Introduction
Wilms tumor (WT) is the most common type of renal
cancer in children between the ages 0 and 14 years, with an
incidence rate of 9 in 1 million children, based on November 2017
SEER data submission (1,2). As such, close attention is being paid
to this major global public health issue (3,4). The
incidence of WT in children <15 years of age in the United
States of America is ~7–8 cases per million, with ~500 new cases
per year (5). Although the prognosis
of children with WT has improved due to multimodal therapies, WT
continue to recur even five years post-diagnosis (6–8). Thus,
considering the disease's severity, the early diagnosis of WT and
investigation of the molecular mechanisms associated with the
development of the disease are of great importance. Effective
biomarkers for the early detection of WT are urgently needed to
improve the quality of life and survival of patients with WT.
Technological advances in genomic and transcriptomic
analysis have led to the identification of various types of
non-coding RNAs (ncRNAs) (9,10). Long non-coding RNAs (lncRNAs), which
consist of >200 nucleotides, account for a large number of
ncRNAs (11). Emerging evidence
indicates that lncRNAs are important in genetic,
post-transcriptional and epigenetic regulation (12,13). In
addition, a growing number of studies suggest that lncRNAs are
involved in cell proliferation, migration and apoptosis (14,15).
Therefore, lncRNAs may serve as potential diagnostic and prognostic
biomarkers for WT (16,17). However, knowledge of the association
between lncRNAs and prognosis in patients with WT is limited
(18). The present study
investigated differential lncRNA expression patterns between WT and
normal tissues, and revealed a three-lncRNA signature that may
predict the survival time of patients with WT.
Materials and methods
Acquisition of therapeutically
applicable research to generate effective treatments (TARGET)
data
Raw lncRNA expression data and corresponding
clinical information were downloaded from the TARGET database
(ocg.cancer.gov/programs/target) Version: December,
2018. Samples obtained from patients with an overall survival (OS)
of more than one month were included in the present study.
Additionally, the patient clinical information, differentially
expressed lncRNAs and prognosis information were downloaded. A
total of 136 samples, including 130 WT tissues and six normal
tissues, were investigated in the present study. The detailed
clinical characteristics of the patients are presented in Table I. R language package (edgeR, Release
3.9; http://www.bioconductor.org/packages/release/bioc/html/edgeR.html)
was used to interpret lncRNA sequencing data and the limma package
(Release 3.9; http://www.bioconductor.org/packages/release/bioc/html/limma.html)
was used to analyze differentially expressed lncRNAs between WT and
normal tissues. Fold-changes (FCs) in the expression of individual
lncRNAs were calculated, and differentially expressed lncRNAs with
log2|FC| >1.0 and P<0.05 were considered
significant.
 | Table I.Baseline characteristics of the
subjects. |
Table I.
Baseline characteristics of the
subjects.
|
| All subjects
(n=136) |
|---|
|
|
|
|---|
| Variable | No. | % |
|---|
| Gender |
|
|
|
Female | 76 | 55.9 |
| Male | 60 | 44.1 |
| Age at diagnosis |
|
|
|
<4 | 62 | 45.6 |
| ≥4 | 74 | 54.4 |
| Ethnicity |
|
|
|
Caucasian | 98 | 72.1 |
|
African-American | 20 | 14.7 |
| Not
available | 18 | 13.2 |
| Stage |
|
|
| I+II | 73 | 53.7 |
|
III+IV+V | 63 | 46.3 |
Patient prognosis and target gene
prediction
Differentially expressed lncRNA profiles were
normalized following log2 transformation. The Kaplan-Meier method
and the log-rank test were used to evaluate the prognostic value of
each differentially expressed lncRNA. Three lncRNAs that were
significantly associated with OS were identified as prognostic
lncRNAs and were subjected to receiver operating characteristic
(ROC) and Cox regression analyses. The Cox model was used to
investigate the association between the expression of each lncRNA
and OS according to age, ethnicity, gender and disease stage.
lncRNAs with hazard ratios (HRs) <1 were defined as protective,
while those with HRs >1 were defined as high-risk. Eventually, a
prognostic lncRNA signature was constructed, and patients with WT
were classified into low- and high-risk groups using the median
risk score as the cut-off value. Kaplan-Meier analysis and log-rank
test were used to evaluate differences in patient survival between
the two groups. ROC analysis was performed to compare the
sensitivity and specificity of the survival prediction based on the
lncRNA risk score. Protein-coding genes (PCGs) genes correlated
with the differentially expressed lncRNAs were identified using the
co-expression method. The PCGs with a Pearson's correlation
coefficient >0.40 and P<0.01 were considered to be associated
with the lncRNAs. Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway (https://david.ncifcrf.gov/) and Gene
Ontology (GO; http://david.ncifcrf.gov/) enrichment analyses were
subsequently performed for the target genes. P<0.05 were
considered to indicate a statistically significant difference.
Statistical analysis
Univariate and multivariate Cox proportional hazard
regression and Kaplan-Meier survival analyses with log-rank test
were used to compare each lncRNA (low vs. high expression level)
and their prognostic signatures (low vs. high risk). Hazard ratio
(95% CI) was expressed in the Cox regression analysis. The data for
categorical variables were presented as percentages (%). Pearson's
correlation test and log-rank test were also used in this study.
P<0.05 was considered to indicate a statistically significant
difference. Statistical analyses were performed using R (version
3.5.1; www.r-project.org) (19) and SPSS software (version 17.0; SPSS
Inc.).
Results
Characteristics of the differentially
expressed lncRNAs
The present study investigated 136 samples,
including 130 WT and 6 normal tissues. The detailed clinical
characteristics, including gender, ethnicity, age at diagnosis and
disease stage, are presented in Table
I. A total of 1,833 differentially expressed lncRNAs, including
1,087 upregulated and 746 downregulated lncRNAs, were identified
between WT and normal tissues (Fig.
1).
Association between the three lncRNAs
and OS in patients with WT
The Kaplan-Meier method and log-rank test were used
to evaluate the association between OS and lncRNA expression
patterns. As depicted in Fig. 2, two
upregulated lncRNAs (RP11-93B14.6 and RP11554F20.1) and one
downregulated lncRNA (DLGAP1-AS2), were significantly associated
with OS rate (all P<0.05).
Prognostic value of the three-lncRNA
signature risk scores for WT
A prognostic signature was identified by integrating
three lncRNA expression profiles and the corresponding estimated
regression coefficient. The 130 patients investigated in the
present study were subsequently divided into high- and low-risk
groups (n=65 per group) according to the median risk score.
Patients in the high-risk group had a significantly worse OS than
those in the low-risk group (P<0.001; Fig. S1). The prognostic ability of the
three-lncRNA signature was evaluated using ROC curve analysis.
Results revealed that the area under the ROC curve of the
three-lncRNA signature was 0.80 (Fig.
S2).
Univariate and multivariate Cox regression analyses
were used to determine the effects of the three-lncRNA signature
(high vs. low risk) on OS (Tables
II and III). In univariate
models, gender (HR=1.94; P=0.015), disease stage (HR=2.65;
P<0.001), DLGAP1-AS2 signature (HR=0.60; P<0.001),
RP11-93B14.6 signature (HR=1.37; P<0.001) and RP11-554F20.1
signature (HR=1.92; P<0.001) were all independent factors
associated with OS in patients with WT. In multivariate models,
gender (HR=3.10; P<0.001), stage (HR=3.36; P<0.001),
DLGAP1-AS2 signature (HR=0.60; P=0.001), RP11-93B14.6 signature
(HR=1.30; P=0.006) and RP11-554F20.1 signature (HR=2.26;
P<0.001) were associated with OS in patients with WT.
 | Table II.Univariate and multivariate Cox
proportional hazard models in patients with Wilm's tumor. |
Table II.
Univariate and multivariate Cox
proportional hazard models in patients with Wilm's tumor.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Gender |
|
|
|
|
| Male vs.
female | 1.94 (1.14–3.31) | 0.015 | 3.10 (1.71–5.60) | <0.001 |
| Age at
diagnosis |
|
|
|
|
| ≥4 vs.
<4 | 1.09
(0.64–1.86) | 0.749 | 0.61
(0.33–1.14) | 0.122 |
| Race |
|
|
|
|
|
Caucasian vs.
othersa | 0.90
(0.50–1.61) | 0.717 | 0.68
(0.36–1.28) | 0.233 |
| Stage |
|
|
|
|
|
III+IV+V vs. I+II | 2.65
(1.53–4.59) | <0.001 | 3.36
(1.89–5.96) | <0.001 |
| DLGAP1-AS2
signature |
|
|
|
|
| High
risk vs. low risk | 0.60
(0.46–0.78) | <0.001 | 0.60
(0.45–0.80) | 0.001 |
| RP11-93B14.6
signature |
|
|
|
|
| High
risk vs. low risk | 1.37
(1.15–1.64) | <0.001 | 1.30
(1.08–1.56) | 0.006 |
| RP11-554F20.1
signature |
|
|
|
|
| High
risk vs. low risk | 1.92
(1.39–2.66) | <0.001 | 2.26
(1.60–3.18) | <0.001 |
 | Table III.Three lncRNA signatures in unadjusted
and adjusted models. |
Table III.
Three lncRNA signatures in unadjusted
and adjusted models.
|
| Unadjusted | Mode 1 | Mode 2 | Mode 3 |
|---|
|
|
|
|
|
|
|---|
| Model | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| DLGAP1-AS2
signature | 0.60
(0.46–0.78) | <0.001 | 0.61
(0.46–0.80) | <0.001 | 0.60
(0.46–0.80) | <0.001 | 0.60
(0.45–0.80) | 0.001 |
| RP11-93B14.6
signature | 1.37
(1.15–1.64) | <0.001 | 1.25
(1.04–1.49) | 0.017 | 1.27
(1.05–1.52) | 0.012 | 1.30
(1.08–1.56) | 0.006 |
| RP11-554F20.1
signature | 1.92
(1.39–2.66) | <0.001 | 2.42
(1.67–3.49) | <0.001 | 2.38
(1.65–3.44) | <0.001 | 2.26
(1.60–3.18) | <0.001 |
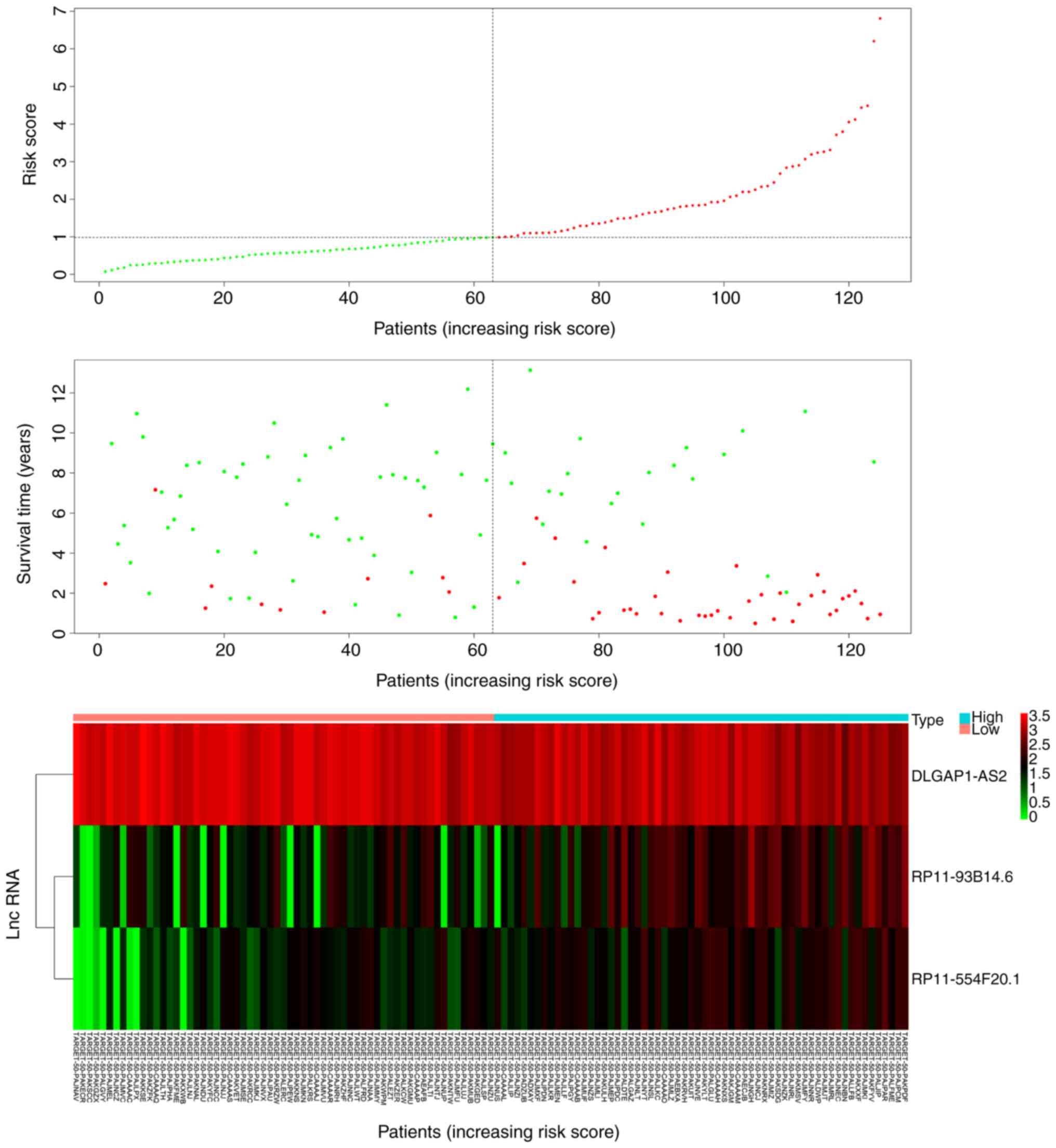
Patients in the low-risk group expressed increased
levels of the protective lncRNA (DLGAP1-AS2) compared with patients
in the high-risk group (P<0.05; Fig.
3). Patients in the high-risk group expressed increased levels
of RP11-93B14.6 and RP11-554F20.1 compared with the low-risk group
(P<0.05; Fig. 3). In addition,
patients in the low-risk group had a significantly longer survival
time than those in the high-risk group (P<0.05; Fig. 3).
lncRNA target prediction and
functional GO and KEGG analysis
A co-expression method was used to predict the
potential target of the three lncRNAs of interest. GO enrichment
and KEGG pathway analyses were performed to elucidate the
biological functions of the associated target genes (Fig. 4). GO biological process, molecular
function and cellular component terms were mainly enriched in
‘coenzyme binding’, ‘cofactor binding’, ‘flavin adenine
dinucleotide binding’, ‘acetylgalactosaminyltransferase activity’
and ‘steroid binding’ (P<0.05). In addition, KEGG pathways were
significantly enriched in ‘fatty acid metabolism’, ‘histidine
metabolism’, ‘peroxisome’, ‘AGE-RAGE signaling pathway in diabetic
complication’ and ‘insulin resistance’ (P<0.05).
Discussion
WT is currently the most common primary malignant
renal tumor in children (20).
Although outcomes have improved due to multidisciplinary
treatments, chemotherapeutic drugs and radiotherapy, the incidence
and recurrence of WT remains high, and presents a heavy burden for
patients, families and medical institutions (21–23). The
prognosis of patients with WT may be considerably improved if the
characteristics of the tumor, including clinical symptoms and
genetic phenotypes, are reliably predicted at the time of initial
diagnosis (18). Therefore, there is
a requirement for the identification of prognostic biomarkers as
well as the investigation of the molecular mechanisms underlying
the development of WT.
A number of previously studies have revealed that
genetic factors may contribute to the development of WT (2,18).
Furthermore, studies have indicated that altered lncRNA expression
levels are associated with disease development; however, their
prognostic values have not been thoroughly investigated (18,24).
Therefore, the present study identified three differentially
expressed lncRNAs, which were correlated with OS in patients with
WT. The lncRNAs, including DLGAP1-AS2, RP11-93B14.6 and
RP11554F20.1, were validated as independent prognostic factors for
patients with WT. Additionally, the target genes of the three
lncRNAs, as well as their enrichment pathways and biological
functions, were investigated using bioinformatics. The results
suggested that these three lncRNAs may participate in the molecular
pathogenesis, clinical progression and prognosis of WT,
highlighting the potential of lncRNA profiling to improve the
clinical prognosis in patients with WT.
Previous studies have demonstrated that functional
lncRNA expression plays an important role in carcinogenesis and
strongly correlates with gene expression and pathway regulation
(25,26). lncRNAs promote tumor formation,
progression and metastasis by regulating various major pathways,
including cancer cell proliferation, cell cycle arrest, apoptosis,
differentiation, adhesion, migration, invasion and survival
(27,28). Zhu et al (24) reported that upregulated expression of
long intergenic non-protein coding RNA 473 was associated with the
molecular pathogenesis of WT via the miR-195/IKK complex α
signaling pathway. However, the sample size, sample types and
number of lncRNAs assessed were limited.
The present study analyzed high-throughput data and
identified two upregulated lncRNAs (RP11-93B14.6 and RP11554F20.1)
and one downregulated lncRNA (DLGAP1-AS2) associated with the
clinical outcomes of patients with WT. Therefore, the expression
levels of these three lncRNAs may serve as prognostic markers for
WT. Furthermore, to the best of our knowledge, the functions of
these lncRNAs have not been previously investigated. The results of
the present study revealed that RP11-93B14.6, RP11554F20.1 and
DLGAP1-AS2 were enriched in ‘coenzyme binding’, ‘cofactor binding’,
‘fatty acid metabolism’, ‘histidine metabolism’, ‘peroxisome’,
‘AGE-RAGE signaling pathway in diabetic complication’ and ‘insulin
resistance’, all of which are classical signaling pathways closely
associated with tumorigenesis and the progression of malignancies
(29,30). However, further molecular
investigations in patients with WT are required to validate these
results and to inform new therapeutic interventions.
The present study had several limitations. Firstly,
the mechanisms underlying the prognostic values of the three
lncRNAs were not investigated and warrant experimental studies.
Secondly, a single published dataset with a small sample size was
used. Therefore, the results obtained may differ from the general
population. Thirdly, this study is not an intervention and
treatment experiment, therefore data on the therapeutic effect of
WT cannot be obtained. Finally, the WT cohort had a relatively high
censor rate, which may have affected the reliability of the
Kaplan-Meier estimates. Thus, a larger and multicenter clinical
cohort study is required to validate the results obtained in the
present study.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available in the TARGET repository, https://ocg.cancer.gov/programs/target.
Authors' contributions
PR contributed to the conception, design and final
approval of the submitted version. MH contributed to the
interpretation of data and completion of figures and tables. Both
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participant
The study was performed in accordance with the
guidelines of the Declaration of Helsinki. The study protocol was
approved by the TARGET publication guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
WT
|
Wilms' tumor
|
|
TARGET
|
Therapeutically Applicable Research to
Generate Effective Treatments
|
|
ncRNAs
|
non-coding RNAs
|
|
PCGs
|
protein-coding genes
|
|
ROC
|
receiver operating characteristic
|
|
AGE-RAGE
|
advanced glycation end
product-receptor of advanced glycation end product
|
|
OS
|
overall survival
|
|
FC
|
fold-change
|
|
HR
|
hazard ratio
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GO
|
Gene Ontology
|
References
|
1
|
Noone A, Howlader N, Krapcho M, Miller D,
Brest A and Yu M: SEER cancer statistics review, 1975–2015, based
on November 2017 SEER data submission, posted to the SEER website,
April 2018.
|
|
2
|
Scott RH, Stiller CA, Walker L and Rahman
N: Syndromes and constitutional chromosomal abnormalities
associated with Wilms tumour. J Med Genet. 43:705–715. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dome JS, Graf N, Geller JI, Fernandez CV,
Mullen EA, Spreafico F, Van den Heuvel-Eibrink M and
Pritchard-Jones K: Advances in Wilms tumor treatment and biology:
Progress through international collaboration. J Clin Oncol.
33:2999–3007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
D'Angelo P, Di Cataldo A, Terenziani M,
Bisogno G, Collini P, Di Martino M, Melchionda F, Mosa C, Nantron
M, Perotti D, et al: Factors possibly affecting prognosis in
children with Wilms' tumor diagnosed before 24 months of age: A
report from the Associazione Italiana Ematologia Oncologia
Pediatrica (AIEOP) Wilms tumor working group. Pediatr Blood Cancer.
642017.doi: 10.1002/pbc.26644. PubMed/NCBI
|
|
5
|
Bernstein L, Linet M and Smith M: Renal
tumorsNational Cancer Institute, SEER Program. Bethesda, MD:
National Institutes of Health; pp. 79–90. 1999
|
|
6
|
Mitchell C, Pritchard-Jones K, Shannon R,
Hutton C, Stevens S, Machin D, Imeson J, Kelsey A, Vujanic GM,
Gornall P, et al: Immediate nephrectomy versus preoperative
chemotherapy in the management of non-metastatic Wilms' tumour:
Results of a randomised trial (UKW3) by the UK Children's Cancer
Study Group. Eur J Cancer. 42:2554–2562. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malogolowkin M, Spreafico F, Dome JS, van
Tinteren H, Pritchard-Jones K, van den Heuvel-Eibrink MM, Bergeron
C, de Kraker J and Graf N; COG Renal Tumors Committee and the SIOP
Renal Tumor Study Group, : Incidence and outcomes of patients with
late recurrence of Wilms' tumor. Pediatr Blood Cancer. 6:1612–1615.
2013. View Article : Google Scholar
|
|
8
|
Venkatramani R, Chi YY, Coppes MJ,
Malogolowkin M, Kalapurakal JA, Tian J and Dome JS: Outcome of
patients with intracranial relapse enrolled on national Wilms tumor
study group clinical trials. Pediatr Blood Cancer. 642017.doi:
10.1002/pbc.26406. PubMed/NCBI
|
|
9
|
Clark MB, Johnston RL, Inostroza-Ponta M,
Fox AH, Fortini E, Moscato P, Dinger ME and Mattick JS: Genome-wide
analysis of long noncoding RNA stability. Genome Res. 22:885–898.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tani H, Imamachi N, Mizutani R, Imamura K,
Kwon Y, Miyazaki S, Maekawa S, Suzuki Y and Akimitsu N: Genome-wide
analysis of long noncoding RNA turnover. Methods Mol Biol.
1262:305–320. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carpenter S: Long noncoding RNA: Novel
links between gene expression and innate immunity. Virus Res.
212:137–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li CH and Chen Y: Insight into the role of
long noncoding RNA in cancer development and progression. Int Rev
Cell Mol Biol. 326:33–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moran VA, Perera RJ and Khalil AM:
Emerging functional and mechanistic paradigms of mammalian long
non-coding RNAs. Nucleic Acids Res. 40:6391–6400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong X, Chen R, Lin H, Lin T and Pan S:
LncRNA BG981369 inhibits cell proliferation, migration, and
invasion, and promotes cell apoptosis by SRY-related high-mobility
group box 4 (SOX4) signaling pathway in human gastric cancer. Med
Sci Monit. 24:718–726. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lian Y, Xiao C, Yan C, Chen D, Huang Q,
Fan Y, Li Z and Xu H: Knockdown of pseudogene derived from LncRNA
DUXAP10 inhibits cell proliferation, migration, invasion, and
promotes apoptosis in pancreatic cancer. J Cell Biochem.
119:3671–3682. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin MT, Song HJ and Ding XY: Long
non-coding RNAs involved in metastasis of gastric cancer. World J
Gastroenterol. 24:3724–3737. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rönnau CG, Verhaegh GW, Luna-Velez MV and
Schalken JA: Noncoding RNAs as novel biomarkers in prostate cancer.
Biomed Res Int. 2014:5917032014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu KR, Sun QF and Zhang YQ: Long
non-coding RNA LINP1 induces tumorigenesis of Wilms' tumor by
affecting Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol
Sci. 23:5691–5698. 2019.PubMed/NCBI
|
|
19
|
R Core Team, . A language and environment
for statistical computingVienna, Austria: R Foundation for
Statistical Computing; 2014
|
|
20
|
Rivera MN and Haber DA: Wilms' tumour:
Connecting tumorigenesis and organ development in the kidney. Nat
Rev Cancer. 5:699–712. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hamilton TE and Shamberger RC: Wilms
tumor: Recent advances in clinical care and biology. Semin Pediatr
Surg. 21:15–20. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Irtan S, Ehrlich PF and Pritchard-Jones K:
Wilms tumor: ‘State-of-the-art’ update, 2016. Semin Pediatr Surg.
25:250–256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nakayama DK and Bonasso PC: The history of
multimodal treatment of Wilms' tumor. Am Surg. 82:487–492.
2016.PubMed/NCBI
|
|
24
|
Zhu S, Fu W, Zhang L, Fu K, Hu J, Jia W
and Liu G: LINC00473 antagonizes the tumour suppressor miR-195 to
mediate the pathogenesis of Wilms tumour via IKKα. Cell Prolif.
512018.doi: 10.1111/cpr.12416.
|
|
25
|
Mitra SA, Mitra AP and Triche TJ: A
central role for long non-coding RNA in cancer. Front Genet.
3:172012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang M, Zhou L, Yu F, Zhang Y, Li P and
Wang K: The functional roles of exosomal long non-coding RNAs in
cancer. Cell Mol Life Sci. 76:2059–2076. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
de Fatima Silva F, Ortiz-Silva M, Galia
WBS, Cassolla P, de Silva FG, Graciano MFR, Carpinelli AR and de
Souza HM: Effects of metformin on insulin resistance and metabolic
disorders in tumor-bearing rats with advanced cachexia. Can J
Physiol Pharmacol. 96:498–505. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hage-Sleiman R, Herveau S, Matera EL,
Laurier JF and Dumontet C: Tubulin binding cofactor C (TBCC)
suppresses tumor growth and enhances chemosensitivity in human
breast cancer cells. BMC Cancer. 10:1352010. View Article : Google Scholar : PubMed/NCBI
|