Introduction
Lung cancer is the leading cause of
cancer-associated mortality globally. Although progress has been
made in the treatment of lung cancer, the survival of patients with
lung cancer remains poor with a 5-year survival rate of only 17%
(1,2). The characteristics of lung cancer are
uncontrolled proliferation and metastasis of tumor cells.
Therefore, understanding the regulatory mechanisms underlying lung
cancer carcinogenesis and progression is necessary for tumor
therapy.
Non-coding RNAs, including microRNAs (miRNAs/miRs)
and long non-coding RNAs (lncRNAs) are considered to be potential
biomarkers and candidate targets for the treatment of numerous
cancer types (3). Certain lncRNAs
have key functions in a variety of biological processes, including
proliferation, apoptosis, stem cell properties, differentiation and
metastasis (4,5). To date, numerous lncRNAs have been
reported to be involved in the genesis of lung cancer. The lncRNA
activated by transforming growth factor-β was identified to be
overexpressed in lung cancer tissues and to promote the
proliferation and metastasis of tumor cells by activating the p38
signaling pathway (6).
Salt-inducible kinase (SIK)1-LNC, a type of lncRNA adjacent to SIK,
was reported to be downregulated in lung cancer tissues and to
repress the proliferation, migration and invasion of lung cancer
cells (7). LncRNA KCNK15 and WISP2
antisense RNA 1 (KCNK15-AS1) was determined to be overexpressed in
lung cancer tissues, and the higher expression of KCNK15-AS1 was
associated with a shorter survival (8). However, the functional roles and
underlying mechanisms of KCNK15-AS1 in the genesis of lung cancer
remain largely elusive.
miR-202 and miR-370 have been previously reported to
be decreased in lung cancer (9–11).
miR-202 induces cell cycle arrest and apoptosis by targeting cyclin
D1 (CCND1) and inhibits cell proliferation, migration and invasion
via targeting signal transducer and activator of transcription
(STAT3) in lung cancer (12,13). miR-370 has a tumor suppressive
function in lung cancer by targeting tumor necrosis factor
receptor-associated factor (TRAF4) and epidermal growth factor
receptor (EGFR) (11,14).
In the present study, the regulatory functions of
KCNK15-AS1 in lung cancer development, in addition to the
associated molecular mechanisms, were investigated.
Materials and methods
Patients and samples
Fresh lung adenocarcinoma (LAD) and adjacent normal
tissue samples from 40 patients were collected at the Department of
Thoracic Surgery of the First People's Hospital of Yunnan (Kunming,
China) between June 2014 and September 2015 and immediately stored
at −70°C. All patients with LAD were treated using radical surgery
and no patients received any pre-operative treatment. All samples
were residual specimens following diagnostic sampling, and all
patients provided written informed consent for sampling and
molecular analysis separately. The present study was ethically
approved by the Institutional Ethics Committee of the First
People's Hospital of Yunnan Province (Kunming, China).
Paracancerous tissue samples were collected at a 2-cm distance from
the tumor edge as previously described (15), and the normal tissues were
pathologically confirmed. The samples were graded by the AJCC
staging classification system (8th edition) (16). The mean age of the patients was 62
years old (range, 38–79), and 65% of the patients (n=40) were male.
Overall survival (OS) was defined as the time from surgery
treatment to mortality or to the last follow-up. The
clinical-pathological characteristics of the samples are presented
in Table I.
 | Table I.Association between KCNK15-AS1
expression and clinicopathological characteristics. |
Table I.
Association between KCNK15-AS1
expression and clinicopathological characteristics.
|
|
| KCNK15-AS1
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | n | High (n=19) | Low (n=21) | P-value |
|---|
| Sex |
|
|
| 0.7475 |
|
Female | 14 | 6 | 8 |
|
| Male | 26 | 13 | 13 |
|
| Age, years |
|
|
| 0.5266 |
|
<50 | 15 | 6 | 9 |
|
| ≥50 | 25 | 13 | 12 |
|
| Degree of
differentiation |
|
|
| 0.0309 |
| Well or
moderate | 18 | 5 | 13 |
|
| Poor or
undifferentiated | 22 | 14 | 8 |
|
| Clinical stage |
|
|
| 0.0270 |
|
I–II | 16 | 4 | 12 |
|
|
III–IV | 24 | 15 | 9 |
|
| T-stage |
|
|
| 0.7518 |
|
T1-T2 | 21 | 9 | 12 |
|
|
T3-T4 | 19 | 10 | 9 |
|
| N
classification |
|
|
| 0.0309 |
|
N0-N1 | 18 | 5 | 13 |
|
|
N2-N3 | 22 | 14 | 8 |
|
| M
classification |
|
|
| 0.5962 |
| M0 | 37 | 17 | 20 |
|
| M1 | 3 | 2 | 1 |
|
Cell culture
A549 (cat. no. CCL-185) and H460 cells (cat. no.
HTB-177) were purchased from the American Type Culture Collection
(Manassas, VA, USA). All cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml
streptomycin in an incubator at 37°C with 5% CO2.
Small interfering (si)RNA
transfection
Cancer cells (2×105) were seeded in
six-well plates and cultured with DMEM. KCNK15-AS1 siRNAs, miR-202
inhibitor, miR-370 inhibitor and a negative control were purchased
from Shanghai GenePharma Co., Ltd. (Shanghai, China). siRNAs or
inhibitors (50 nM) were transfected into cells using
Lipofectamine® RNAiMAX Transfection reagent for 48 h
according to the manufacturer's protocol (Thermo Fisher Scientific,
Inc.). The sequences of the KCNK15-AS1 siRNAs and negative control
were as follows: siRNA-1 sense, 5′-GUCAUCACUACCAUCGGUGATT-3′ and
antisense, 5′-UCACCGAUGGUAGUGAUGACTT-3′; siRNA-2 sense,
5′-GUCCGAGGCGGAAAGCGGTT-3′ and antisense,
5′-CCGCUUUCCGCCUCGGACTT-3′; negative control sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-202 inhibitor,
5′-CAAAGAAGUAUAUGCAUAGGAA-3′; miR-370 inhibitor,
5′-GUAACUGCAGAGACGUGACCUG-3′; inhibitor negative control,
5′-CAGUACUUUUGUGUAGUACAA-3′. The miR-202 inhibitor and miR-370
inhibitor were used at a final concentration of 20 nM.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was isolated from cells using RNAiso Plus
(Takara Bio, Inc., Otsu, Japan). RT-qPCR was used to determine the
expression levels of KCNK15-AS1, CCND1, EGFR and GAPDH. PCR was
performed in a total volume of 20 µl, including 10 µl PowerUp™
SYBR™ Green Mix (Thermo Fisher Scientific, Inc.), 2 µl
complementary DNA and 1 µl primer mix (10 µM each). PCR was
performed in an ABI 7300 real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) as follows: Initial denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C
for 1 min. The PCR data were normalized to GAPDH and the relative
expression of each gene was calculated using the 2−∆∆Cq
method (17). The following primers
were used: KCNK15-AS1 forward, 5′-AGCAGATGCAGAGAACCCAAA-3′ and
reverse, 5′-TTGCAAGGCAGGTGTTTGTTC-3′; CCND1 forward,
5′-GCTGCGAAGTGGAAACCATC-3′ and reverse,
5′-CCTCCTTCTGCACACATTTGAA-3′; EGFR forward,
5′-AGGCACGAGTAACAAGCTCAC-3′ and reverse,
5′-ATGAGGACATAACCAGCCACC-3′; GAPDH forward,
5′-AAATCCCATCACCATCTTCCAG-3′ and reverse,
5′-GAGTCCTTCCACGATACCAAAGTTG-3′.
Western blot analysis
Cells were lysed using radioimmunoprecipitation
assay buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl and 1% Nonidet
P-40]. Following incubation on ice for 45 min, the homogenates were
centrifuged at 13,000 × g for 15 min at 4°C. The concentrations of
samples were detected using the Bradford method (Beyotime Institute
of Biotechnology, Haimen, China). Proteins (10–20 µg/lane) were
separated by 12% SDS-PAGE and then transferred onto a 0.45-µm
polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA,
USA). Subsequently, the membranes were blocked with 5% skimmed milk
for 30 min at room temperature. The membranes were incubated with
primary antibody at 4°C overnight. Prior to incubation with
secondary antibodies [anti-mouse immunoglobulin G (IgG),
horseradish peroxidase (HRP)-linked antibody, cat no. 7075,
1:10,000 dilution, Cell Signaling Technology, Inc., Danvers, MA,
USA; anti-rabbit IgG, HRP-linked antibody, cat no. 7074, 1:10,000
dilution, Cell Signaling Technology, Inc.] at room temperature for
1 h, the membranes were washed with Tris-buffered saline containing
0.3% Tween-20. The protein signals were visualized using the
Enhanced Chemiluminescence Detection reagent (cat no. PE0020;
Beijing Solarbio Bioscience & Technology Co., Ltd., Beijing,
China). The primary antibodies and dilution ratios were as follows:
CCND1 (cat no. sc-450; 1:1,000 dilution; Santa Cruz Biotechnology,
Inc.); EGFR (cat no. 4267, 1:1,000 dilution; Cell Signaling
Technology, Inc., Dallas, TX, USA); phosphorylated protein kinase B
(AKT; cat no. 4060, 1:1,000 dilution; Cell Signaling Technology,
Inc.); AKT (cat no. 9272, 1:1,000 dilution; Cell Signaling
Technology, Inc.) and β-actin (cat no. 4970, 1:5,000 dilution; Cell
Signaling Technology, Inc.).
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was used to quantify the
proliferation of A549 and H460 cells according to the
manufacturer's protocol. Cells were cultured at 1,000 cells/well in
96-well plates and transfected for 24 h. Following incubation for
24, 48, 72, 96 or 120 h at 37°C, 10 µl CCK-8 reagent was added to
each well, followed by incubation at 37°C for 1 h. The absorbance
of each well was read at 450 nm on a microplate spectrophotometer
(SPECTRAMax 190; Molecular Devices, LLC, Sunnyvale, CA, USA). Three
independent experiments were performed.
Statistical analysis
Data were presented as the mean ± standard
deviation, and statistical analysis was performed using GraphPad
Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). The
expression levels of KCNK15-AS1 between LAD tissues and
paracancerous normal tissues were analyzed using a Student's t-test
(two-tailed, paired). Other data were analyzed using a Student's
t-test (two-tailed, unpaired) between two groups and one-way
analysis of variance followed by a Tukey's post-hoc test for
multiple comparisons. The association between KCNK15-AS1 expression
and clinicopathological characteristics were analyzed using a
χ2 test. Estimation of survival time distribution was
performed using the Kaplan-Meier method. P<0.05 was considered
to indicate a statistically significant difference. All experiments
were independently performed at least three times.
Results
Silencing of KCNK15-AS1 inhibits the
proliferation and decreases the expression of CCND1 in lung cancer
cells
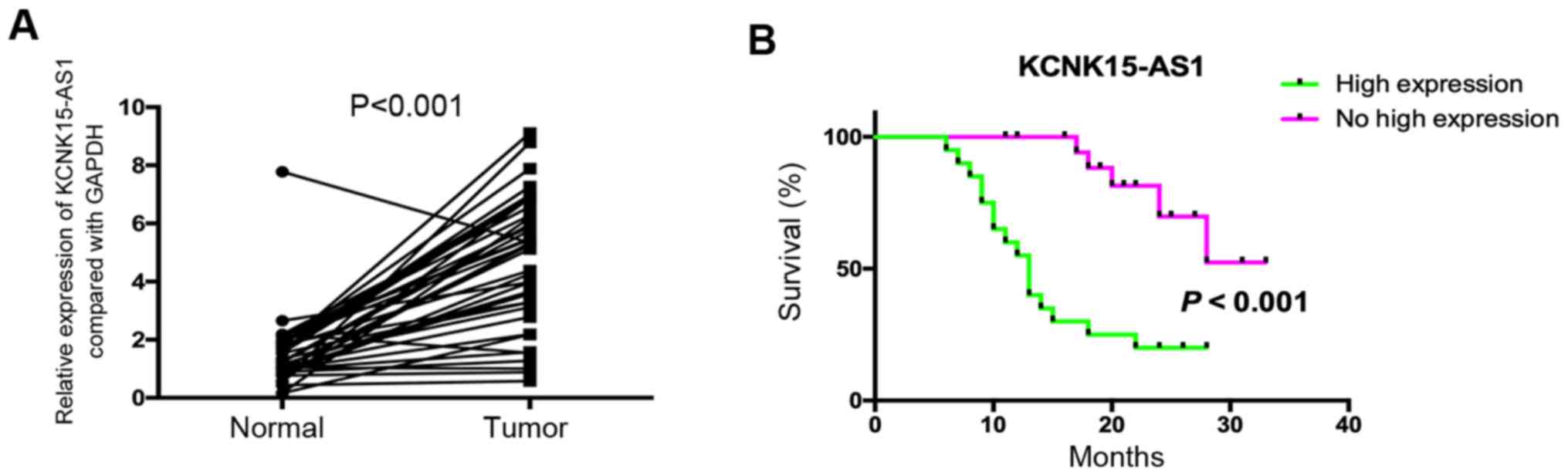
A previous study indicated that KCNK15-AS1 was
overexpressed in lung cancer and that the high expression of
KCNK15-AS1 was associated with poor prognosis (8). In the present study, 40 patients with
lung cancer were assessed to confirm these results, and the
analysis suggested that KCNK15-AS1 was significantly overexpressed
in lung cancer compared with paracancerous tissues (P<0.001) and
that its high expression was associated with poor overall survival
(Fig. 1A and B). The high expression
of KCNK15-AS1 was significantly associated with a poor degree of
differentiation, advanced clinical stage and lymph node metastasis
(P<0.05; Table I). In order to
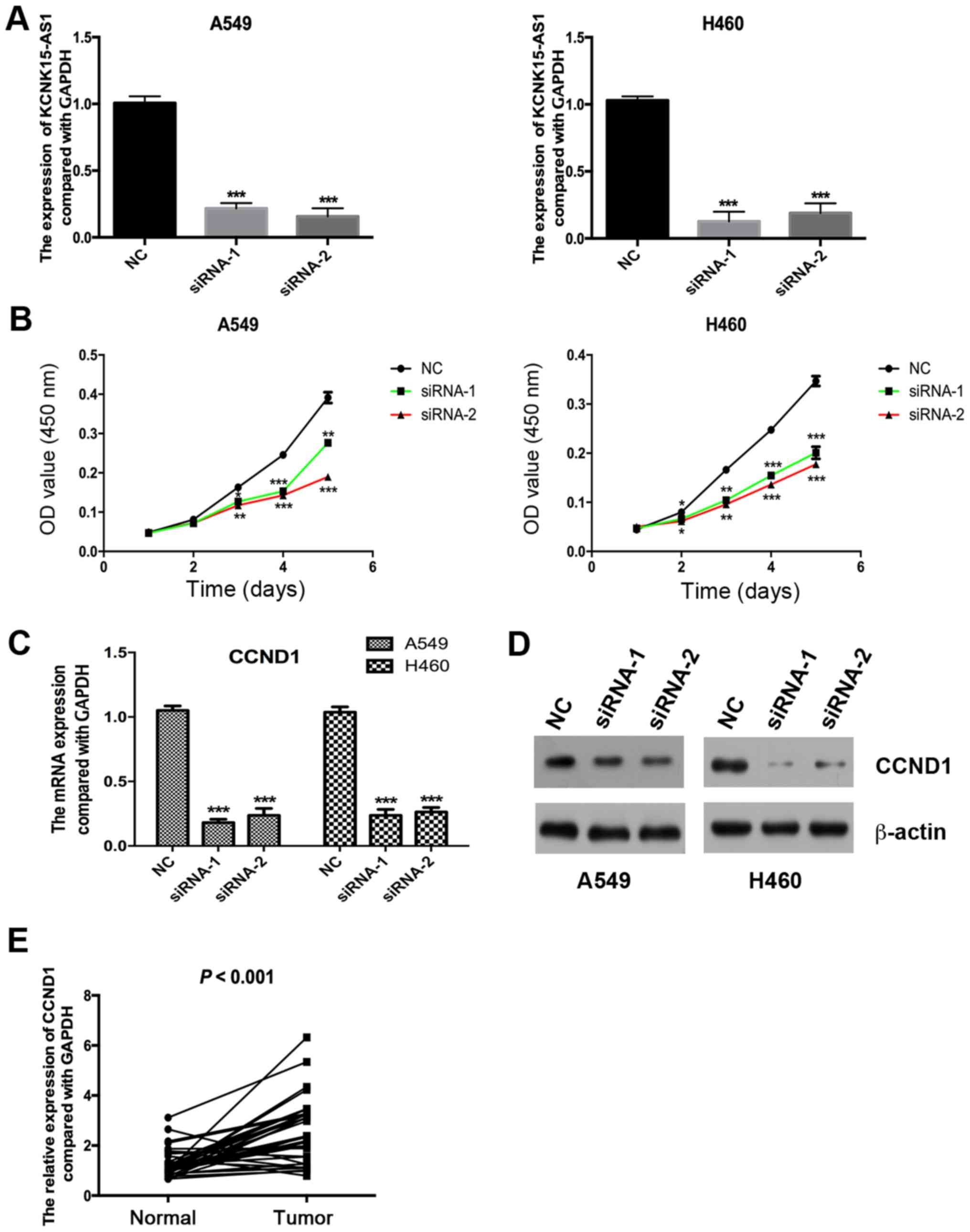
investigate the function of KCNK15-AS1, it was knocked down using
siRNAs (Fig. 2A). The results
indicated that the silencing of KCNK15-AS1 significantly inhibited
the proliferation of A549 and H460 lung cancer cells (P<0.05;
Fig. 2B). CCND1 is involved in the
regulation of the cell cycle (18).
Notably, the knockdown of KCNK15-AS1 caused a significant
downregulation of the expression of CCND1 at the mRNA levels
(P<0.001; Fig. 2C) and
downregulation at the protein level (Fig. 2D). Furthermore, the expression of
CCND1 in lung cancer tissues was significantly higher compared with
that in paracancerous normal tissues (P<0.001; Fig. 2E).
Silencing of KCNK15-AS1 decreases the
expression of EGFR and inhibits the phosphorylation of AKT
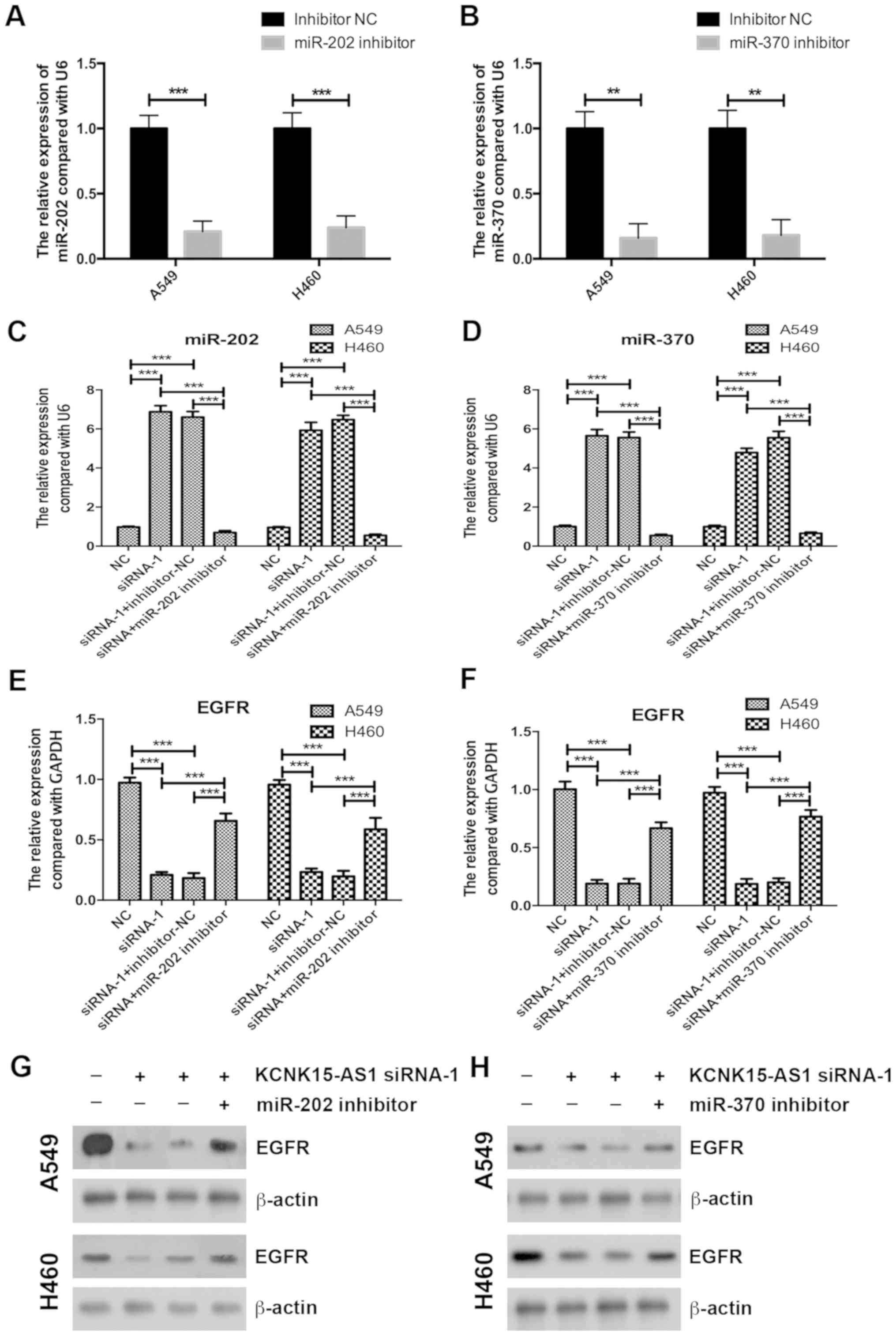
The EGFR/AKT axis has important functions in cancer
cell proliferation (19,20); therefore, the present study assessed
whether the silencing of KCNK15-AS1 affects EGFR/AKT signaling.
RT-qPCR and western blot analyses indicated that the silencing of
KCNK15-AS1 reduced the protein expression and significantly reduced
the mRNA levels of EGFR (P<0.001) and inhibited the
phosphorylation of AKT (Fig. 3A and
B).
In tumors, EGFR has been reported to be targeted and
regulated by miR-202 and miR-370 (14,21,22). The
results of the present study suggested that the silencing of
KCNK15-AS1 caused a significant upregulation of the expression of
miR-202 and miR-370 in A549 and H460 lung cancer cells (P<0.001;
Fig. 3C and D).
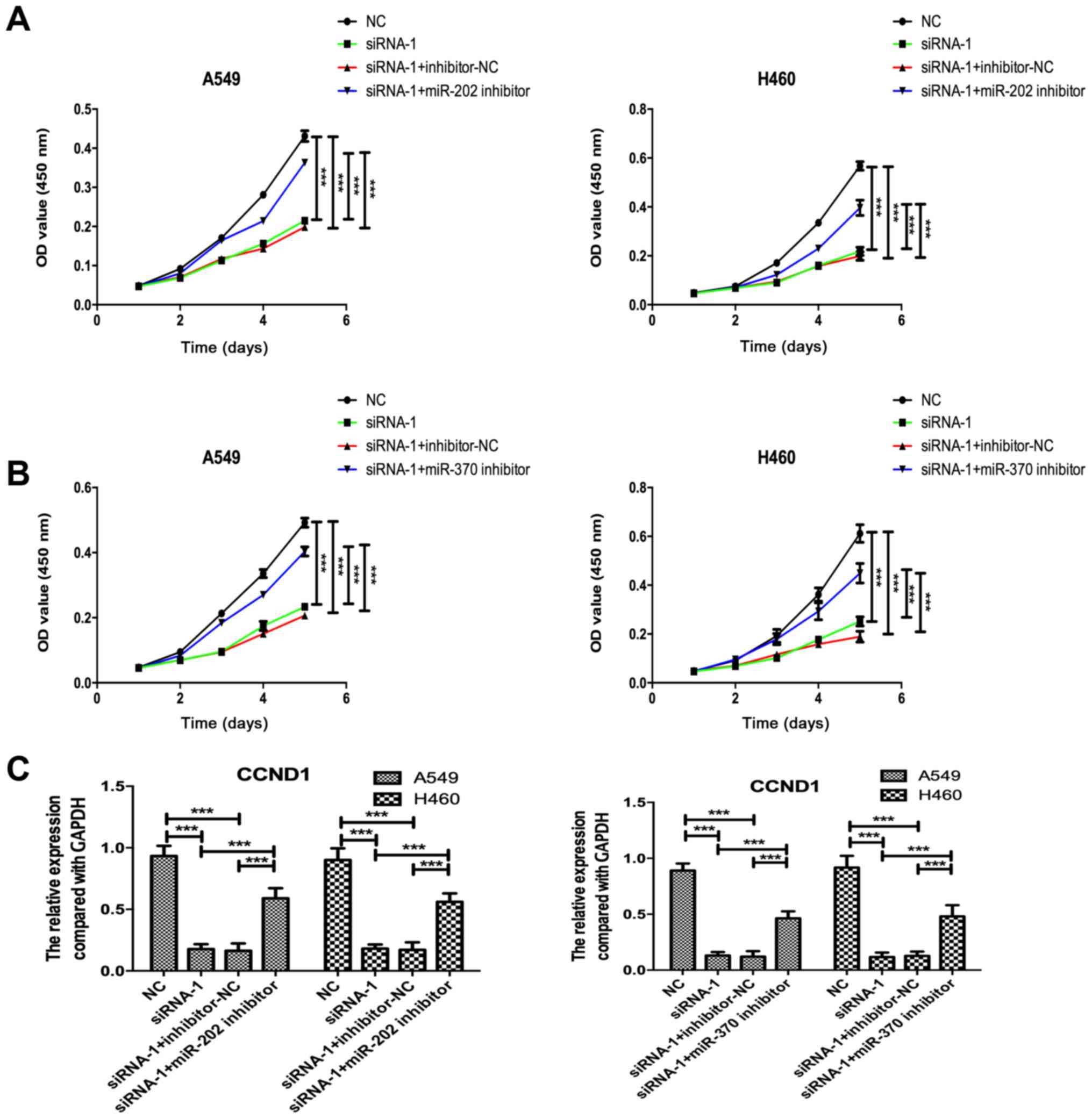
Inhibition of miR-202 or miR-370
partially recovers the inhibitory effect of KCNK15-AS1 knockdown on
the proliferation of lung cancer cells
To confirm the involvement of miR-202 or miR-370 in
the regulatory effect of KCNK15-AS1 on lung cancer cell
proliferation, initially, miR-202 and miR-370 were knocked-down
using inhibitors in A549 and H460 cells. A RT-qPCR assay confirmed
the efficiency of miR-202 and miR-370 silencing (Fig. 4A and B). miR-202 and miR-370
inhibitors also significantly decreased the expression levels of
miR-202 and miR-370 in the KCNK15-AS1-silenced lung cancer cells
compared with the negative control (P<0.001; Fig. 4C and D). Inhibition of miR-202 or
miR-370 partially recovered the EGFR expression levels in the
KCNK15-AS1 silenced lung cancer cells (Fig. 4E-H). Furthermore, the knockdown of
miR-202 or miR-370 partially recovered the cell proliferation
ability and CCND1 expression in the KCNK15-AS1-silenced lung cancer
cells (Fig. 5A-C).
Discussion
LncRNAs, a class of RNAs with a length of >200
nucleotides, have important functions in numerous biological
processes, including tumor formation and development (23). A previous study indicated that
KCNK15-AS1 was overexpressed in lung cancer tissues, and the higher
expression of KCNK15-AS1 was associated with poor survival
(8). On the other hand, KCNK15-AS1
was reported to be decreased in pancreatic cancer tissues, and to
inhibit the migration and invasion of pancreatic cancer cells
(24). Furthermore, the expression
of KCNK15-AS1 was regulated by the m6A eraser AlkB homolog 5, RNA
demethylase (24). Therefore,
KCNK15-AS1 may have different functions in different types of
cancer, and elucidation of the mechanisms associated with
KCNK15-AS1 overexpression is urgently required.
The results of the present study initially confirmed
the overexpression and association with the poor prognosis of
KCNK15-AS1 in lung cancer. Furthermore, it was determined that the
knockdown of KCNK15-AS1 significantly inhibited the proliferation
of lung cancer cells and caused the downregulation of the cell
cycle-associated gene CCND1 at the mRNA and protein levels.
The EGFR/AKT signaling pathway has important
functions in lung carcinogenesis and certain miRNAs are known to
regulate this signaling pathway. miR-133a was reported to decrease
the expression of EGFR and inhibit the phosphorylation of AKT in
human non-small cell lung cancer cells (25). miR-145 was reported to induce
apoptosis and inhibit the migratory ability of non-small cell lung
cancer cells by inhibiting the EGFR/phosphoinositide-3-kinase/AKT
signaling pathway (26). The present
study indicated that KCNK15-AS1 activated the EGFR/AKT signaling
pathway via reducing miR-202 and miR-370.
Previous studies have indicated that miR-202
exhibits tumor suppressive functions and is downregulated in
numerous types of cancer, including prostate cancer, breast cancer,
osteosarcoma and lung cancer (12,27–29).
miR-202 has been reported to be significantly downregulated in
bladder cancer tissues and cell lines, and the overexpression of
miR-202 inhibited cell proliferation, colony formation, invasion
and migration in vitro, as well as suppressed tumor growth
in vivo; of note, miR-202 exerted its tumor suppressive
functions via targeting EGFR (21).
In lung cancer, miR-202 was decreased, and the overexpression of
miR-202 enhanced the sensitivity of lung cancer cells to cisplatin
by inactivating the Ras/mitogen-activated protein kinase signaling
pathway (30). miR-202 was also
demonstrated to have a tumor suppressive function in lung cancer by
targeting STAT3 and CCND1 (12,13).
miR-370 was downregulated in lung cancer tissues and the
overexpression of miR-370 inhibited the proliferation, colony
formation, migration and invasion of lung cancer cells.
Furthermore, miR-370 was indicated to target EGFR and regulate its
expression, and also to inhibit AKT phosphorylation (14). miR-370 also inhibited the CCND1/CCND
kinase (CDK)4/CDK6 pathway by increasing p21 expression in lung
cancer cells, and suppressing the progression of non-small cell
lung cancer via targeting and reducing the expression of TRAF4
(11,31). In gastric cancer, the expression of
miR-370 was negatively associated with EGFR, and the overexpression
of miR-370 suppressed the proliferation and migration of gastric
cancer cells by targeting EGFR (22). The present study indicated that
KCNK15-AS1 knockdown inhibited lung cancer cell proliferation,
reduced CCND1 expression and inactivated the EGFR/AKT axis via
upregulating miR-202 and miR-370 by a rescue experiment.
In conclusion, the results of the present study
suggested that KCNK15-AS1 promoted lung cancer cell proliferation
and inactivated the EGFR/AKT signaling pathway via downregulating
miR-202 and miR-370. Future studies should focus on the mechanisms
of how KCNK15-AS1 regulates miR-202 and miR-370, in addition to the
diagnostic and therapeutic potential of KCNK15-AS1 in lung
cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
JP and HP designed the study. JP, XC, HC, ZX, HW,
ZS, JL and XN performed the experiments. JP, XC and HP analyzed the
data. JP and HP wrote the paper.
Ethics approval and consent to
participate
The present study was ethically approved by the
Institutional Ethics Committee of the First People's Hospital of
Yunnan Province (Kunming, China). All samples were residual
specimens following diagnostic sampling, and all patients provided
written informed consent for sampling and molecular analysis
separately.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
miRNA
|
microRNA
|
|
KCNK15-AS1
|
KCNK15 and WISP2 antisense RNA 1
|
|
EGFR
|
epidermal growth factor receptor
|
References
|
1
|
Hao Y, Yang X, Zhang D, Luo J and Chen R:
Long noncoding RNA LINC01186, regulated by TGF-β/SMAD3, inhibits
migration and invasion through Epithelial-Mesenchymal-Transition in
lung cancer. Gene. 608:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang
C, Dou C, Xu M, Liu Q and Tu K: Long non-coding RNA CASC2
suppresses epithelial-mesenchymal transition of hepatocellular
carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer.
16:1232017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie
Y, Wang K, Jia W, Chu WM and Sun B: The long noncoding RNA lnc-EGFR
stimulates T-regulatory cells differentiation thus promoting
hepatocellular carcinoma immune evasion. Nat Commun. 8:151292017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu YY, Chen ZH, Peng JJ, Wu JL, Yuan YJ,
Zhai ET, Cai SR, He YL and Song W: Up-regulation of long non-coding
RNA XLOC_010235 regulates epithelial-to-mesenchymal transition to
promote metastasis by associating with Snail1 in gastric cancer.
Sci Rep. 7:24612017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei L, Wu T, He P, Zhang JL and Wu W:
LncRNA ATB promotes the proliferation and metastasis of lung cancer
via activation of the p38 signaling pathway. Oncol Lett.
16:3907–3912. 2018.PubMed/NCBI
|
|
7
|
Yang L, Xie N, Huang J, Huang H, Xu S,
Wang Z and Cai J: SIK1-LNC represses the proliferative, migrative,
and invasive abilities of lung cancer cells. Onco Targets Ther.
11:4197–4206. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Chi Q and Zhao Z: Up-regulation
of long non-coding RNA SPRY4-IT1 promotes tumor cell migration and
invasion in lung adenocarcinoma. Oncotarget. 8:51058–51065.
2017.PubMed/NCBI
|
|
9
|
Wang R, Chen XF and Shu YQ: Prediction of
non-small cell lung cancer metastasis-associated microRNAs using
bioinformatics. Am J Cancer Res. 5:32–51. 2014.PubMed/NCBI
|
|
10
|
Nymark P, Guled M, Borze I, Faisal A,
Lahti L, Salmenkivi K, Kettunen E, Anttila S and Knuutila S:
Integrative analysis of microRNA, mRNA and aCGH data reveals
asbestos- and histology-related changes in lung cancer. Genes
Chromosomes Cancer. 50:585–597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen T, Gao F, Feng S, Yang T and Chen M:
MicroRNA-370 inhibits the progression of non-small cell lung cancer
by downregulating oncogene TRAF4. Oncol Rep. 34:461–468. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Z, Lv B, Zhang L, Zhao N and Lv Y:
miR-202 functions as a tumor suppressor in non-small cell lung
cancer by targeting STAT3. Mol Med Rep. 16:2281–2289. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang J, Huang J, Wang XR and Quan YH:
MicroRNA-202 induces cell cycle arrest and apoptosis in lung cancer
cells through targeting cyclin D1. Eur Rev Med Pharmacol Sci.
20:2278–2284. 2016.PubMed/NCBI
|
|
14
|
Liu X, Huang YG, Jin CG, Zhou YC, Chen XQ,
Li J, Chen Y, Li M, Yao Q, Li K, et al: MicroRNA-370 inhibits the
growth and metastasis of lung cancer by down-regulating epidermal
growth factor receptor expression. Oncotarget. 8:88139–88151.
2017.PubMed/NCBI
|
|
15
|
Yong-Hao Y, Xian-Guo W, Ming X and
Jin-Ping Z: Expression and clinical significance of miR-139-5p in
non-small cell lung cancer. J Int Med Res. 47:867–874. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
In H, Solsky I, Palis B, Langdon-Embry M,
Ajani J and Sano T: Validation of the 8th edition of the AJCC TNM
staging system for gastric cancer using the National Cancer
Database. Ann Surg Oncol. 24:3683–3691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen C, Zhang Z, Li J and Sun Y: SNHG8 is
identified as a key regulator in non-small-cell lung cancer
progression sponging to miR-542-3p by targeting CCND1/CDK6. Onco
Targets Ther. 11:6081–6090. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu F, Shangli Z and Hu Z: CAV2 promotes
the growth of renal cell carcinoma through the EGFR/PI3K/Akt
pathway. Onco Targets Ther. 11:6209–6216. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Qiu X, Xi G, Liu H, Zhang F, Lv T
and Song Y: Downregulation of GSDMD attenuates tumor proliferation
via the intrinsic mitochondrial apoptotic pathway and inhibition of
EGFR/Akt signaling and predicts a good prognosis in non-small cell
lung cancer. Oncol Rep. 40:1971–1984. 2018.PubMed/NCBI
|
|
21
|
Zhang L, Xu J, Yang G, Li H and Guo X:
miR-202 inhibits cell proliferation, migration, and invasion by
targeting epidermal growth factor receptor in human bladder cancer.
Oncol Res. 26:949–957. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ning T, Zhang H, Wang X, Li S, Zhang L,
Deng T, Zhou L, Liu R, Wang X, Bai M, et al: miR-370 regulates cell
proliferation and migration by targeting EGFR in gastric cancer.
Oncol Rep. 38:384–392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zou Y, Zhang B, Mao Y, Zhang H and Hong W:
Long non-coding RNA OECC promotes cell proliferation and metastasis
through the PI3K/Akt/mTOR signaling pathway in human lung cancer.
Oncol Lett. 18:3017–3024. 2019.PubMed/NCBI
|
|
24
|
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P,
Liu D, Tian L, Yin J, Jiang K and Miao Y: ALKBH5 inhibits
pancreatic cancer motility by decreasing long non-coding RNA
KCNK15-AS1 methylation. Cell Physiol Biochem. 48:838–846. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo N, Zhao Y, Zhang W, Li S, Li S and Yu
J: MicroRNA-133a downregulated EGFR expression in human non-small
cell lung cancer cells via AKT/ERK signaling. Oncol Lett.
16:6045–6050. 2018.PubMed/NCBI
|
|
26
|
Li B, Ding CM, Li YX, Peng JC, Geng N and
Qin WW: MicroRNA145 inhibits migration and induces apoptosis in
human non-small cell lung cancer cells through regulation of the
EGFR/PI3K/AKT signaling pathway. Oncol Rep. 40:2944–2954.
2018.PubMed/NCBI
|
|
27
|
Zhang S, Cai J, Xie W, Luo H and Yang F:
miR-202 suppresses prostate cancer growth and metastasis by
targeting PIK3CA. Exp Ther Med. 16:1499–1504. 2018.PubMed/NCBI
|
|
28
|
Gao S, Cao C, Dai Q, Chen J and Tu J:
miR-202 acts as a potential tumor suppressor in breast cancer.
Oncol Lett. 16:1155–1162. 2018.PubMed/NCBI
|
|
29
|
Li C, Ma D, Yang J, Lin X and Chen B:
miR-202-5p inhibits the migration and invasion of osteosarcoma
cells by targeting ROCK1. Oncol Lett. 16:829–834. 2018.PubMed/NCBI
|
|
30
|
Sun W, Ping W, Tian Y, Zou W, Liu J and Zu
Y: miR-202 enhances the anti-tumor effect of cisplatin on non-small
cell lung cancer by targeting the Ras/MAPK pathway. Cell Physiol
Biochem. 51:2160–2171. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li C, Ge Q, Liu J, Zhang Q, Wang C, Cui K
and Chen Z: Effects of miR-1236-3p and miR-370-5p on activation of
p21 in various tumors and its inhibition on the growth of lung
cancer cells. Tumour Biol. 39:10104283177108242017. View Article : Google Scholar : PubMed/NCBI
|