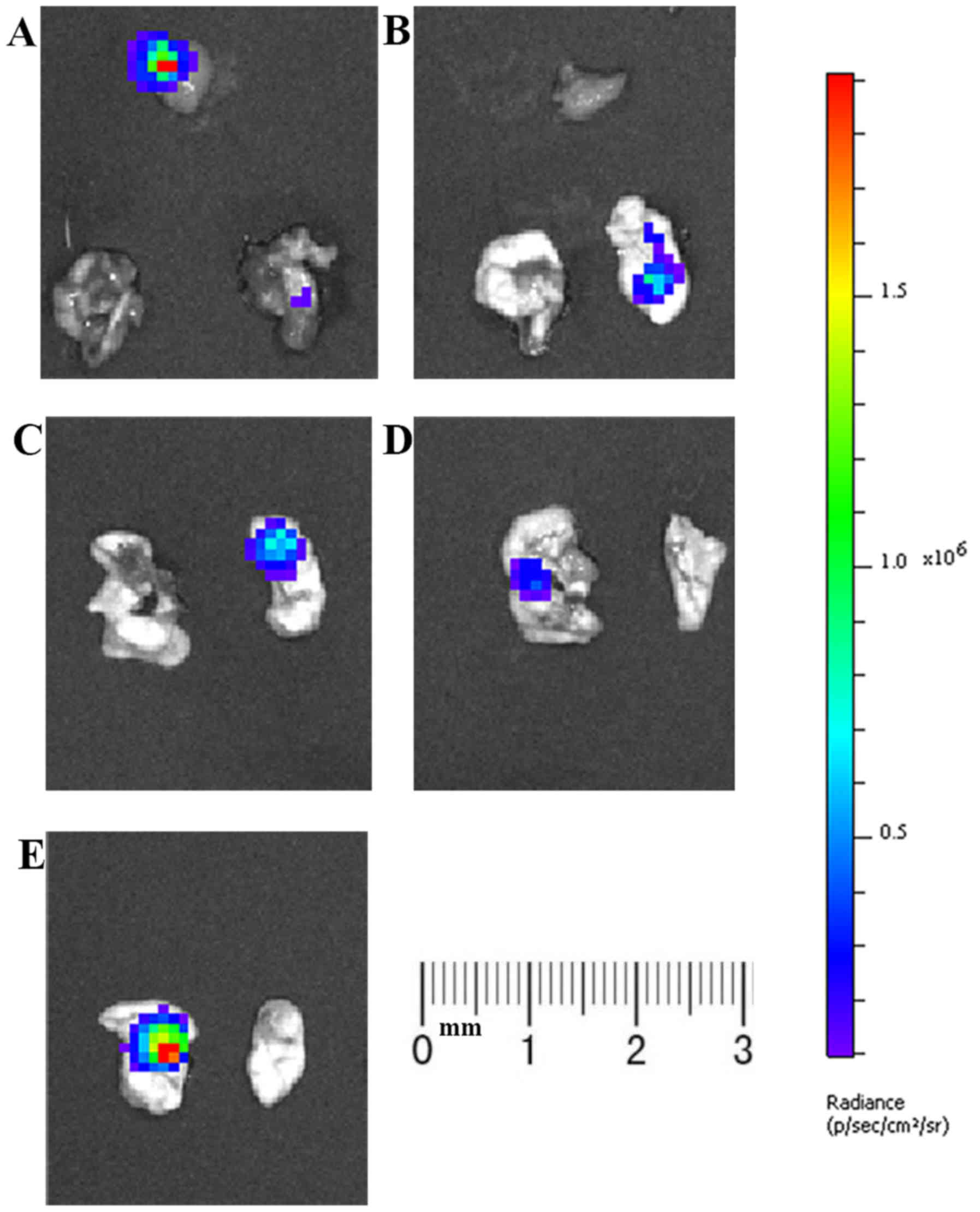
At week 4, both lungs and other tissues with
fluorescent foci were resected for ex vivo bioluminescence
imaging to confirm the metastasis of tumors. Fluorescent foci were
detected in one or both lungs of all mice (Fig. 7), suggesting the lung metastasis of
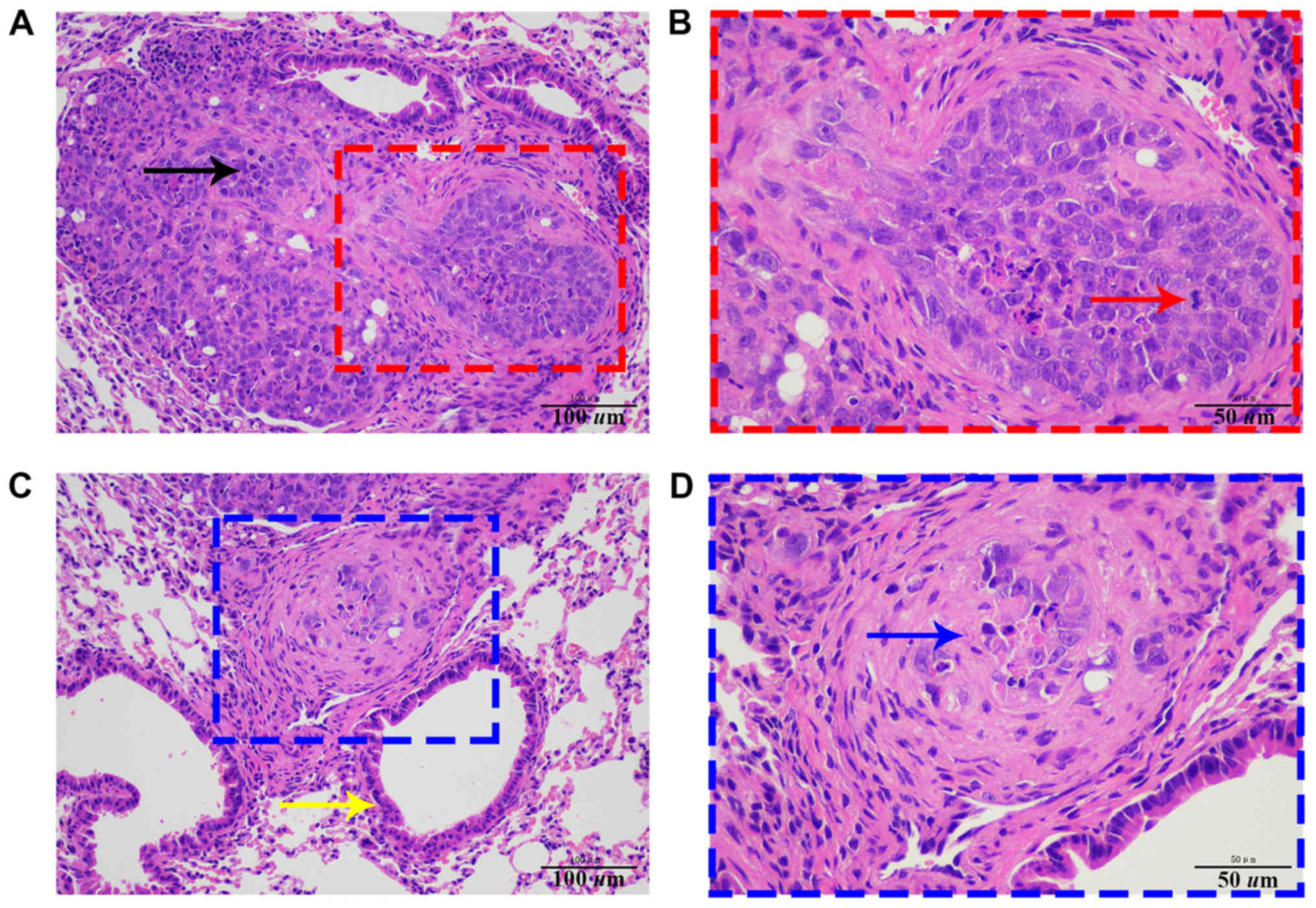
tumor cells. Subsequent histopathological analysis further
confirmed this result (Fig. 8). In
mice 2, 4 and 5, a small number of tumor cells escaped during tail
vein injection, resulting in strong fluorescence foci in the lower
abdomen. In order to eliminate interference from unrelated
fluorescent foci, opaque black paper was used to cover these areas.
In addition, a fluorescent focus was also detected in the
submandibular gland tissue of a mouse, and pathological results
confirmed tumor metastasis.
Discussion
Clinical studies have demonstrated that thyroid
cancer is a malignant tumor with a 5-year survival rate of >89%;
however, if metastasis occurs, the survival rate of patients is
reduced to 51% (18). Therefore,
early diagnosis and prognostic assessment of thyroid cancer are of
great significance. However, an accurate early diagnosis and a
complete prognostic assessment are relatively difficult due to the
inconspicuous clinical symptoms and limitation of fine needle
aspiration biopsy, including the the size and location of nodules
and the unavailability of an experienced thyroid surgeon in
grassroots hospitals in China. Molecular markers are effectively
applied in this area, especially in thyroid nodules with
cytological uncertainty (19,20). The
classical oncogenic genetic alterations commonly observed in
thyroid cancer include BRAF V600E, TERT promoter and Ras mutation,
RET/PTC rearrangements and PAX8-peroxisome proliferator-activated
receptor γ fusion oncogene (21–24),
each of which has benefits and limitations for the early diagnosis
and prognostic assessment. Therefore, combined genetic detection
may be applied to obtain a more accurate and complete assessment
for individualized treatment of tumors (25). New generation technology in gene
detection has enabled the simultaneous detection of multiple genes
(26), but the application of gene
detection in the diagnosis and treatment of thyroid cancer remains
limited, since our understanding about the pathogenesis of thyroid
cancer is still insufficient and the molecular markers are limited.
Therefore, it is necessary to continuously explore the complex
molecular pathways and pathogenesis-related processes of thyroid
cancer.
CCDC67 gene is a tumor-suppressor gene in papillary
thyroid cancer, and our previous in vitro study has
demonstrated that CCDC67 is a reliable molecular marker with
potential to predict the malignant biological properties of thyroid
cancers (12). However, these
results were not validated in the complex physiological environment
in vivo. A stable and controllable animal model is an
essential factor in in vivo experiments, and the
construction of an animal model is based on a mature and reliable
cell tool. In order to generate a suitable cell line for further
in vivo research, not only a successful intervention
(silencing or overexpression) of the target gene needs to be
achieved, but also an applicable fluorescent marker and antibiotic
resistance gene are required, which may facilitate the dynamic
monitoring of tumor cell growth. Classical fluorescent markers,
such as green, red and blue fluorescent protein, are stable,
hypotoxic and easily visualized, but for in vivo experiments
(27–29), the luciferase reporting system is
preferable, as it is non-toxic, sensitive and exhibits a high
signal-to-noise ratio (30). In
addition, numerous antibiotic resistance genes exist for screening,
and PAC is one of the common types used in lentiviral vectors,
which may be related to certain biological characteristics of
lentiviral vectors. In the present study, a thyroid cancer cell
line expressing CCDC67 gene, luciferase reporter gene and puromycin
acetyltransferase gene simultaneously was generated and
identified.
The construction of the cell line
TPC-1-Luc-Puromycin-CCDC67 may provide a convenient tool for the
establishment of a lung metastasis and orthotopic models of thyroid
cancer, which enables further evaluation of the tumor inhibition
effect and mechanism of CCDC67 in vivo. The lentiviral
vector constructed in the present study has the potential to be a
useful tool for transforming other tumor cell lines.
Acknowledgements
The authors would like to thank Dr Peng Youmei
(Henan Key Laboratory for Pharmacology of Liver Diseases) for
guidance on animal experiments and Dr Wang Ning (Henan Key
Laboratory for Pharmacology of Liver Diseases) for assisting with
in vivo imaging.
Funding
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no. 81372863),
College Scientific and Technological Innovation Team Project of
Henan Province (grant no. 19IRTSTHN002) and Thousand Talents
Program of Central China (grant no. ZYQR201810015).
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ and DY conceived and designed the experiments.
LZ, FY, CL and RM performed the experiments. ML and LW collected
and analyzed the data. RM and ML provided suggestions and technical
support on the project. DY wrote the manuscript and supervised the
project. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal experiments in this study were approved
by the Ethical Committee of Zhengzhou University (Zhengzhou,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yu Y, Yu X, Fan C, Wang H, Wang R, Feng C
and Guan H: Targeting glutaminase-mediated glutamine dependence in
papillary thyroid cancer. J Mol Med (Berl). 96:777–790. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hardin H, Helein H, Meyer K, Robertson S,
Zhang R, Zhong W and Lloyd RV: Thyroid cancer stem-like cell
exosomes: Regulation of EMT via transfer of lncRNAs. Lab Invest.
98:1133–1142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Raue F and Frank-Raue K: Thyroid cancer:
Risk-stratified management and individualized therapy. Clin Cancer
Res. 22:5012–5021. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nikitski AV, Rominski SL, Wankhede M,
Kelly LM, Panebianco F, Barila G, Altschuler DL and Nikiforov YE:
Mouse model of poorly differentiated thyroid carcinoma driven by
STRN-ALK fusion. Am J Pathol. 188:2653–2661. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang J, Li LF, Zhang XM, Xu Q, Zhang J,
Weng WW and Dong MJ: Unusual synchronous skeletal muscle and lung
metastasis in papillary thyroid cancer: A case report and review of
the literature. Oncol Lett. 9:727–730. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bruglia M, Palmonella G, Silvetti F,
Rutigliano P, Criante P, Marmorale C, Boscaro M and Taccaliti A:
Skin and thigh muscle metastasis from papillary thyroid cancer.
Singapore Med J. 50:e61–e64. 2009.PubMed/NCBI
|
|
7
|
Luo Q, Luo QY, Sheng SW, Chen LB, Yu YL,
Lu HK and Zhu RS: Localization of concomitant metastases to kidney
and erector spinae from papillary thyroid carcinoma using
(131)I-SPECT and CT. Thyroid. 18:663–664. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
American Thyroid Association (ATA)
Guidelines Taskforce on Thyroid Nodules and Differentiated Thyroid
Cancer, ; Cooper DS, Doherty GM, Haugen BR, Kloos RT, Lee SL,
Mandel SJ, Mazzaferri EL, McIver B, Pacini F, et al: Revised
American Thyroid Association management guidelines for patients
with thyroid nodules and differentiated thyroid cancer. Thyroid.
19:1167–1214. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park SJ, Jang HR, Kim M, Kim JH, Kwon OH,
Park JL, Noh SM, Song KS, Kim SY, Kim YH and Kim YS: Epigenetic
alteration of CCDC67 and its tumor suppressor function in gastric
cancer. Carcinogenesis. 33:1494–1501. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu YP, Ding Y, Chen Z, Liu S,
Michalopoulos A, Chen R, Gulzar ZG, Yang B, Cieply KM, Luvison A,
et al: Novel fusion transcripts associate with progressive prostate
cancer. Am J Pathol. 184:2840–2849. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen ZH, Yu YP, Zuo ZH, Nelson JB,
Michalopoulos GK, Monga S, Liu S, Tseng G and Luo JH: Targeting
genomic rearrangements in tumor cells through Cas9-mediated
insertion of a suicide gene. Nat Biotechnol. 35:543–550. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yin DT, Xu J, Lei M, Li H, Wang Y, Liu Z,
Zhou Y and Xing M: Characterization of the novel tumor-suppressor
gene CCDC67 in papillary thyroid carcinoma. Oncotarget.
7:5830–5841. 2016.PubMed/NCBI
|
|
13
|
Sekiguchi Y, Owada J, Oishi H, Katsumata
T, Ikeda K, Kudo T and Takahashi S: Noninvasive monitoring of
β-cell mass and fetal β-cell genesis in mice using bioluminescence
imaging. Exp Anim. 61:445–451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tu SH, Hsieh YC, Huang LC, Lin CY, Hsu KW,
Hsieh WS, Chi WM and Lee CH: A rapid and quantitative method to
detect human circulating tumor cells in a preclinical animal model.
BMC Cancer. 17:4402017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng L, Jia X, Zhu M, Chen Y and Shi F:
Chemoprevention by Prunella vulgaris L. extract of non-small cell
lung cancer via promoting apoptosis and regulating the cell cycle.
Asian Pac J Cancer Prev. 11:1355–1358. 2010.PubMed/NCBI
|
|
16
|
Kato H, Wakabayashi H, Naito Y, Kato S,
Nakagawa T, Matsumine A and Sudo A: Anti-tumor necrosis factor
therapy inhibits lung metastasis in an osteosarcoma cell line.
Oncology. 88:139–146. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Boivin GP, Bottomley MA, Schiml PA, Goss L
and Grobe N: Physiologic, behavioral, and histologic responses to
various euthanasia methods in C57BL/6NTac male mice. J Am Assoc Lab
Anim Sci. 56:69–78. 2017.PubMed/NCBI
|
|
18
|
Randle RW, Balentine CJ, Leverson GE,
Havlena JA, Sippel RS, Schneider DF and Pitt SC: Trends in the
presentation, treatment, and survival of patients with medullary
thyroid cancer over the past 30 years. Surgery. 161:137–146. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pstrag N, Ziemnicka K, Bluyssen H and
Wesoły J: Thyroid cancers of follicular origin in a genomic light:
In-depth overview of common and unique molecular marker candidates.
Mol Cancer. 17:1162018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bath SC, Pop VJ, Furmidge-Owen VL, Broeren
MA and Rayman MP: Thyroglobulin as a functional biomarker of iodine
status in a cohort study of pregnant women in the United Kingdom.
Thyroid. 27:426–433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xing M: BRAF mutation in thyroid cancer.
Endocr Relat Cancer. 12:245–262. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu X, Bishop J, Shan Y, Pai S, Liu D,
Murugan AK, Sun H, El-Naggar AK and Xing M: Highly prevalent TERT
promoter mutations in aggressive thyroid cancers. Endocr Relat
Cancer. 20:603–610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Basolo F, Pisaturo F, Pollina LE,
Fontanini G, Elisei R, Molinaro E, Iacconi P, Miccoli P and Pacini
F: N-ras mutation in poorly differentiated thyroid carcinomas:
Correlation with bone metastases and inverse correlation to
thyroglobulin expression. Thyroid. 10:19–23. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dwight T, Thoppe SR, Foukakis T, Lui WO,
Wallin G, Höög A, Frisk T, Larsson C and Zedenius J: Involvement of
the PAX8/peroxisome proliferator-activated receptor gamma
rearrangement in follicular thyroid tumors. J Clin Endocrinol
Metab. 88:4440–4445. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marini F, Falchetti A, Del Monte F,
Carbonell Sala S, Tognarini I, Luzi E and Brandi ML: Multiple
endocrine neoplasia type 2. Orphanet J Rare Dis. 1:452006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Wang S, Hu B, Zhao F, Xiang P, Ji
D, Chen F, Liu X, Yang F, Wu Y, et al: Direct detection of
Helicobacter pylori in biopsy specimens using a
high-throughput multiple genetic detection system. Future
Microbiol. 11:1521–1534. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng L, Du C, Murray D, Tong X, Zhang YA,
Chen BP and Hawley RG: A GFP reporter system to assess gene
transfer and expression in human hematopoietic progenitor cells.
Gene Ther. 4:1013–1022. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fischer M, Haase I, Simmeth E, Gerisch G
and Müller-Taubenberger A: A brilliant monomeric red fluorescent
protein to visualize cytoskeleton dynamics in Dictyostelium.
FEBS Lett. 577:227–232. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kao TH, Chen Y, Pai CH, Chang MC and Wang
AH: Structure of a NADPH-dependent blue fluorescent protein
revealed the unique role of Gly176 on the fluorescence enhancement.
J Struct Biol. 174:485–493. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cissell KA, Rahimi Y, Shrestha S, Hunt EA
and Deo SK: Bioluminescence-based detection of microRNA, miR21 in
breast cancer cells. Anal Chem. 80:2319–2325. 2008. View Article : Google Scholar : PubMed/NCBI
|