Introduction
Lung cancer has emerged as the most common malignant
tumor and accounts for ~25% of cancer-associated mortalities
worldwide according to the latest cancer statistics report
(1). The main pathological types of
lung cancer include small cell lung cancer and non-small cell lung
cancer, the latter of which predominantly comprising of lung
adenocarcinoma (LUAD) cases (2).
Despite advances in diagnostic tools, surgical approaches and
chemotherapy, the 5-year survival rate of patients with lung cancer
is reported to be <10%, potentially due to the lack of effective
biomarkers (3,4). Therefore, the identification of novel
and reliable diagnostic and prognostic biomarkers is required.
The human brain-expressed X-linked (BEX) family
consists of five members, BEX1-5, located on the Xq22 chromosome
(5). Members of the BEX family are
widely expressed in several types of tissues, and are closely
associated with the development of neurons (6). Previous studies have demonstrated that
BEX members play important roles in regulating important cellular
processes, including the cell cycle and apoptosis (7–9).
Increasing evidence revealed that BEX members are aberrantly
expressed in different types of cancer, suggesting that they may
serve roles in tumorigenesis (10–22).
BEX1 is implicated in leukemogenesis, and aberrant downregulation
of BEX1 has been observed in acute and chronic myeloid leukemia
(10,11). Furthermore, BEX1 is associated with
the development of other types of cancer, including pediatric
intracranial ependymoma and esophageal squamous cell cancer
(12,13). BEX2 may serve as an oncogene in
different types of cancer, including glioma, breast and colorectal
cancer (14–16). By contrast, BEX3 inhibits the
formation of breast tumors and serves a pro-apoptotic role
(17,18). Previous studies revealed that BEX4
serves as a tumor suppressor in ovarian cancer and as an oncogene
in oral squamous cell carcinoma (19,20). To
the best of the authors' knowledge, the role of BEX5 in cancer
remains unreported. The aforementioned studies highlighted an
association between the BEX family and various types of cancer.
However, few studies have investigated the association between the
BEX family and lung cancer. A previous study reported that high
expression levels of BEX2 and BEX4 were significantly associated
with a favorable prognosis in patients with LUAD (21). Furthermore, overexpression of BEX4
may promote LUAD cell proliferation and growth in vitro
(22). The potential significance of
the BEX family in LUAD therefore warrants further
investigation.
The present study investigated the specific
expression patterns of BEX members as well as their clinical
significance in LUAD by mining databases and performing in
vitro experiments. The results obtained in the present study
revealed that the BEX family was downregulated in LUAD samples and
tumor cell lines compared with normal samples and human bronchial
epithelial cells, respectively, and may be involved in LUAD
pathogenesis. The association between the expression levels of BEX
members and the clinicopathological parameters of patients with
LUAD was therefore further investigated and the diagnostic and
prognostic values of the BEX family in patients with LUAD were
assessed.
Materials and methods
Data mining
Publicly available LUAD gene expression data and the
corresponding The Cancer Genome Atlas (TCGA; cancergenome.nih.gov/) clinical data were obtained
from the University of California Santa Cruz Xena repository
(xenabrowser.net). The expression data was
obtained from the files entitled
‘TCGA_LUAD_sampleMap/HiSeqV2/PANCAN’. Additionally, the clinical
phenotype information was selected from
‘TCGA_LUAD_sampleMap/LUAD_clinicalMatrix’. Based on the criteria,
the gene expression data from the TCGA was transformed by log2(x+1)
and normalized across all cohorts, then extracted and matched with
the clinical data as reported in (23).
A total of 574 cases (515 LUAD samples and 59 normal
samples from heathy controls) with BEX family expression data and
482 LUAD samples with gene expression data and clinicopathological
information were included in the current study. In order to
investigate the expression levels of BEX members in clinical LUAD
samples, a cohort of 59 pairs of LUAD and adjacent non-cancerous
tissues were taken from the 574 LUAD cases. The cancer samples were
staged according to the American Joint Committee on Cancer staging
system (24).
The mRNA expression of BEX members in LUAD was
investigated using the Oncomine database (oncomine.org), a gene expression array database used
for the analysis of the transcription levels of genes in several
types of cancer. The datasets used in the present study included
Garber Lung Statistics (25),
Okayama Lung Statistics (26),
Bhattacharjee Lung Statistics (27),
Hou Lung Statistics (28), Selamat
Lung Statistics (29) and Landi Lung
Statistics (30). P<0.01 and a
1.5-fold change was set as the threshold. Genes in the present
study were not limited by their rank. The final datasets collected
are presented in Table I.
 | Table I.Changes in the BEX family
transcription levels between lung adenocarcinoma and normal tissues
obtained from the ONCOMINE database. |
Table I.
Changes in the BEX family
transcription levels between lung adenocarcinoma and normal tissues
obtained from the ONCOMINE database.
| A, BEX1 |
|---|
|
|---|
| Reporter | Fold change | P-value | t-test | Oncomine
dataset |
|---|
| 218332_at | −3.869 |
1.53×10−11 | −10.401 | Okayama lung
statistics |
| IMAGE:341706 | −1.502 | 0.003 | −3.193 | Garber lung
statistics |
| ILMN_2234697 | −1.941 |
1.22×10−10 | −7.024 | Selamat lung
statistics |
|
| B, BEX2 |
|
|
Reporter | Fold
change | P-value | t-test | Oncomine
dataset |
|
| 224367_at | −1.723 |
2.39×10−4 | −3.727 | Hou lung
statistics |
|
| C, BEX3 |
|
|
Reporter | Fold
change | P-value | t-test | Oncomine
dataset |
|
| ILMN_1729208 | −1.508 |
2.02×10−5 | −4.276 | Selamat lung
statistics |
|
| D, BEX4 |
|
|
Reporter | Fold
change | P-value | t-test | Oncomine
dataset |
|
| 40916_at | −3.440 |
4.62×10−5 | −4.828 | Bhattacharjee lung
statistics |
| ILMN_2351638 | −2.339 |
3.68×10−17 | −9.910 | Selamat lung
statistics |
| 215440_s_at | −1.756 |
1.55×10−9 | −6.562 | Landi lung
statistics |
| 215440_s_at | −1.960 |
4.20×10−7 | −5.594 | Hou lung
statistics |
|
| E, BEX5 |
|
|
Reporter | Fold
change | P-value | t-test | Oncomine
dataset |
|
| ILMN_1806473 | −1.624 |
1.82×10−11 | −7.327 | Selamat lung
statistics |
| 229963_at | −1.868 |
1.33×10−4 | −3.885 | Hou lung
statistics |
Data on the methylation status of BEX family was
downloaded from MethHC (methhc.mbc.nctu.edu.tw/), a database of DNA
methylation status and gene expression in human cancer. Data on 464
tissue samples, 435 LUAD tissues and 29 normal tissues from healthy
controls, with methylation values were obtained.
Mutational and copy number variations (CNVs)
analyses of the BEX family members in LUAD samples were performed
using cBioPortal for Cancer Genomics (cbioportal.org/) as described previously (31). Data from the following studies were
included: Lung Adenocarcinoma (Broad, Cell 2012) (32), Lung Adenocarcinoma (TCGA, Nature
2014) (33), Lung Adenocarcinoma
(TCGA, Provisional) and Lung Adenocarcinoma (TCGA, PanCancer Atlas)
(34).
Cell lines and culture
Human bronchial epithelial (HBE) cells and the lung
cancer cell lines H520, H1975, H358, H460, A549, 95D and SPC-A-1
were obtained from the Cell Bank of the Chinese Academy of Science.
HBE cells were maintained in DMEM (HyClone) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
the lung cancer cell lines were cultured in RPMI-1640 medium
(HyClone) supplemented with 10% FBS. All cells were maintained at
37°C with 5% CO2.
RNA extraction and
reverse-transcription quantitative polymerase chain reaction
(RT-qPCR)
RNA extraction and RT-qPCR were performed as
previously described (35,36). The primers used in the present study
are presented in Table SI.
Receiver operating characteristic
(ROC) curve analysis
The diagnostic value of the expression levels of the
BEX family in LUAD was investigated by analyzing the expression
data from 515 LUAD and 59 normal tissues. Specificity and
sensitivity were plotted on the x- and y-axes, respectively. The
area under curve (AUC) was calculated to assess the ability of the
expression levels of BEX members to predict the outcome of patients
with LUAD.
Survival analysis
The association between expression of the BEX family
members and the overall survival (OS) of patients with LUAD was
assessed using a log-rank test and Kaplan-Meier analysis. The
median expression value of each gene was used as the cut-off value
to divide patients into high- and low-expression groups. The
multivariate Cox regression model was applied for prognostic
analysis.
Statistical analysis
A two-tailed paired t-test with a Bonferroni's
correction was used to analyze the gene expression differences in
tumor and adjacent non-cancerous tissues. The Mann Whitney U test
was used to compare tumor samples and unpaired normal control
samples. The associations between the expression levels of BEX
members and patient clinical characteristics were determined using
a χ2 test. One-way ANOVA followed by the Dunnett's
multiple comparisons test was used to analyze the expression level
of BEX members in HBE cells compared with lung cancer cell lines.
SPSS software (version 20; IBM Corp.) was used to perform all the
statistical analysis. P<0.05 was used to indicate a
statistically significant difference.
Results
Expression level of BEX members is
significantly downregulated in LUAD samples and tumor cell
lines
The expression level of each BEX member was
significantly downregulated in 59 LUAD tissues compared with the
paired adjacent non-cancerous tissues (P<0.01; Fig. 1A-E). Similarly, the expression level
of BEX members was significantly downregulated in the remaining 515
tumor samples compared with the unpaired normal control samples
(P<0.001; Fig. S1A-E). As
presented in Table I, the expression
results were successfully validated in independent cohorts from the
Oncomine database. RT-qPCR revealed that the mRNA levels of BEX
members were significantly decreased in the majority of the tumor
cell lines, particularly in H1975, H358, SPC-A-1 and H520, compared
with the HBE cells (Fig. 2A-E).
 | Figure 2.Expression levels of the BEX family
were significantly downregulated in the majority of the lung
adenocarcinoma cell lines investigated. The mRNA expression of (A)
BEX1, (B) BEX2, (C) BEX3, (D) BEX4 and (E) BEX5 in HBE, H520,
H1975, H358, H460, A549, 95D and SPC-A-1 cells was determined using
reverse-transcription quantitative polymerase chain reaction.
*P<0.05 and **P<0.01 vs. HBE cells. BEX, brain-expressed
X-linked; HBE, human bronchial epithelial. |
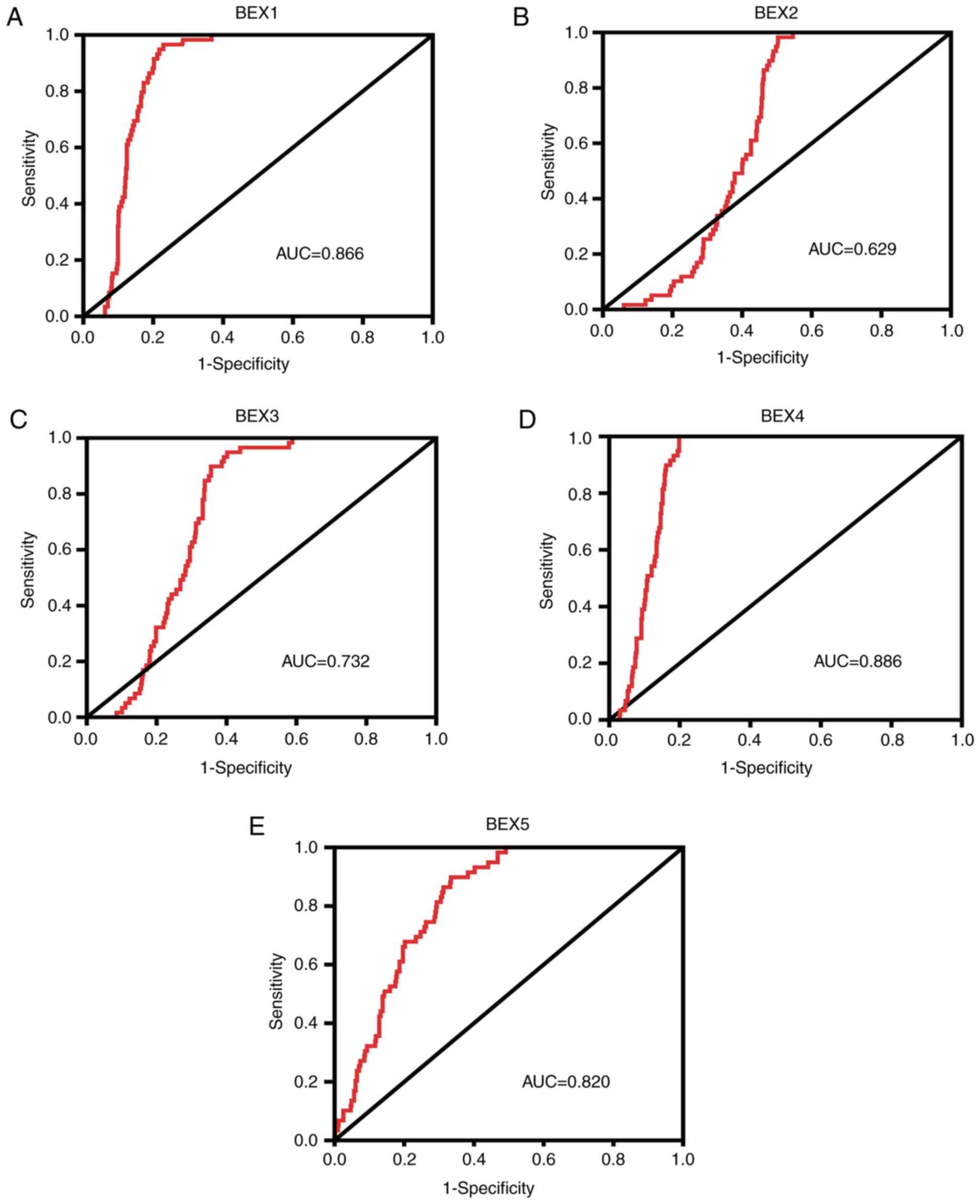
ROC analysis of BEX expression in
patients with LUAD
Since the BEX family was significantly downregulated
in LUAD samples compared with controls, the current study explored
whether BEX members may serve as potential diagnostic biomarkers in
LUAD. ROC curves and AUC analysis were performed to evaluate the
overall prognostic performance. BEX1 [AUC, 0.866; 95% CI
(confidence interval), 0.837–0.895], BEX2 (AUC, 0.629; 95% CI,
0.584–0.674), BEX3 (AUC, 0.732; 95% CI, 0.690–0.773), BEX4 (AUC,
0.886; 95% CI, 0.859–0.912) and BEX5 (AUC, 0.820; 95% CI,
0.779–0.860) had high sensitivity and specificity, suggesting that
the BEX family may have diagnostic value in patients with LUAD
(Fig. 3A-E).
Associations between the expression of
the BEX family and clinicopathological characteristics of patients
with LUAD
The associations between the expression levels of
BEX members and the clinicopathological characteristics of 482
patients with LUAD were investigated. Low expression levels of BEX4
and BEX5 were significantly associated with an advanced American
Joint Committee on Cancer (AJCC) clinical stage (P=0.004 and
P=0.027, respectively; Tables II
and III, respectively) and
advanced pathological T stage (P=0.002 and P=0.003, respectively;
Tables II and III, respectively). Lower expression level
of BEX4 was significantly associated with pathological N stage
(P=0.033, Table II). Decreased mRNA
levels of BEX1 was significantly associated with advanced
pathological T stage (P=0.023; Tables
SII). The expression level of BEX2 and BEX4 was strongly
associated with sex (P=0.003; P=0.005, respectively; Tables SIII and II, respectively). The expression level of
BEX4 was significantly associated with age (P=0.019; Table II). There was no significant
association between the expression of BEX3 and any
clinicopathological characteristic in patients with LUAD (Table SIV).
 | Table II.Association between the BEX4
expression level and patient clinical characteristics in The Cancer
Genome Atlas lung adenocarcinoma cohort. |
Table II.
Association between the BEX4
expression level and patient clinical characteristics in The Cancer
Genome Atlas lung adenocarcinoma cohort.
|
|
| BEX4
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | n | High | Low | χ2 | P-value |
|---|
| Age at diagnosis
(years) |
|
|
| 5.545 | 0.019 |
|
<60 | 131 | 54 | 77 |
|
|
|
≥60 | 351 | 187 | 164 |
|
|
| Sex |
|
|
| 8.030 | 0.005 |
|
Male | 221 | 95 | 126 |
|
|
|
Female | 261 | 146 | 115 |
|
|
| Clinical stage |
|
|
| 8.288 | 0.004 |
|
I+II | 378 | 202 | 176 |
|
|
|
III+IV | 104 | 39 | 65 |
|
|
| T (Primary
tumor) |
|
|
| 15.226 | 0.002 |
| T1 | 165 | 101 | 64 |
|
|
| T2 | 252 | 116 | 136 |
|
|
| T3 | 44 | 15 | 29 |
|
|
| T4 | 18 | 7 | 11 |
|
|
| TX | 3 | 2 | 1 |
|
|
| N (Regional lymph
nodes) |
|
|
| 4.549 | 0.033 |
| N0 | 312 | 166 | 146 |
|
|
|
N1-3 | 161 | 69 | 92 |
|
|
| NX | 9 | 6 | 3 |
|
|
| M (Distant
metastases) |
|
|
| 0.552 | 0.457 |
| M0 | 319 | 158 | 161 |
|
|
| M1 | 24 | 10 | 14 |
|
|
| MX | 139 | 73 | 66 |
|
|
 | Table III.Associations between the BEX5
expression level and patient clinical characteristics in The Cancer
Genome Atlas lung adenocarcinoma cohort. |
Table III.
Associations between the BEX5
expression level and patient clinical characteristics in The Cancer
Genome Atlas lung adenocarcinoma cohort.
|
|
| BEX5
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | n | High | Low | χ2 | P-value |
|---|
| Age at diagnosis
(years) |
|
|
| 1.268 | 0.260 |
|
<60 | 131 | 60 | 71 |
|
|
|
≥60 | 351 | 181 | 170 |
|
|
| Sex |
|
|
| 3.017 | 0.082 |
|
Male | 221 | 101 | 120 |
|
|
|
Female | 261 | 140 | 121 |
|
|
| Clinical stage |
|
|
| 4.904 | 0.027 |
|
I+II | 378 | 199 | 179 |
|
|
|
III+IV | 104 | 42 | 62 |
|
|
| T (Primary
tumor) |
|
|
| 14.117 | 0.003 |
| T1 | 165 | 102 | 63 |
|
|
| T2 | 252 | 109 | 143 |
|
|
| T3 | 44 | 21 | 23 |
|
|
| T4 | 18 | 8 | 10 |
|
|
| TX | 3 | 1 | 2 |
|
|
| N (Regional lymph
nodes) |
|
|
| 2.204 | 0.138 |
| N0 | 312 | 162 | 150 |
|
|
|
N1-3 | 161 | 72 | 89 |
|
|
| NX | 9 | 7 | 2 |
|
|
| M (Distant
metastases) |
|
|
| 0.041 | 0.840 |
| M0 | 319 | 153 | 166 |
|
|
| M1 | 24 | 11 | 13 |
|
|
| MX | 139 | 77 | 62 |
|
|
Prognostic value of the BEX family in
patients with LUAD
The Kaplan-Meier method was used to assess the
prognostic significance of the BEX family in patients with LUAD in
the TCGA database. The expression levels of BEX1-3 were not
significantly associated with OS (Fig.
4A-C). However, the downregulation of BEX4 and BEX5 expression
levels were significantly associated with poor OS (P<0.001 and
P=0.001, respectively; Fig. 4D-E).
The association between the expression levels of the BEX family and
the outcomes of patients with LUAD using clinical stage subgroup
analysis revealed that BEX4 and BEX5 were associated with the OS of
patients with stage I–II (P<0.001 and P=0.018, respectively;
Fig. S2D-E), but not of patients
with stage III–IV (P=0.261 and P=0.140, respectively; Fig. S3D-E). There was no significant
difference between BEX1, BEX2 or BEX3 expression and OS of patients
with stage I–II or stage III–IV (Figs.
S2A-C and 3A-C).
Characteristics such as age, sex, AJCC stage and the
expression level of BEX members were analyzed using a multivariate
Cox regression model. In addition to clinical stage [hazard ratio
(HR)=2.290; P<0.001], the expression level of the BEX family was
an independent prognostic factor (HR=1.468 and P=0.030 for BEX3;
HR=0.469 and P<0.001 for BEX4; HR=0.723 and P=0.043 for BEX5)
for patients with LUAD (Table IV).
In addition, as shown in Table IV,
BEX4 expression was significantly associated with OS at stages I–II
(HR=0.476; P=0.002), while BEX3 (HR=2.450 and P=0.014), BEX4
(HR=0.288; P=0.001) and BEX5 (HR=0.569 and P=0.047) were associated
with OS at stages III–IV.
 | Table IV.Multivariate analyses of overall
survival in patients with lung adenocarcinoma. |
Table IV.
Multivariate analyses of overall
survival in patients with lung adenocarcinoma.
| A, All
patients |
|---|
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Parameter | P-value | Hazard ratio | 95% confidence
interval |
|---|
| Age (≥60
vs.<60) | 0.549 | 1.108 | 0.792–1.549 |
| Sex (female vs.
male) | 0.543 | 1.081 | 0.797–1.466 |
| Clinical stage
(III/IV vs. I/II) | <0.001 | 2.290 | 1.665–3.149 |
| BEX1 expression
(high vs. low) | 0.394 | 1.146 | 0.838–1.567 |
| BEX2 expression
(high vs. low) | 0.452 | 1.167 | 0.781–1.745 |
| BEX3 expression
(high vs. low) | 0.030 | 1.468 | 1.038–2.077 |
| BEX4 expression
(high vs. low) | <0.001 | 0.469 | 0.320–0.686 |
| BEX5 expression
(high vs. low) | 0.043 | 0.723 | 0.528–0.989 |
|
| B, Patients with
stage I+II |
|
|
| Multivariate
analysis |
|
|
|
|
Parameter | P-value | Hazard
ratio | 95% confidence
interval |
|
| Age (≥60 vs.
<60) | 0.216 | 1.321 | 0.850–2.051 |
| Sex (female vs.
male) | 0.912 | 0.979 | 0.675–1.421 |
| BEX1 expression
(high vs. low) | 0.519 | 1.134 | 0.773–1.664 |
| BEX2 expression
(high vs. low) | 0.915 | 0.974 | 0.604–1.572 |
| BEX3 expression
(high vs. low) | 0.341 | 1.220 | 0.810–1.835 |
| BEX4 expression
(high vs. low) | 0.002 | 0.476 | 0.299–0.757 |
| BEX5 expression
(high vs. low) | 0.265 | 0.798 | 0.537–1.186 |
|
| C, Patients with
stage III+IV |
|
|
| Multivariate
analysis |
|
|
|
|
Parameter | P-value | Hazard
ratio | 95% confidence
interval |
|
| Age (≥60
vs.<60) | 0.892 | 1.040 | 0.592–1.827 |
| Sex (female vs.
male) | 0.126 | 1.587 | 0.878–2.869 |
| BEX1 expression
(high vs. low) | 0.662 | 1.139 | 0.635–2.042 |
| BEX2 expression
(high vs. low) | 0.173 | 1.696 | 0.793–3.628 |
| BEX3 expression
(high vs. low) | 0.014 | 2.450 | 1.197–5.015 |
| BEX4 expression
(high vs. low) | 0.001 | 0.288 | 0.134–0.618 |
| BEX5 expression
(high vs. low) | 0.047 | 0.569 | 0.326–0.992 |
Methylation, mutation and CNV analysis
of BEX family members in patients with LUAD
The total alteration frequency of gene mutations,
amplifications and deletions in patients with LUAD was <2%
(Fig. S4) according to the
sequencing data in datasets obtained from cBioPortal. BEX1 and BEX3
were hypermethylated in tumor tissues compared with normal tissues,
whereas BEX2, BEX4 and BEX5 did not exhibit any statistically
significant difference in methylation (Fig. S5).
Discussion
Despite improvement in diagnostic and treatment
strategies, the prognosis of patients with lung cancer remains poor
(37), potentially due to a lack of
effective predictive markers at the early stages of the disease
(38,39). Therefore, novel biomarkers for the
detection and prognosis evaluation of LUAD are required. Previous
studies have revealed that the BEX family may serve roles as
predictive biomarkers in tumors, such as esophageal squamous cell
cancer and acute myeloid leukemia (12,40). The
potential clinical value of the BEX family members in LUAD warrants
further research.
The present study aimed to investigate the role of
the BEX family in LUAD. The expression levels of BEX1-5 in LUAD
were examined by mining the TCGA and Oncomine databases. The
results obtained in the current study revealed that the expression
levels of BEX members in carcinoma tissues were significantly
decreased compared with normal tissues. Similarly, experimental
in vitro data revealed that the mRNA levels of BEX members
were decreased in the majority of the lung cancer cell lines
investigated compared with HBE cells. The abnormal expression of
the BEX family has been implicated in various types of cancer, such
as breast cancer and oral squamous cell carcinoma (16,20). The
expression levels of BEX1 and BEX4 were significantly reduced in
oral squamous cell carcinoma tissues compared with paired normal
epithelial (20). Foltz et al
(41) reported that BEX1 and BEX2
had differentially methylated CpG sites and increased methylation
was associated with a decreased expression level in malignant
glioma tissues compared with paired normal specimens. Previous
studies revealed that several factors are involved in gene
regulation, including mutations, copy number variations (CNVs) and
DNA methylation (42–46). To further explore the results of
differential expression, gene methylation status, mutation status
and CNVs were analyzed. As the total alteration frequency of gene
mutations, amplifications and deletions was <2%, the expression
levels of the BEX family were likely not associated with mutations
or CNVs. The methylation status of BEX1 and BEX3 may partially
explain the low expression of them in LUAD. However, the mechanisms
underlying the downregulation of BEX family requires further
investigation.
Gene expression signatures have been used for the
clinical diagnosis of various types of cancer (47). The differential expression of the BEX
family in LUAD and normal tissues revealed in the present study
suggested that that the BEX family may have diagnostic value in
LUAD. Furthermore, the high AUC values obtained in the present
study indicated that BEX1-5 exhibited good diagnostic performance.
Whether the combination of BEX members provides higher sensitivity
and specificity compared the individual use of each BEX family
member requires further investigation.
A previous study reported that low expression of
BEX1 was associated with clinicopathological characteristics such
as tumor volume, T stage and clinical stage in esophageal squamous
cell cancer (12). The present study
investigated the association between the expression of the BEX
family and the clinicopathological features of patients with LUAD.
The low expression levels of BEX1, BEX4 and BEX5 were significantly
associated with advanced clinical stage. The downregulation of BEX4
was significantly associated with regional lymph node metastasis.
These results suggested the BEX family may be involved in tumor
invasion and metastasis, which may result in less favorable
outcomes in patients with LUAD. Additional studies are required to
investigate the effect of the BEX family on tumor metastasis.
The present study revealed that BEX4 and BEX5 were
associated with the prognosis of LUAD patients. The low expression
levels of BEX4 and BEX5 were associated with poor prognosis. The
results obtained in the current study suggested that members of the
BEX family may serve as biomarkers for predicting poor prognosis in
patients with LUAD.
The present study is limited by the lack of
sufficient experimental data and validation at the protein level.
Therefore, further studies using large, independent clinical cancer
cohorts are required to corroborate the results obtained in the
present study. Additionally, the roles and underlying mechanisms of
members of the BEX family in the development of LUAD remain unclear
and warrant further investigation. In summary, the present study
demonstrated that the BEX family was significantly downregulated in
LUAD tissues compared with normal tissues, and that this expression
pattern may serve as a potential biomarker for the diagnosis and
prognosis prediction of patients with LUAD.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81573114).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request. The datasets generated and/or analyzed during the present
study are available from The Cancer Genome Atlas (cancer.gov/tcga).
Authors' contributions
JC and WL conceived and designed the study. ZZ and
ZL analyzed the data and drafted the manuscript. FH and JL
participated in data interpretation and manuscript revision. HC
performed the RT-qPCR. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BEX
|
brain-expressed X-linked
|
|
LUAD
|
lung adenocarcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
RT-qPCR
|
reverse-transcription quantitative
polymerase chain reaction
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the curve
|
|
AJCC
|
American Joint Committee on Cancer
|
|
OS
|
overall survival
|
|
CNV
|
copy number variations
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ridge CA, McErlean AM and Ginsberg MS:
Epidemiology of lung cancer. Semin Intervent Radiol. 30:93–98.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang L: Adaptive evolution and frequent
gene conversion in the brain expressed X-linked gene family in
mammals. Biochem Genet. 46:293–311. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alvarez E, Zhou W, Witta SE and Freed CR:
Characterization of the Bex gene family in humans, mice, and rats.
Gene. 357:18–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao Q, Hu Y, Liu Y, Wang Z, Geng H, Hu L,
Xu D, Wang K, Zheng L, Zheng S and Ding K: BEX1 promotes
imatinib-induced apoptosis by binding to and antagonizing BCL-2.
PLoS One. 9:e917822014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vilar M, Murillo-Carretero M, Mira H,
Magnusson K, Besset V and Ibáñez CF: Bex1, a novel interactor of
the p75 neurotrophin receptor, links neurotrophin signaling to the
cell cycle. EMBO J. 25:1219–1230. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou X, Meng Q, Xu X, Zhi T, Shi Q, Wang Y
and Yu R: Bex2 regulates cell proliferation and apoptosis in
malignant glioma cells via the c-Jun NH2-terminal kinase pathway.
Biochem Biophys Res Commun. 427:574–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lindblad O, Li T, Su X, Sun J, Kabir NN,
Levander F, Zhao H, Lu G, Rönnstrand L and Kazi JU: BEX1 acts as a
tumor suppressor in acute myeloid leukemia. Oncotarget.
6:21395–21405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ding K, Su Y, Pang L, Lu Q, Wang Z, Zhang
S, Zheng S, Mao J and Zhu Y: Inhibition of apoptosis by
downregulation of hBex1, a novel mechanism, contributes to the
chemoresistance of Bcr/Abl+ leukemic cells. Carcinogenesis.
30:35–42. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Geng HT, Cheng ZW, Cao RJ, Wang ZB, Xing
SZ, Guo C, Wang F, Liu CM, Chen SS and Cheng YF: Low expression of
BEX1 predicts poor prognosis in patients with esophageal squamous
cell cancer. Oncol Rep. 40:2778–2787. 2018.PubMed/NCBI
|
|
13
|
Karakoula K, Jacques TS, Phipps KP,
Harkness W, Thompson D, Harding BN, Darling JL and Warr TJ:
Epigenetic genome-wide analysis identifies BEX1 as a candidate
tumour suppressor gene in paediatric intracranial ependymoma.
Cancer Lett. 346:34–44. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu Y, Xiao Q, Chen H, He J, Tan Y, Liu Y,
Wang Z, Yang Q, Shen X, Huang Y, et al: BEX2 promotes tumor
proliferation in colorectal cancer. Int J Biol Sci. 13:286–294.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nie E, Zhang X, Xie S, Shi Q, Hu J, Meng
Q, Zhou X and Yu R: B-catenin is involved in Bex2 down-regulation
induced glioma cell invasion/migration inhibition. Biochem Biophys
Res Commun. 456:494–499. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Naderi A, Teschendorff AE, Beigel J,
Cariati M, Ellis IO, Brenton JD and Caldas C: BEX2 is overexpressed
in a subset of primary breast cancers and mediates nerve growth
factor/nuclear factor-kappaB inhibition of apoptosis in breast
cancer cell lines. Cancer Res. 67:6725–6736. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tong X, Xie D, Roth W, Reed J and Koeffler
HP: NADE (p75NTR-associated cell death executor) suppresses
cellular growth in vivo. Int J Oncol. 22:1357–1362.
2003.PubMed/NCBI
|
|
18
|
Yoon K, Jang HD and Lee SY: Direct
interaction of Smac with NADE promotes TRAIL-induced apoptosis.
Biochem Biophys Res Commun. 319:649–654. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chien J, Staub J, Avula R, Zhang H, Liu W,
Hartmann LC, Kaufmann SH, Smith DI and Shridhar V: Epigenetic
silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene.
24:5089–5100. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao W, Li JZ, Chen SQ, Chu CY, Chan JY and
Wong TS: Decreased brain-expressed X-linked 4 (BEX4) expression
promotes growth of oral squamous cell carcinoma. J Exp Clin Canc
Res. 35:922016. View Article : Google Scholar
|
|
21
|
Zhu XF, Zhu BS, Wu FM and Hu HB: DNA
methylation biomarkers for the occurrence of lung adenocarcinoma
from TCGA data mining. J Cell Physiol. 233:6777–6784. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao Z, Li J, Tan F, Gao S and He J: mTOR
up-regulation of BEX4 promotes lung adenocarcinoma cell
proliferation by potentiating OCT4. Biochem Bioph Res Commun.
500:302–309. 2018. View Article : Google Scholar
|
|
23
|
Goldman M, Craft B, Hastie M, Repečka K,
Kamath A, McDade F, Rogers D, Brooks AN, Zhu Z and Haussler D: The
UCSC Xena platform for public and private cancer genomics data
visualization and interpretation. bioRxiv. 3264702019.
|
|
24
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bhattacharjee A, Richards WG, Staunton J,
Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et
al: Classification of human lung carcinomas by mRNA expression
profiling reveals distinct adenocarcinoma subclasses. Proc Natl
Acad Sci USA. 98:13790–13795. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han F, Zhang MQ, Liu WB, Sun L, Hao XL,
Yin L, Jiang X, Cao J and Liu JY: SOX30 specially prevents
Wnt-signaling to suppress metastasis and improve prognosis of lung
adenocarcinoma patients. Respir Res. 19:2412018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han F, Liu W, Jiang X, Shi X, Yin L, Ao L,
Cui Z, Li Y, Huang C, Cao J and Liu J: SOX30, a novel epigenetic
silenced tumor suppressor, promotes tumor cell apoptosis by
transcriptional activating p53 in lung cancer. Oncogene.
34:4391–4402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heckman-Stoddard BM: Oncology biomarkers:
Discovery, validation, and clinical use. Semin Oncol Nurs.
28:93–98. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goossens N, Nakagawa S, Sun X and Hoshida
Y: Cancer biomarker discovery and validation. Transl Cancer Res.
4:256–269. 2015.PubMed/NCBI
|
|
40
|
Fischer C, Drexler HG, Reinhardt J,
Zaborski M and Quentmeier H: Epigenetic regulation of brain
expressed X-linked-2, a marker for acute myeloid leukemia with
mixed lineage leukemia rearrangements. Leukemia. 21:374–377. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Foltz G, Ryu GY, Yoon JG, Nelson T, Fahey
J, Frakes A, Lee H, Field L, Zander K, Sibenaller Z, et al:
Genome-wide analysis of epigenetic silencing identifies BEX1 and
BEX2 as candidate tumor suppressor genes in malignant glioma.
Cancer Res. 66:6665–6674. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Williams JL, Greer PA and Squire JA:
Recurrent copy number alterations in prostate cancer: An in silico
meta-analysis of publicly available genomic data. Cancer Genet.
207:474–488. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
De Carvalho DD, Sharma S, You JS, Su SF,
Taberlay PC, Kelly TK, Yang X, Liang G and Jones PA: DNA
methylation screening identifies driver epigenetic events of cancer
cell survival. Cancer Cell. 21:655–667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Black JC, Manning AL, Van Rechem C, Kim J,
Ladd B, Cho J, Pineda CM, Murphy N, Daniels DL, Montagna C, et al:
KDM4A lysine demethylase induces site-specific copy gain and
rereplication of regions amplified in tumors. Cell. 154:541–555.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Licht JD: DNA methylation inhibitors in
cancer therapy: The immunity dimension. Cell. 162:938–939. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xiao Q, Zhou D, Rucki AA, Williams J, Zhou
J, Mo G, Murphy A, Fujiwara K, Kleponis J, Salman B, et al:
Cancer-associated fibroblasts in pancreatic cancer are reprogrammed
by tumor-induced alterations in genomic DNA methylation. Cancer
Res. 76:5395–5404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kamel HFM and Al-Amodi HSAB: Exploitation
of gene expression and cancer biomarkers in paving the path to era
of personalized medicine. Genomics Proteomics Bioinformatics.
15:220–235. 2017. View Article : Google Scholar : PubMed/NCBI
|