Introduction
Jumonji domain-containing protein 6 (JMJD6)
is a member of the Jumonji C domain-containing metalloenzymes
(1,2). For the past 17 years, studies have
indicated that JMJD6 as a multidimensional protein in biological
processes that interacts with multiple protein substrates in
distinct molecular signaling pathways [such as U2 small nuclear RNA
auxiliary factor 2 and p21 (RAC1) activated kinase 1] (3,4).
According to the current understanding, JMJD6 acts as either a
histone demethylase, which removes methyl moieties on histone H3 at
arginine 2 and H4 at arginine 3, or as a lysyl hydroxylase that
targets the RNA splicing factor U2 small nuclear RNA auxiliary
factor 2 (2,5,6). In
summary, JMJD6 serves diverse roles at the transcriptional,
splicing, posttranscriptional and biochemical levels (7).
The upregulation and functional role of JMJD6 in
cancer areas have been recently demonstrated (8). In breast cancer, overexpression of
JMJD6 has been reported to be associated with poorer outcomes and
more aggressive characteristics, via modulating cell proliferation
and motility (3,7). In hepatocellular carcinoma, JMJD6
promotes progression of malignancy via regulation of CDK4 by
directly targeting its promoter (9).
In melanoma, a positive feedback loop (JMJD6-
serine/threonine-protein kinase PAK 1-mitogen-activated protein
kinase 1-JMJD6) associated with the promotion of melanogenesis,
cell proliferation, invasion and angiogenesis has been investigated
(10). In oral squamous cell
carcinoma (OSCC), JMJD6 has been demonstrated to be a novel
regulator of cancer stem cells via upregulation of interleukin 4
expression (11).
However, little is known regarding the clinical
significance of JMJD6 in head and neck squamous cell carcinoma
(HNSCC), particularly in terms of large-cohort data. The Cancer
Genome Atlas (TCGA) project provides an opportunity for the
comprehensive understanding of human cancer, based on powerful and
detailed information from a large cohort (12–14). In
the present study, bioinformatics analysis was conducted using two
online tools, the University of California Santa Cruz (UCSC) Xena
Browser and Gene Expression Profiling Interactive Analysis 2
(GEPIA2), based on the TCGA primary HNSCC cohort data. The
expression patterns, clinical significance and biological roles of
JMJD6 in HNSCC were systematically illustrated based on the TCGA
cohort and a validation cohort. Subsequent in vitro assays
were performed to investigate the biological roles of JMJD6 in
HNSCC.
Materials and methods
Bioinformatics analysis and
validation
Bioinformatics analysis was performed based on the
TCGA HNSCC cohort using the web-based tools GEPIA2 (http://gepia2.cancer-pku.cn) and the UCSC Xena Browser
(http://xena.ucsc.edu) (12–14).
Only cases of primary HNSCC were included in further analyses of
the expression patterns of JMJD6, snail family
transcriptional repressor 1 (SNAI1), vimentin (VIM),
twist family bHLH transcription factor 1 (TWIST1), cadherin
2 (CDH2) and cadherin 1 (CDH1). Heatmaps of defined
gene sets were generated online (https://xenabrowser.net/heatmap), and detailed data
were downloaded for further analysis (12–14).
A retrospectively validation HNSCC cohort was
constructed, based on 98 primary HNSCC cases admitted to the
Department of Oral Maxillofacial-Head and Neck Oncology, Shanghai
9th People's Hospital between January 2010 and December 2017. The
clinicopathological characteristics of each case were collected for
further analysis. All patients involved in the present study
provided written informed consent, and the study was approved by
the Medical Ethics Committee of the 9th People's Hospital, Shanghai
Jiao Tong University School of Medicine (Shanghai, China)
(SH9H-2019-0421).
Immunohistochemistry staining and
scoring
Tissues were fixed in 10% formalin for 24 h at room
temperature, embedded in paraffin and cut into 5-µm sections.
Following dewaxing (with xylene twice for 10 min), rehydration
(with 100% ethanol twice for 5 min, 95% ethanol twice for 2 min,
and 85% ethanol twice for 2 min at room temperature) and antigen
retrieval (with citrate buffer at 98°C for 15 min), the endogenous
peroxidase activity (with 3% hydrogen peroxide for 10 min at room
temperature) of each section was quenched. Each slide was incubated
with the primary antibody (rabbit polyclonal antibody against
JMJD6; 1:250 dilution; cat no. ab96795; Abcam) overnight at 4°C.
Subsequently, the samples were incubated with a biotinylated
secondary antibody for 50 min at 37°C, and stained with a
3′-diaminobenzidine kit (GT Vision Ltd.) for 1 min at room
temperature. Consecutive sections were counterstained with
hematoxylin and eosin (hematoxylin staining for 5 min and 0.5%
eosin staining for 1 min, at room temperature).
The immunoreactivity score of JMJD6 staining was
recorded by two independent observers under a light microscope
(Olympus Corporation) at ×100 and ×200 magnification, according to
staining intensity and percentage of positive cancer cells. The
staining intensity was scored as follows: Weak, scored 1; moderate,
scored 2; and intense, scored 3. For the percentage of positive
cancer cells, the score was defined as follows: ≤25%, scored 1;
>25%, ≤50%, scored 2; >50, ≤-75%, scored 3; >75%, scored
4. Finally, a total score (ranging between 1 and 12) was acquired
by multiplying the scores for staining intensity and for the
percentage of positive cancer cells. A score of 1–6 was considered
to indicate low expression, whereas a score of 7–12 was considered
to indicate high expression.
Cell culture and plasmid
transfection
The Cal27 and HN12 HNSCC cell lines were cultured in
DMEM (Shanghai Basal Media Technologies Co., Ltd.) supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 µg/ml streptomycin at 37°C with 5%
CO2.
Cal27 cells function as a classical HNSCC cell line,
and HN12 cells possess an increased epithelial-mesenchymal
transition (EMT) potential. In the present study, JMJD6 was
overexpressed in Cal27 cells and knocked down in HN12 cells. The
mammalian expression vector used was pcDNA3.1(+)-myc-JMJD6 and
pcDNA3.1(+) was used as an empty vector control (Genomeditech). The
plasmids (2 µg) were transfected into cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) diluted in OPTI-MEM® I (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Small
interfering RNAs (siRNAs) were constructed (Genomeditech) and
validated at the mRNA and protein levels. Validated small
interfering RNAs (siRNAs, 50 nM) specific to JMJD6 were transfected
into HN12 cells. The sequence of the siRNA targeting JMJD6 was
5′-GAGGGAACCAGCAAGACGA-3′, and the sequence for the scrambled siRNA
was 5′-UUCUCCGAACGUGUCACGUdTdT-3′ (Genomeditech, Co., Ltd.). For
the subsequent analysis, RNA was extracted after 24 h transfection,
and protein was extracted 48 h after transfection.
Western blot analysis
Cellular extracts were acquired by using whole-cell
lysis buffer (Yeasen Biotechnology, Co., Ltd.) containing a
proteinase inhibitor cocktail (1 mM PMSF, Yeasen Biotechnology,
Co., Ltd.; cat. no. 20104ES03). The protein concentration was
determined using BCA assays (Thermo Fisher Scientific, Inc.). After
subjecting the lysates (30 µg protein per lane) to 10% SDS-PAGE,
proteins were transferred to a PVDF membrane by electroblotting.
Subsequently, the membranes were blocked in 5% skimmed milk for 1 h
at room temperature and incubated overnight at 4°C with specific
primary antibodies against JMJD6 (1:1,000; cat. no. ab50720), Snai1
(1:1,000; cat. no. ab53519), Vimentin (1:1,000; cat. no. ab16700),
Twist1 (1:1,000; cat. no. ab175430), N-cadherin (1:1,000; cat. no.
ab98952), E-cadherin (1:1,000; cat. no. ab40772) (all from Abcam)
and GAPDH (1:5000; cat. no. BM3876; Wuhan Boster Biological
Technology, Ltd.). Specific secondary antibodies (horseradish
peroxidase-conjugated goat anti-mouse IgG or horseradish
peroxidase-conjugated goat anti-rabbit IgG; both 1:5,000; cat. nos.
33201ES60 or 34301ES60, respectively; both from Yeasen
Biotechnology, Co., Ltd.;) were incubated in room temperature for 1
h. Specific antibody-bound protein bands were detected with ECL
Plus reagent (EMD Millipore) using an Amersham Imager 600 (GE
Healthcare).
Reverse transcription-quantitative
PCR
Total RNA was extracted from treated HNSCC cells
using lysis buffer (Takara Bio, Inc.) according to the
manufacturer's protocol. Then, reverse transcription was performed
by using the PrimeScript RT reagent kit (Takara Bio, Inc.) with the
following RT protocol: 15 min at 37°C and 5 sec at 85°C. The cDNA
was subjected to PCR detection using a SYBR Green Premix kit
(Takara Bio, Inc.) with the following protocols: 95°C for 30 sec;
and followed by 40 cycles, 95°C for 5 sec and 60°C for 34 sec.
Relative expression was calculated using the 2−ΔΔCq
method by using GAPDH as reference (15). The primers for each gene are listed
in Table I.
 | Table I.Primer sequences for each gene. |
Table I.
Primer sequences for each gene.
| Gene name | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| JMJD6 |
TTGGACCCGGCACAACTACTA |
TCTGCCCTTTCCACGTTATCC |
| SNAI1 |
TCGGAAGCCTAACTACAGCGA |
AGATGAGCATTGGCAGCGAG |
| VIM |
GACGCCATCAACACCGAGTT |
CTTTGTCGTTGGTTAGCGGT |
| TWIST1 |
GTCCGCAGTCTTACGAGGAG |
GCTTGAGGGTCTGAATCTGCT |
| CDH1 |
ATTTTTCCCTCGACACCCGAT |
TCCCAGGCGTAGACCAAGA |
| CDH2 |
AGCCAACCTTAACTGAGGAGT |
GGCAAGTTGATTGGAGGGATG |
| GAPDH |
AACGTGTCAGTGGTGGACCTG |
AGTGGGTGTCGCTGTTGAGT |
Transwell assays
For cellular migration, 2×104 cells
suspended in 200 µl fresh DMEM were plated in Millicell chambers
(8-µm pore size; EMD Millipore), and 600 µl DMEM containing 10% FBS
was plated in the lower chamber. Cells that migrated through the
membrane were fixed with 4% paraformaldehyde for 15 min at room
temperature and stained with 0.1% crystal violet for 15 min at room
temperature. For cellular invasion assays, the chamber was coated
with 50 µl Matrigel for 1 h at room temperature, which was then
hydrated for 2 h at 37°C (1:8 dilution; BD Biosciences) prior to
the assay. Images of the membrane were captured and the number of
migratory or invasive cells was observed under a light microscope
(Olympus Corporation) and counted from five fields of view
(magnification, ×100).
Colony formation assay
A cellular suspension was diluted and seeded at
1×103 cells/well on 6-well plates in triplicate. The
cells were cultured with culture medium (DMEM supplemented with 10%
FBS, 100 U/ml penicillin and 100 µg/ml streptomycin) for 10 days,
and then harvested and fixed with 4% paraformaldehyde for 15 min at
room temperature. The cell colonies were observed by staining with
Coomassie brilliant blue for 30 min at room temperature. The number
of large colonies (consisting of ≥50 cells) was counted and
analyzed under a light microscope, at ×100 magnification (Olympus
Corporation).
Cell viability analysis
MTT assays were performed to examine the viability
of cells. Briefly, cells were diluted and seeded at
1×104 cells/100 µl in 96-well plates for 12 h for
attachment, and then incubated with 5-fluorouracil (5-FU; Selleck
Chemicals) or cisplatin (Selleck Chemicals) at various
concentrations for 48 h at 37°C with 5% CO2. Absorbance
measurements following DMSO resolution were performed at a
wavelength of 490 nm (Bio-Rad Laboratories, Inc.).
Statistical analysis
All statistical analyses were conducted using SPSS
v16.0 software (SPSS, Inc.). Statistical comparisons of the
transcriptional level of JMJD6 among different stages were made
using one-way ANOVA and least significant difference post hoc test
(LSD). The different Kaplan-Meier curves were compared by using the
log-rank test. Bioinformatically, based on the detailed data of
each biomarker, all cases involved were divided equally into two
groups; the high-expression group and the low-expression group (the
median expression levels were used as cut-off values for each
biomarker: 9.318 for JMJD6, 6.433 for SNAI1, 13.53 for vimentin,
7.299 for Twist1, 5.103 for CDH2, and 13.23 for CDH1). To
illustrate the underlying association between JMJD6 and EMT
markers, crosstabs analyses were performed, and the χ2
test and Pearson's R correlation coefficient test were used to
assess the associations between the gene expression levels of JMJD6
and the gene expression levels of EMT markers (16). Significant differences between two
groups were determined using unpaired Student's t-test. The
IC50 values were calculated with GraphPad Prism (version
6; GraphPad Software, Inc.). All experiments were performed in
triplicate and representative results are presented. The values
presented correspond to the mean ± standard deviation. P<0.05
was considered to indicate a statistically significant
difference.
Results
JMJD6 is highly expressed in HNSCC
tissues
Based on the bioinformatics results from GEPIA2, the
mRNA expression levels of JMJD6 were significantly
upregulated in HNSCC tissues compared with normal tissues in the
head and neck area (Fig. 1A and B).
However, when further prognostic analysis was performed, no obvious
significant differences were observed in overall survival (OS) or
disease-free survival (DFS) times in HNSCC for different JMJD6
expression levels (Fig. 1C and D).
Therefore, the exact roles of highly expressed JMJD6 in HNSCC
remain to be determined. In the present study, bioinformatics
analysis based on TCGA HNSCC database revealed a significant
association between the expression level of JMJD6 and the
pathological stage of HNSCC (Fig.
1E; Table II). These results
indicated that JMJD6 could predict HNSCC cases with an advanced
stage but had no predictive value for prognosis.
 | Table II.Pathological characteristics of head
and neck squamous cell carcinoma cases, based on The Cancer Genome
Atlas cohort, according to JMJD6 expression. |
Table II.
Pathological characteristics of head
and neck squamous cell carcinoma cases, based on The Cancer Genome
Atlas cohort, according to JMJD6 expression.
|
|
| JMJD6 expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Patients, n | ≤9.318, n | >9.318, n | P-value |
|---|
| All cases | 566 | 283 | 283 |
|
| Pathologic T |
|
|
| 0.005 |
|
T1+T2 | 208 | 119 | 89 |
|
|
T3+T4 | 295 | 131 | 164 |
|
| Pathologic N |
|
|
| 0.013 |
| N0 | 193 | 107 | 86 |
|
|
N1+ | 260 | 113 | 147 |
|
| Pathologic M |
|
|
| / |
| M0 | 190 | 85 | 105 |
|
| M1 | 1 | 1 | 0 |
|
| Pathologic
stage |
|
|
| <0.001 |
|
I+II | 119 | 77 | 42 |
|
|
III+IV | 374 | 167 | 207 |
|
| Neoplasm histologic
grade |
|
|
| 0.143 |
|
G1+G1 | 398 | 206 | 192 |
|
|
G3+G4 | 142 | 63 | 79 |
|
| Lymphovascular
invasion present |
|
|
| 0.052 |
|
Yes | 136 | 54 | 82 |
|
| No | 239 | 121 | 118 |
|
| Pathological nodal
extracapsular spread |
|
|
| <0.001 |
|
Yes | 118 | 36 | 82 |
|
| No | 262 | 136 | 126 |
|
| Perineural invasion
present |
|
|
| 0.613 |
|
Yes | 186 | 83 | 103 |
|
| No | 208 | 99 | 109 |
|
High JMJD6 expression acts as a risk
factor for the malignant progression of patients with HNSCC
Based on 566 cases from the TCGA primary HNSCC
cohort, increased JMJD6 expression was significantly associated
with pathologic tumor size, pathologic lymph node status,
pathologic stage and pathological nodal extracapsular spread (the
AJCC Cancer Staging manual for head and neck cancer 8th Edition,
2017) (17) (Table II). In the retrospectively
validation cohort, based on 98 HNSCC cases, all cases were divided
into two group under the expression level of JMJD6 (Fig. 2). Besides, significant associations
between increased JMJD6 expression and tumor size and nodal status
were observed (Table III). These
results suggested that JMJD6 may promote the malignant progression
of HNSCC.
 | Table III.Clinicopathological characteristics
of patients divided by JMJD6 expression based on a validation head
and neck squamous cell carcinoma cohort. |
Table III.
Clinicopathological characteristics
of patients divided by JMJD6 expression based on a validation head
and neck squamous cell carcinoma cohort.
|
|
| JMJD6
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | Patients, n | IRS=1-6, n | IRS=7-12, n | P-value |
|---|
| All cases | 98 | 48 | 50 |
|
| Age, years |
|
|
| 0.837 |
|
<60 | 59 | 28 | 31 |
|
|
≥60 | 39 | 20 | 19 |
|
| Sex |
|
|
| 0.843 |
|
Male | 52 | 26 | 26 |
|
|
Female | 46 | 22 | 24 |
|
| Smoking |
|
|
| 0.999 |
|
Yes | 51 | 25 | 26 |
|
| No | 47 | 23 | 24 |
|
| Alcohol |
|
|
| 0.999 |
|
Yes | 48 | 24 | 24 |
|
| No | 50 | 24 | 26 |
|
| Histological
grade |
|
|
| 0.110 |
|
Well/moderate-differentiation | 53 | 30 | 23 |
|
|
Poor-differentiation | 45 | 18 | 27 |
|
| Tumor size |
|
|
| 0.017 |
|
T1+T2 | 51 | 31 | 20 |
|
|
T3+T4 | 47 | 17 | 30 |
|
| Nodal status |
|
|
| 0.003 |
| N0 | 52 | 33 | 19 |
|
|
N1+ | 46 | 15 | 31 |
|
| Metastasis |
|
|
| / |
| M0 | 98 | 48 | 50 |
|
| M1 | 0 | 0 | 0 |
|
JMJD6 regulates the EMT behavior of
HNSCC
JMJD6 has been demonstrated to induce EMT and
promote malignant migration and invasion in breast cancer (18). In the present study, bioinformatics
analysis based on the TCGA primary HNSCC cohort was performed to
analyze the association between JMJD6 expression and EMT
status (based on the expression of SNAI1, VIM, TWIST1, CDH2
and CDH1). As shown in Fig.
3A, significant positive correlations between JMJD6 and
SNAI1, TWIST1 and CDH2 expression and a significant
negative correlation between JMJD6 and CDH1
expression were observed. Subsequently, a JMJD6 overexpression
model in Cal27 cells and a JMJD6 knockdown model in HN12 cells were
constructed (Fig. 3B).
Overexpression of JMJD6 in Cal27 cells resulted in increased
expression levels of SNAI1, VIM, TWIST1 and CDH2, and
significantly decreased the expression levels of CDH1
(Fig. 3B and D). Similarly, JMJD6
was knocked down in HN12 cells, which resulted in decreased
expression levels of SNAI1, VIM, TWIST1 and CDH2,
while significantly increased expression levels of CDH1 were
observed (Fig. 3C and D).
Morphologically, Cal27 cells exhibited an epithelial multiangle
shape, and a long, spindle shape was observed in Cal27 cells
overexpressing JMJD6 (Fig. 3E). In
HN12 cells, knockdown of JMJD6 resulted in a morphological
alteration from a long, spindle shape to a multiangle shape
(Fig. 3E). Therefore, increased
JMJD6 expression could regulate the EMT status of HNSCC cells.
 | Figure 3.Effects of JMJD6 expression on the
epithelial-mesenchymal transition status of HNSCC. (A) Heat map
from the UCSC Xena Browser, based on the primary HNSCC cohort from
The Cancer Genome Atlas revealing the associations between
JMJD6 and SNAI1, VIM, TWIST1, CDH2 and CDH1
gene expression. B A significant correlation with JMJD6 expression
was observed for SNAI1, TWIST1 (Positive correlation) and
CDH1 (negative correlation). (B) mRNA expression levels of
JMJD6, SNAI1, VIM, TWIST1, CDH2 and CDH1 in Cal27
cells transfected with Myc-JMJD6 for 72 h. (C) mRNA expression
levels of JMJD6, SNAI1, VIM, TWIST1, CDH2 and CDH1 in
HN12 cells transfected with siRNA-JMJD6 for 72 h. (D) Western blot
analysis of the expression of JMJD6 in transiently transfected
Cal27 and HN12 cells. The upper band of JMJD6 for Cal27 cells
indicated the exogenous myc-JMJD6. (E) Representative morphological
images of Cal27 cells transfected with Myc-JMJD6 and HN12 cells
transfected with si-JMJD6 for 4 days. Magnification, ×200.
*P<0.05. **P<0.01. CDH1, cadherin 1; CDH2, cadherin 2; JMJD6,
Jumonji domain-containing protein 6; HNSCC, head and neck squamous
cell carcinoma; siRNA, small interfering RNA; SNAI1, snail family
transcriptional repressor 1; TWIST1, twist family bHLH
transcription factor 1; VIM, vimentin. |
JMJD6 overexpression promotes the
colony formation, and migration and invasion abilities of HNSCC
cells
Clinicopathological analysis based on the TCGA
primary HNSCC cohort and the validation cohort indicated roles of
JMJD6 in the regulation of the metastatic ability of HNSCC.
Furthermore, additional roles of JMJD6 in the regulation of EMT
were demonstrated in the present study. Colony formation assays
indicated markedly increased colony-forming abilities in
JMJD6-overexpression Cal27 cells, whereas the knockdown of JMJD6 in
HN12 cells significantly decreased their colony-forming abilities
(Fig. 4A and B). Transwell assays
were performed to investigate the effects of JMJD6 on the migration
and invasion ability of HNSCC cells. Overexpression of JMJD6 in
Cal27 cells resulted in significantly increased migration and
invasion (Fig. 4C and D). By
contrast, knockdown of JMJD6 in HN12 cells resulted in
significantly decreased migration and invasion (Fig. 4C and D).
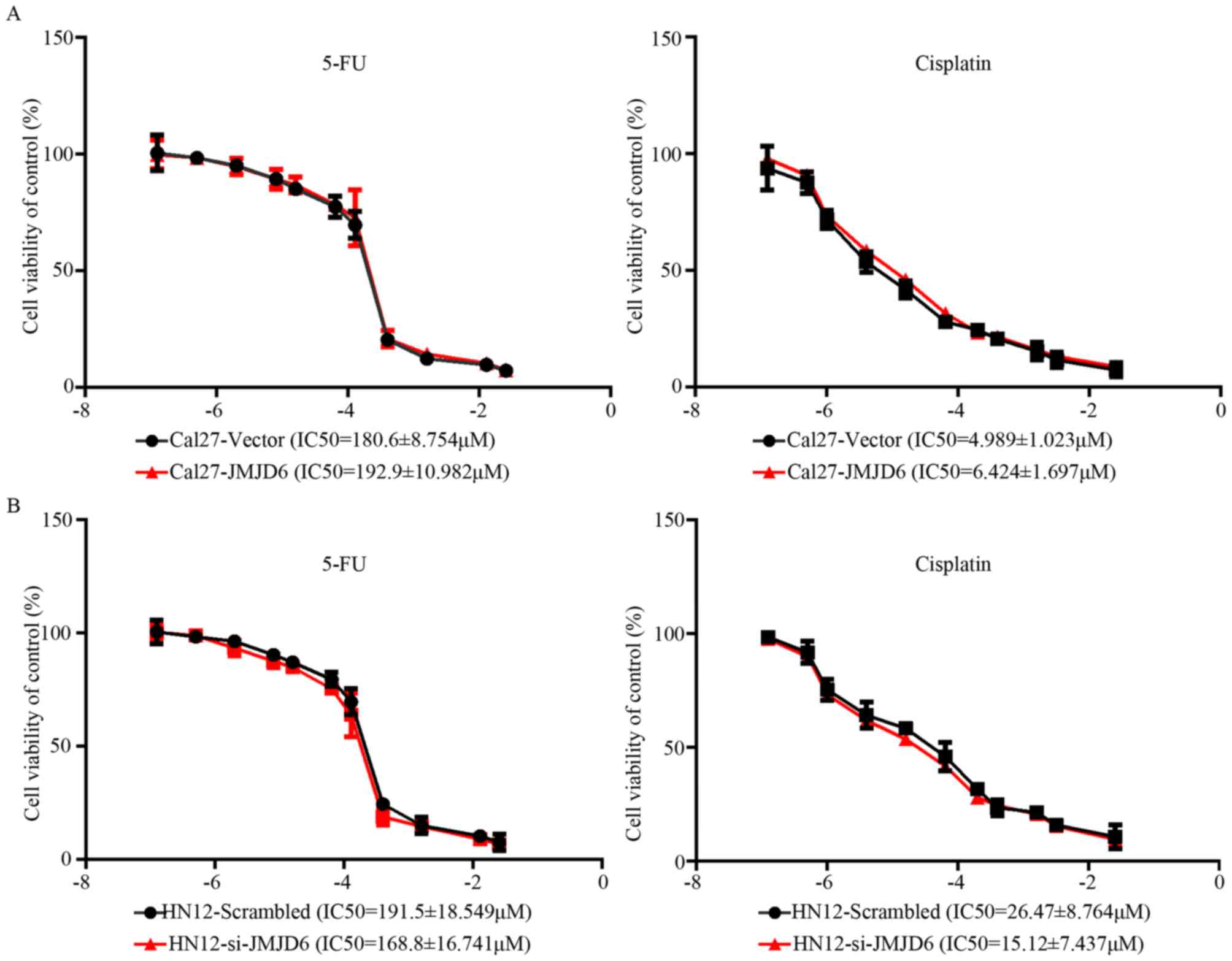
JMJD6 has no effect on the sensitivity
of HNSCC to drugs
The aforementioned data indicated that JMJD6 is
significantly associated with malignant behaviors but not with the
clinical outcomes of HNSCC. The viability of Cal27 and HN12 cells
with different JMJD6 expression levels in response to treatment
with 5-FU or cisplatin was investigated, and the IC50
values were calculated. No obvious differences were observed in the
IC50 values of HNSCC cells treated with 5-FU or
cisplatin, in association with the different expression levels of
JMJD6 (Fig. 5A and B). Therefore,
HNSCC with high JMJD6 expression may exhibit a more advanced stage;
however, to the best of our knowledge, this is not associated with
responsiveness to chemotherapy.
Discussion
The dysregulation of JMJD6 has been reported to
contribute to several types of human cancer by controlling a wide
range of biological functions (3,4). To the
best of our knowledge, the biological roles and clinical
significance of JMJD6 in HNSCC remain uncertain. In the present
study, the clinical significance of JMJD6 based on the TCGA primary
HNSCC cohort and a validation HNSCC cohort was investigated.
Subsequently, the roles of JMJD6 in HNSCC were revealed through a
series of cellular assays. Consequently, it can be concluded that
JMJD6 contributes to the malignant progression of HNSCC by
regulating EMT, whereas JMJD6 overexpression exerts no significant
effects on the drug responsiveness and prognosis of HNSCC.
The data from the present study demonstrated that
JMJD6 was significantly upregulated in HNSCC samples compared with
normal samples. Overexpression of JMJD6 did not affect the
prognosis (OS and DFS times) of patients with HNSCC; however, it
was strongly associated with advanced pathological stage
(particularly in terms of tumor size, nodal status and nodal
extracapsular spread), based on the TCGA cohort and a validation
cohort. Previous studies have demonstrated that the upregulation of
JMJD6 indicates aggressive phenotypes and a poor prognosis in
multiple types of human cancer (3,11). The
present study suggested that patients with HNSCC and high JMJD6
expression are liable to develop advanced-stage disease but
exhibited no difference in drug sensitivity. Further studies are
required to analyze the expression of JMJD6 across different HNSCC
subtypes.
Based on the clinical significance of JMJD6, it was
speculated that JMJD6 serves key roles in the regulation of
migration/invasion behaviors in HNSCC. Invasion and migration are
the main characteristics of the EMT process (19). In the present study, bioinformatics
analysis based on the TCGA primary HNSCC cohort was carried out in
order to detect the association between JMJD6 expression and EMT
status (based on the expression of SNAI1, VIM, TWIST1, CDH2
and CDH1). As expected, JMJD6 was associated with EMT status
in HNSCC. Additionally, overexpression of JMJD6 in HNSCC cells
resulted in increased migration and invasion rates. Thus, JMJD6
promoted the malignant progression of HNSCC by regulating the EMT
process. In lung cancer, JMJD6 has been reported to cooperate with
c-Myc to enhance the EMT process (18). In HNSCC, the underlying mechanisms
will be further investigated in subsequent studies.
Based on the results of IC50 experiments, the
present study revealed that overexpression of JMJD6 had no effect
on the drug responses of HNSCC to chemotherapy. These data may
illustrate that JMJD6 contributed to a more advanced stage, however
not a poorer prognosis. To the best our knowledge, the present
study was the first to demonstrate that high JMJD6 expression
promoted malignant progression but not the development of drug
resistance in HNSCC. Treatment strategies for HNSCC with an
advanced stage should be based on combined and sequential therapy.
For patients with advanced HNSCC, radical surgeries always lead to
poor quality of life without a markedly improved prognosis
(20). According to the data
obtained in the present study, rational chemotherapy may be
beneficial for patients with advanced HNSCC who exhibit high JMJD6
expression.
In conclusion, the present study demonstrated the
clinical significance of JMJD6 in HNSCC, and that high JMJD6
expression was significantly associated with an advanced stage,
with no effect on drug responsiveness or prognosis. However,
additional studies are required to investigate the underlying
mechanisms in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Excellent
Young Doctor Scholarship of the Shanghai 9th People's Hospital
(grant no. 20170912) and the college fund of Shanghai Jiao Tong
University School of Medicine (grant no. 13XJ10049).
Availability of data and materials
The datasets analysed during the present study are
available in the TCGA-HNSC repository (https://xenabrowser.net), or available from the
corresponding author upon reasonable request.
Authors' contributions
BG, YS and CM conceived and designed the study. BG
and LW performed the experiments. BG and XQ compiled and analyzed
the data. BG, LW and XQ contributed to writing and revising the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All patients involved in the present study provided
written informed consent, and the study was approved by the Medical
Ethics Committee of the 9th People's Hospital, Shanghai Jiao Tong
University School of Medicine (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kwok J, O'Shea M, Hume DA and Lengeling A:
JMJD6, a JMJC dioxygenase with many interaction partners and
pleiotropic functions. Front Genet. 8:322017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang F, He L, Huangyang P, Liang J, Si W,
Yan R, Han X, Liu S, Gui B, Li W, et al: JMJD6 promotes colon
carcinogenesis through negative regulation of p53 by hydroxylation.
PLoS Biol. 12:e10018192014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu Y, Long YH, Wang SQ, Zhang YY, Li YF,
Mi JS, Yu CH, Li DY, Zhang JH and Zhang XJ: JMJD6 regulates histone
H2A.X phosphorylation and promotes autophagy in triple-negative
breast cancer cells via a novel tyrosine kinase activity. Oncogene.
38:980–997. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wan J, Xu W, Zhan J, Ma J, Li X, Xie Y,
Wang J, Zhu WG, Luo J and Zhang H: PCAF-mediated acetylation of
transcriptional factor HOXB9 suppresses lung adenocarcinoma
progression by targeting oncogenic protein JMJD6. Nucleic Acids
Res. 44:10662–10675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Unoki M, Masuda A, Dohmae N, Arita K,
Yoshimatsu M, Iwai Y, Fukui Y, Ueda K, Hamamoto R, Shirakawa M, et
al: Lysyl 5-hydroxylation, a novel histone modification, by Jumonji
domain containing 6 (JMJD6). J Biol Chem. 288:6053–6062. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yi J, Shen HF, Qiu JS, Huang MF, Zhang WJ,
Ding JC, Zhu XY, Zhou Y, Fu XD and Liu W: JMJD6 and U2AF65
co-regulate alternative splicing in both JMJD6 enzymatic activity
dependent and independent manner. Nucleic Acids Res. 45:3503–3518.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee YF, Miller LD, Chan XB, Black MA, Pang
B, Ong CW, Salto-Tellez M, Liu ET and Desai KV: JMJD6 is a driver
of cellular proliferation and motility and a marker of poor
prognosis in breast cancer. Breast Cancer Res. 14:R852012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Konuma T, Yu D, Zhao C, Ju Y, Sharma R,
Ren C, Zhang Q, Zhou MM and Zeng L: Structural mechanism of the
oxygenase JMJD6 recognition by the extraterminal (ET) domain of
BRD4. Sci Rep. 7:162722017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wan J, Liu H, Yang L, Ma L, Liu J and Ming
L: JMJD6 promotes hepatocellular carcinoma carcinogenesis by
targeting CDK4. Int J Cancer. 144:2489–2500. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu X, Si W, Liu X, He L, Ren J, Yang Z,
Yang J, Li W, Liu S, Pei F, Yang X and Sun L: JMJD6 promotes
melanoma carcinogenesis through regulation of the alternative
splicing of PAK1, a key MAPK signaling component. Mol Cancer.
16:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee CR, Lee SH, Rigas NK, Kim RH, Kang MK,
Park NH and Shin KH: Elevated expression of JMJD6 is associated
with oral carcinogenesis and maintains cancer stemness properties.
Carcinogenesis. 37:119–128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu J, Sanborn JZ, Benz S, Szeto C, Hsu F,
Kuhn RM, Karolchik D, Archie J, Lenburg ME, Esserman LJ, et al: The
UCSC cancer genomics browser. Nat Methods. 6:239–240. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goldman M, Craft B, Swatloski T, Cline M,
Morozova O, Diekhans M, Haussler D and Zhu J: The UCSC cancer
genomics browser: Update 2015. Nucleic Acids Res. 43:D812–D817.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao M, Zhang J and Chen W and Chen W:
M1-like tumor-associated macrophages activated by
exosome-transferred THBS1 promote malignant migration in oral
squamous cell carcinoma. J Exp Clin Cancer Res. 37:1432018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lydiatt WM, Patel SG, O'Sullivan B,
Brandwein MS, Ridge JA, Migliacci JC, Loomis AM and Shah JP: Head
and neck cancers-major changes in the American Joint Committee on
cancer eighth edition cancer staging manual. CA Cancer J Clin.
67:122–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aprelikova O, Chen K, El Touny LH,
Brignatz-Guittard C, Han J, Qiu T, Yang HH, Lee MP, Zhu M and Green
JE: The epigenetic modifier JMJD6 is amplified in mammary tumors
and cooperates with c-Myc to enhance cellular transformation, tumor
progression, and metastasis. Clin Epigenetics. 8:382016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miyauchi S, Kim SS, Pang J, Gold KA,
Gutkind JS, Califano JA, Mell LK, Cohen EEW and Sharabi AB: Immune
modulation of head and neck squamous cell carcinoma and the tumor
microenvironment by conventional therapeutics. Clin Cancer Res.
25:4211–4423. 2019. View Article : Google Scholar : PubMed/NCBI
|