Introduction
The BCR-ABL1 gene is a molecular marker of
chronic myeloid leukemia (CML), and its transcript level can
accurately reflect tumor burden (1).
Molecular monitoring refers to the detection of BCR-ABL1
transcripts in the peripheral blood through the use of reverse
transcription quantitative-PCR (RT-qPCR) (2,3). In
October 2005, an International Standard was proposed for molecular
testing, which included changing BCR-ABL1 gene detection in
every laboratory via a conversion factor (4). Therefore, BCR-ABL1 gene
detection was also converted to the international standard value
BCR-ABL1IS in the present study (5,6).
Quantitative monitoring of
BCR-ABL1IS via RT-qPCR is currently the gold
standard method of evaluating patient response to tyrosine kinase
inhibitors (TKIs) and subsequent classification into prognostic
subgroups (7). However, RT-qPCR has
a number of shortcomings: The difference in amplification
efficiency between the reference gene and the target gene, and the
difference between platforms and personnel skill level among
different laboratories leads to errors in amplification efficiency
(8). Therefore, a consistent
conversion factor must be established and verified on a regular
basis (9).
Digital PCR (dPCR) is used in the detection and
quantification of nucleic acids (10). First, dPCR evenly distributes the
reaction system into a large number of reaction units, and the
number of nucleic acid sequences of interest conforms to the
Poisson distribution (7). PCR
amplification is then performed independently in each reaction
unit. Following the end of the amplification, the fluorescence
signal of each reaction unit is detected, and finally the copy
number of the nucleic acid sequence of interest is calculated based
on the ratio of the Poisson distribution and the reaction unit that
is positive for the fluorescent signal in all the reaction units
(7). The primary advantage of dPCR
over RT-qPCR is that it can be performed without the requirement
for a calibration curve, therefore offering more straightforward
means of ensuring interlaboratory reproducibility (9,11).
Materials and methods
Patients and samples
A total of 83 patients were included in the present
study, and these were divided into two groups: Groups A and B. All
patients were diagnosed in the Department of Hematology of The
Affiliated Hospital of Xuzhou Medical University between September
10, 2016 and March 4, 2017. A total of 43 patients with
undetectable BCR-ABL1IS result from peripheral
blood were selected as the group A. The median age of patients in
group A was 45 years (age range, 13–72 years), including 22 men and
21 women. Only 25 patients from group A had scokal scores (1). The same blood samples of Group A
patients were tested using the Clarity™ dPCR system (JN Medsys).
Group B comprised of 40 patients who achieved either cytogenetic
response or further response between January 3, 2017 and May 3,
2017. The median age of patients in group B was 49 years (age
range, 10–68 years), including 24 men and 16 women. Only 24
patients from groups B had sokal scores. A RT-qPCR platform (Roche
Diagnostics) and Clarity™ dPCR system was used to detect the
BCR-ABL1 fusion gene within the same peripheral blood
sample, simultaneously. There was no BCR-ABL1 kinase domain
mutation detected in patients enrolled in the present study. The
study protocol was approved by the Ethics Committee of the
Affiliated Hospital of Xuzhou Medical University, and all patients
included in the present study provided written informed
consent.
Quantification of the human BCR-ABL1
fusion gene using the Clarity™ dPCR system
DNA was diluted 10- or 100-fold prior to quantifying
the human BCR-ABL1 fusion gene using the Clarity™ dPCR
system to achieve the expected target concentration range of 0.38
to 2,240 copies/µl reaction mixture. The samples were diluted using
sterile water for injection. The probe and primers used were
supplied by JN Medsys. The sequences of the primers and probes are
listed in Table I. Each sample was a
total of 15 µl, with a mixture of 0.25 µM forward and reverse
primers, 0.25 µM probe, 1×Master Mix, 1×Clarity™ JN solution, 3 µl
DNA sample and PCR grade water. Using the Clarity™ autoloader, the
resultant mixture was then delivered onto the chip where it was
subdivided into 10,000 partitions. The partitions were then sealed
using Clarity™ Sealing Enhancer and 230 µl Clarity™ Sealing Fluid
with the following thermocycling conditions: Initial cycle of 95°C
for 10 min, 40 cycles of 95°C for 50 sec and 58°C for 90 sec (ramp
rate, 1°C/sec). Following amplification via PCR, the tube strips
were transferred to the Clarity™ Reader, which detects fluorescent
signals from each partition simultaneously. The data were analyzed
using Clarity™ software (version 1.0; JN Medsys), and a proprietary
algorithm was used for setting each threshold based on fluorescent
intensities to determine the proportion of positive partitions out
of the total. Based on this information, the software determines
the DNA copies/µl of the dPCR mix using the Poisson statistics. The
mean partition volume of 1.336 nl was used to calculate the copy
number. Each dPCR test was performed twice, and the average value
was taken as the final result.
 | Table I.Oligonucleotides used for
BCR-ABL1 amplification. |
Table I.
Oligonucleotides used for
BCR-ABL1 amplification.
|
Oligonucleotide | Sequence
(5′-3′) |
|---|
| BCR-ABL1
forward |
TCCGCTGACCATCAATAAGGA |
| BCR-ABL1
reverse |
CACTCAGACCCTGAGGCTCAA |
| ABL
forward |
TGGAGATAACACTCTAAGCATAACTAAAGGT |
| ABL
reverse |
GATGTAGTTGCTTGGGACCCA |
| BCR-ABL
probe |
CCCTTCAGCGGCCAGTAGCATCTGA |
| ABL
probe |
CCATTTTTGGTTTGGGCTTCACACCATT |
Statistical analysis
The results of BCR-ABL1 transcripts were
statistically analyzed, with descriptions of the data including the
calculation of mediums, ranges, standard deviations (SDs),
coefficients of variation (CVs). The association between age and
relapse was evaluated using the χ2 test. Age and dPCR
outcome were assessed via Kaplan-Meier analysis and log-rank test.
Linear regression analysis was used to analyze the results of dPCR
and RT-qPCR, and R2 represents the coefficient of
determination. P<0.05 was considered to indicate a statistically
significant difference.
Results
Baseline characteristics of
patients
In the present study, 39 patients were MR4.5
(BCR-ABL1IS ≤0.003 2% or undetectable disease in
cDNA over 32, 000 ABL1 transcripts) when enrolled in Group
A, and 4 patients were MR5. Table
II presents the clinical features of Group A patients. The
median age of the patients at diagnosis was 45 years (range, 13–72
years). The disease state of patients in group A was chronic
phrase, no patients were in accelerated phase or blast phase, and
68.0% of these patients had a low Sokal Score (1). The median white blood cell (WBC) count
at the time of initial diagnosis was 62.7×109/l
(17.8×109/l-263×109/l), median hemoglobin
(HB) level was 94 g/l (53 g/l-145 g/l), median platelet (PLT) count
was 414×109/l
(153×109/l-2886×109/l). Of the total
patients, 37 received hydroxyurea; 41 received TKIs; 3 patients had
been treated with interferon; and four had received a hematopoietic
stem cell transplantation (HSCT). The median duration of TKI
treatment of the patients in Group A was 46.5 months (range,
14.1–149.0 months) prior to enrollment in the present study. When
BCR-ABL1 gene was undetectable by RT-qPCR, the maximum of
the ABL1 control transcripts was 228,110 copies. dPCR was
used to detect peripheral blood samples from patients at the time
that their BCR-ABL1IS results were undetectable.
The % of BCR-ABL1 was between 0.0030 and 9.2390% and the
median was 0.0517%. The BCR-ABL1% was <0.1% in 24
patients, and was >0.1% in other 19 patients.
 | Table II.Baseline patient characteristics of
groups A and B. |
Table II.
Baseline patient characteristics of
groups A and B.
|
| Group A (n=43) | Group B (n=40) |
|---|
|
|
|
|
|---|
| Variables | n | % | n | % |
|---|
| Sex |
|
|
|
|
|
Male | 22 | 51.2 | 24 | 60.0 |
|
Female | 21 | 48.8 | 16 | 40.0 |
| Age (years) |
|
|
|
|
|
≤45 | 24 | 55.8 | 17 | 42.5 |
|
>45 | 19 | 44.2 | 23 | 57.5 |
| Disease status |
|
|
|
|
| Chronic
phase | 43 | 100.0 | 39 | 97.5 |
|
Accelerated phase | 0 | 0.0 | 1 | 2.5 |
| Blast
crisis | 0 | 0.0 | 0 | 0.0 |
| ECOG PS |
|
|
|
|
| 0 | 39 | 90.7 | 37 | 92.5 |
| 1 | 4 | 9.3 | 3 | 7.5 |
| Sokal score
range |
|
|
|
|
|
<0.8 | 17 | 68.0a | 11 | 45.8b |
|
0.8–1.2 | 5 | 20.0 | 10 | 41.7 |
|
>1.2 | 3 | 12.0 | 3 | 12.5 |
| Interferon |
|
|
|
|
| No | 40 | 93.0 | 37 | 92.5 |
|
Yes | 3 | 7.0 | 3 | 7.5 |
| Hydroxyurea |
|
|
|
|
| No | 6 | 14.0 | 1 | 2.5 |
|
Yes | 37 | 86.0 | 39 | 97.5 |
| Treatment
response |
|
|
|
|
|
PCyR | 0 | 0.0 | 2 | 5.0 |
|
CCyR | 0 | 0.0 | 32 | 80.0 |
|
MMR | 0 | 0.0 | 6 | 15.0 |
|
MR4.5 | 39 | 90.7 | 0 | 0.0 |
|
MR5 | 4 | 9.3 | 0 | 0.0 |
There were two patients already at PCyR when they
were enrolled in Group B; 32 patients were CCyR; and 6 patients
were MMR. Table II presents the
clinical features of Group B patients. The median age at diagnosis
was 47 years (range, 10–68 years). The median WBC count at the time
of initial diagnosis was 126.7×109/l
(10.8×109/l-700.7×109/l); HB level was 97 g/l
(70 g/l-140 g/l); PLT count was 445×109/l
(184×109/l-1,536×109/l). Of the total
patients, 39 received hydroxyurea; 34 received TKIs; three received
both TKIs and interferon; 2 patients received a HSCT; and 1 patient
was diagnosed in the accelerated phase of disease at the initial
diagnosis. The median duration of TKI treatment of patients in
Group B was 24.7 months (3.4–127.9 months) prior to enrollment in
the present study. When the BCR-ABL1% results detected via
dPCR achieved PCyR, the PCyR or MMR values were between 0.0260 and
22.5714%, and the median was 0.6237%. The BCR-ABL1% results
detected by dPCR were <1.0% in 25 patients, and was >1.0% in
other 15 patients.
Molecular relapse of Group A
patients
RT-qPCR was used to detect BCR-ABL1 level in
the peripheral blood of patients in Group A every 1–3 months
starting from their initial enrollment date in the present study.
Molecular relapse was defined as a BCR-ABL1 level >0.1%.
Relapsed patients ended their follow-up at the time of recurrence.
Patients who did not relapse were followed up until October 31,
2018. During the follow-up period, 37 patients received TKIs
according to the original protocol; two received both TKIs and
interferon; and 4 patients who received HSCT did not take TKIs
after the transplantation. At the end of the follow-up, depth of
remission in two patients maintained MR5, 21 patients maintained
MR4.5, and 20 patients had a molecular relapse. None of the
patients included in the present study succumbed. No mutations in
the BCR-ABL1 kinase domain were detected. The median time to
molecular relapse was 11.0 months (range, 3.6–18.4 months). The
median follow-up time for all patients was 20.1 months (range,
3.6–25.3 months). Of the 24 patients with BCR-ABL1 level
<0.1% detected by dPCR, seven relapsed. Of the 19 patients with
BCR-ABL1 >0.1%, 13 experienced recurrence. (Fig. 1A; χ2=5.560; P=0.018). The
median BCR-ABL1 level detected by dPCR system in the
relapsed patients was 0.1860% (range, 0.0062–9.2390%), and the
median BCR-ABL1% in patients without recurrence was 0.0537%
(range, 0.0003–1.5323%) (P=0.080). Fig.
1B and Table III present
relapse by age (≤45 years or >45 years) (χ2=1.773;
P=0.183). There was no statistically significant difference in the
Kaplan-Meier survival curve (Fig.
1B; χ2=6.731; P=0.081).
 | Table III.Correlation between age and relapse
status in Group A patients. |
Table III.
Correlation between age and relapse
status in Group A patients.
| Variable | Maintain MMR | Relapse | Total |
|---|
| Age ≤45 years | 15 | 9 | 24 |
| Age >45
years | 8 | 11 | 19 |
| Total | 23 | 20 | 43 |
Comparison of BCR-ABL1 detected by
RT-qPCR and dPCR in Group B patients
Table IV presents
the SD and CV of the BCR-ABL1 gene in Group B patients
detected by two platforms. The SD and CV of dPCR detection results
were lower than RT-qPCR in the two groups with
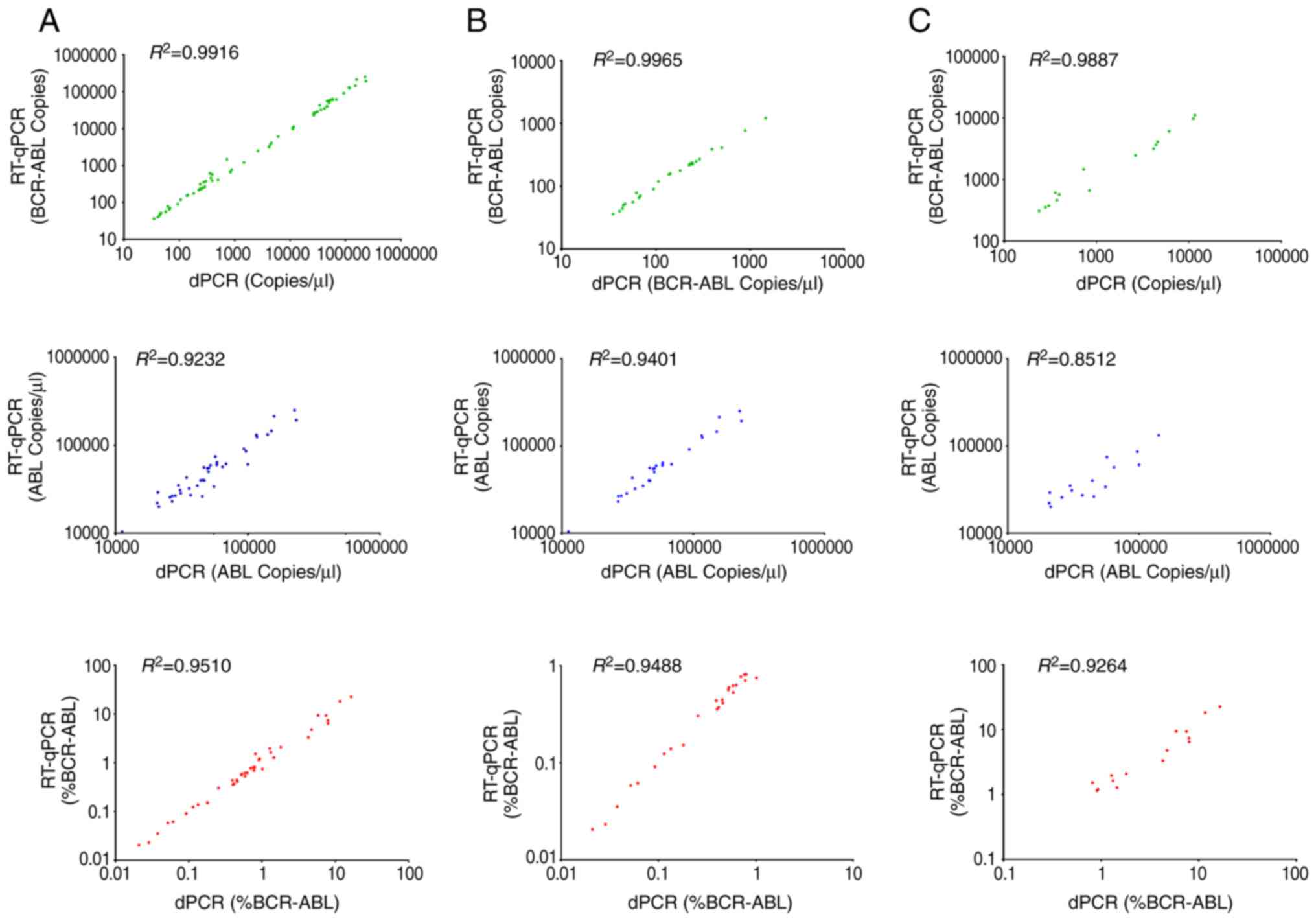
BCR-ABL1IS <1.0% or >1.0%. Scatter plots
demonstrate the linear relationship between the quantification of
BCR-ABL1 transcript copies (green plots), ABL1 (blue
plots) and BCR-ABL1 (red plots) (Fig. 2). Quantification of the cDNA derived
from clinical samples by RT-qPCR was compared with the dPCR
platform. BCR-ABL1 transcript copy numbers, ABL1
transcript copy numbers and BCR-ABL1% measured by dPCR
platform revealed a good association with RT-qPCR across all sample
groups (Fig. 2). Fig. 3 presents BCR-ABL1% of the two
disease levels (BCR-ABL1IS <1.0% or
BCR-ABL1IS >1.0%) and the matched non-CML
controls measured by dPCR and RT-qPCR. The samples for blank
controls were generated with water instead of cDNA. The non-CML
control groups also revealed a positive measurement for
BCR-ABL1 and can be distinguished from the test for patients
with CML.
 | Table IV.Comparison of SD and CV of BCR-ABL1%
detected by RT-qPCR and dPCR in Group B patients. |
Table IV.
Comparison of SD and CV of BCR-ABL1%
detected by RT-qPCR and dPCR in Group B patients.
|
BCR-ABL1IS level | SD | CV |
|---|
|
BCR-ABL1IS <1.0% |
|
|
|
RT-qPCR | 0.276 | 0.705 |
|
dPCR | 0.273 | 0.703 |
|
BCR-ABL1IS >1.0% |
|
|
|
RT-qPCR | 6.541 | 1.061 |
|
dPCR | 4.645 | 0.930 |
Discussion
At present, RT-qPCR is the preferred method for
detecting BCR-ABL1 for the initial diagnosis and ongoing
monitoring of CML (3,9). However, RT-qPCR can lead to errors in
BCR-ABL detection. As such, appropriate correction factors
need to be established and periodically verified (8,9). In the
present study, only dPCR could be applied to the detection of
BCR-ABL1 gene, as dPCR is able detect a lower level of
BCR-ABL1 gene than RT-qPCR (8). dPCR has a higher sensitivity, which is
an advantage when detecting very low levels of the BCR-ABL1
gene (12). For molecular response
monitoring of rare fusion transcripts associated with CML, dPCR is
a very useful tool (13). Patients
who intend to discontinue TKIs must achieve deep molecular
remission, as when the RT-qPCR result is undetectable, a positive
dPCR result may indicate a higher risk of relapse (14). The data from the present study
demonstrated that there was no statistically significant difference
between the BCR-ABL1 level detected by dPCR in relapsed and
non-relapsed groups (P=0.080), but patients with BCR-ABL1
>0.1% were more likely to experience molecular relapse
(P=0.018), which is consistent with a previous study (14). The results from the present study
also demonstrated that patients aged <45 years were more likely
to relapse, but this was not statistically significant and
therefore further larger scale studies are required (14). In the present study, the molecular
relapse rate of patients in Group A was 46.5%, even if they
achieved MR4.5 or MR5, suggesting it is necessary to closely
monitor the BCR-ABL1 gene for patients who achieved further
MR (3,14). An ideal strategy would be to
determine the BCR-ABL1 level every 1–3 months, in order to
expose early molecular relapse (3).
The main advantage of dPCR is that it can be
performed without the need for a calibration curve, therefore
offering a simpler method of ensuring reproducibility between
different laboratories (11). In
addition, dPCR can provide greater confidence in detecting low
BCR-ABL1 copy number concentrations at the limits of current
RT-qPCR technology (12). Goh et
al (15) compared dPCR and
RT-qPCR to detect BCR-ABL1 fusion gene in patients with CML
and revealed that its sensitivity was 3 times higher than RT-qPCR.
The data from the present study revealed that the results of dPCR
for BCR-ABL1 transcription, ABL1 transcription and
BCR-ABL1% were in accordance with those of RT-qPCR, and the
coherence at BCR-ABL1IS <1.0% was better than
that at >1.0%. Normalization using the ABL1 gene appeared
to lead to error in the results, as there was generally a better
agreement between dPCR and RT-qPCR when measuring BCR-ABL1
absolute values than when measuring for ABL1 (7). These results suggest that ABL1
may be a good choice for a reference gene for RT-qPCR, rather than
the ideal internal reference gene for dPCR (14).
In conclusion, the detection results of the
BCR-ABL1 gene in patients with CML using dPCR apply well
with the results obtained by RT-qPCR, particularly in the detection
of low abundance BCR-ABL1 gene (BCR-ABL1IS
<1.0%). dPCR has advantages for patients with CML, who have a
deep molecular response, as the results of dPCR can be applied as a
supplement to RT-qPCR before planning TKIs discontinuation
(16).
Acknowledgements
Not applicable.
Funding
The present study was funded by the following:
National Natural Science Foundation (grant. nos. 81300399 and
81500088); Jiangsu Natural Science Foundation (grant. no.
BK20161178); Key Research & Development Plan of Jiangsu
Province (grant. no. BE2015625); Scientific Research Project of
Jiangsu Province Health and Family Planning Commission (grant. no.
Q201506); Postgraduate Research & Practice Innovation Program
of Jiangsu Province (grant. no. SJCX17_0558).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZY and QS conceived and designed the study. HZ, YH
and QS collected the samples and carried out the research. ZY, HZ,
QS, SZ, KZ, QW, JC, MN, JQ and HC analyzed the data and wrote the
manuscript. JC and MN revised the manuscript. ZY, QS, JQ and HC
participated in the translation of the manuscript. The study was
conducted under the guidance of KX, LZ and ZL. All the authors
accepted the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Clinical Trials of the Affiliated Hospital of Xuzhou
Medical College (approval number, XYFY2015-KL005-01). Certain
patients received hematopoietic stem cell transplantation. No
organs/tissues were obtained from prisoners. Patients donated
hematopoietic stem cells in the Affiliated Hospital of Xuzhou
Medical College. The informed consent was signed by all
participants.
Patient consent for publication
Written informed consent was provided by all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Apperley JF: Chronic myeloid leukaemia.
Lancet. 385:1447–1459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cross NC, White HE, Colomer D, Ehrencrona
H, Foroni L, Gottardi E, Lange T, Lion T, Machova Polakova K,
Dulucq S, et al: Laboratory recommendations for scoring deep
molecular responses following treatment for chronic myeloid
leukemia. Leukemia. 29:999–1003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pallera A, Altman JK, Berman E, Abboud CN,
Bhatnagar B, Curtin P, DeAngelo DJ, Gotlib J, Hagelstrom RT, Hobbs
G, et al: NCCN guidelines insights: Chronic myeloid leukemia,
version 1.2017. J Natl Compr Canc Netw. 14:1505–1512. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Branford S, Cross NC, Hochhaus A, Radich
J, Saglio G, Kaeda J, Goldman J and Hughes T: Rationale for the
recommendations for harmonizing current methodology for detecting
BCR-ABL transcripts in patients with chronic myeloid leukaemia.
Leukemia. 20:1925–1930. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ramsden SC, Daly S, Geilenkeuser WJ,
Duncan G, Hermitte F, Marubini E, Neumaier M, Orlando C, Palicka V,
Paradiso A, et al: EQUAL-quant: An international external quality
assessment scheme for real-time PCR. Clin Chem. 52:1584–1591. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang T, Grenier S, Nwachukwu B, Wei C,
Lipton JH and Kamel-Reid S; Association for Molecular Pathology
Hematopathology Subdivision, : Inter-laboratory comparison of
chronic myeloid leukemia minimal residual disease monitoring:
Summary and recommendations. J Mol Diagn. 9:421–430. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alikian M, Whale AS, Akiki S, Piechocki K,
Torrado C, Myint T, Cowen S, Griffiths M, Reid AG, Apperley J, et
al: RT-qPCR and RT-digital PCR: A comparison of different platforms
for the evaluation of residual disease in chronic myeloid leukemia.
Clin Chem. 63:525–531. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Branford S, Fletcher L, Cross NC, Müller
MC, Hochhaus A, Kim DW, Radich JP, Saglio G, Pane F, Kamel-Reid S,
et al: Desirable performance characteristics for BCR-ABL
measurement on an international reporting scale to allow consistent
interpretation of individual patient response and comparison of
response rates between clinical trials. Blood. 112:3330–3338. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Foskett P, Gerrard G and Foroni L:
Real-time quantification assay to monitor BCR-ABL1 transcripts in
chronic myeloid leukemia. Methods Mol Biol. 1160:115–124. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huggett JF, Cowen S and Foy CA:
Considerations for digital PCR as an accurate molecular diagnostic
tool. Clin Chem. 61:79–88. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Whale AS, Cowen S, Foy CA and Huggett JF:
Methods for applying accurate digital PCR analysis on low copy DNA
samples. PLoS One. 8:e581772013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang WJ, Zheng CF, Liu Z, Tan YH, Chen XH,
Zhao BL, Li GX, Xu ZF, Ren FG, Zhang YF, et al: Droplet digital PCR
for BCR/ABL(P210) detection of chronic myeloid leukemia: A high
sensitive method of the minimal residual disease and disease
progression. Eur J Haematol. 101:291–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zagaria A, Anelli L, Coccaro N, Tota G,
Casieri P, Cellamare A, Impera L, Brunetti C, Minervini A,
Minervini CF, et al: BCR-ABL1 e6a2 transcript in chronic myeloid
leukemia: Biological features and molecular monitoring by droplet
digital PCR. Virchows Arch. 467:357–363. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mori S, Vagge E, le Coutre P, Abruzzese E,
Martino B, Pungolino E, Elena C, Pierri I, Assouline S, D'Emilio A,
et al: Age and dPCR can predict relapse in CML patients who
discontinued imatinib: The ISAV study. Am J Hematol. 90:910–914.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goh HG, Lin M, Fukushima T, Saglio G, Kim
D, Choi SY, Kim SH, Lee J, Lee YS, Oh SM and Kim DW: Sensitive
quantitation of minimal residual disease in chronic myeloid
leukemia using nanofluidic digital polymerase chain reaction assay.
Leuk Lymphoma. 52:896–904. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Taylor SC, Laperriere G and Germain H:
Droplet digital PCR versus RT-qPCR for gene expression analysis
with low abundant targets: From variable nonsense to publication
quality data. Sci Rep. 7:24092017. View Article : Google Scholar : PubMed/NCBI
|