Introduction
Colorectal carcinoma (CRC) is a common digestive
system carcinoma, remaining one of the major factors of tumor
deaths globally (1). As there are no
obvious symptoms for early stage CRC, it is usually diagnosed at
advanced stages (2). Although CRC
patients without metastases could be surgically cured, those in
advanced stage are mainly treated with chemotherapy. Studies above
indicated that chemotherapy has effective roles in preventing tumor
metastasis, reducing the tumor volume and improving the clinic
symptoms (3–6). However, most patients eventually
develop drug resistance after chemotherapy treatment. Therefore,
there is a great need for a continued effort to better understand
the complexity of CRC development and to identify new directions
for CRC therapy.
Overwhelming evidence has shown that abnormal
microRNA (miRNA/miR) expression mediated CRC development by
affecting the expression of the genes which regulated tumor
progression (7). miRNAs are highly
conserved small non-coding RNAs, playing important functions in
multiple biological processes (8,9). In
recent years, miRNAs have been extensively studied in
tumorigenesis, including in osteosarcoma (10), glioma (11) and breast cancer (12) research. These studies indicated that
miRNAs were associated with tumor pathogenesis along with the
potential to develop tumor therapeutics and diagnostics. However,
miR-331-3p expression patterns in human CRC and its biological
mechanism still remained obscure.
Neuropilin-2 (NRP2), a member of the NRPs family, is
a nontyrosine kinase transmembrane glycoprotein (13) and characterized as a receptor for the
vascular semaphorin (SEMA) families and endothelial growth factor
(VEGF) (14). A number of studies
have notably demonstrated that the NRP2 expression is ubiquitous in
various tumor cells such as lung cancer (15), cervical cancer (16) and breast cancer (17). Therefore, it is imperative to
understand the specific effects of NRP2 on tumor progression.
Nevertheless, the expressions and functions of NRP2 in CRC remain
largely unclear. In the present study, we evaluated NRP2 expression
and investigated the correlations between miR-331-3p and NRP2 in
CRC.
Materials and methods
CRC tissue samples
A total of 54 pairs of human CRC tissues and matched
normal tissues were collected from the Linyi Central Hospital
(Linyi, China) between May 2016 and July 2018, with approval from
the institutional Ethics Committee. Specimens were freshly frozen
in liquid nitrogen and stored at −80°C for further assays. Written
informed consent from each patient was received before the samples
were collected.
CRC cell culture
Human CRC cells (SW480 and HCT116) as well as normal
colon cells FHC were purchased from the Chinese Academy of Sciences
Cell Bank of Type Culture Collection (Shanghai, China). The cells
were cultured with RPMI-1640 medium containing FBS (10%),
penicillin (100 U/ml) and streptomycin (100 mg/ml) in a humidified
chamber (37°C, 5% CO2).
Cell transfection
miR-331-3p mimics or inhibitor as well as NRP2 siRNA
and the corresponding controls were purchased from Gene Pharma
(Shanghai, China) and transfected into CRC cell lines by
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in strict accordance with the manufacturer's
instructions.
Reverse transcription (RT)-qPCR
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to isolate the total RNAs from CRC cell
lines or tissue samples according to the manufacturer's guidelines.
Then, the extracted total RNA was used to generate the cDNA with
the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.).
The temperature conditions for reverse transcription were as
follows: 37°C for 15 min and 85°C for 5 sec. Real-time PCR assays
were conducted by SYBR® Premix Ex Taq™ (Takara
Biotechnology Co., Ltd.) on the ABI 7900 Sequence Detection System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
miR-331-3p expression was normalized to U6 while the NRP2 was
normalized to GAPDH. The primers used were as follows: For
miR-331-3p: Forward, 5′-GAGCTGAAAGCACTCCCAA-3′ and reverse
5′-CACACTCTTGATGTTCCAGGA-3′; for U6 forward,
5′-AGAGCCTGTGGTGTCCG-3′ and reverse 5′-CATCTTCAAAGCACTTCCCT-3′; for
NRP2 forward, 5′-CCCCGAACCCAACCAGAAGA-3′ and reverse
5′-GAATGCCATCCCAGATGTCCA-3′; and for GAPDH, forward
5′-GGCACTGAGAAGCGGGGCCG-3′ and reverse 5′-CCCTTGTTTTTTGCTTCCCTT-3′.
The thermocycling conditions were as follows: 95°C for 10 min,
followed by 45 cycles of denaturation at 95°C for 15 sec and
annealing/elongation at 60°C for 15 sec. 2−ΔΔCq method
was used to determine the relative expression of the genes
(18).
Immunohistochemistry (IHC)
IHC was performed to detect the NRP2 expression in
CRC tissues. Samples were fixed, embedded, and sliced into 4 µm
thick tissue sections. The sections were then dewaxed and
rehydrated. For antigen retrieval, the sections were microwaved in
citrate buffer for 15 min. Then, endogenous peroxidase activity was
blocked with 3% H2O2. Subsequently, the
sections were incubated with primary NRP2 antibody (1:100) at 4°C
overnight, and a secondary goat anti-rabbit IgG (1:1,000) (both
from Abcam) labeled by HRP was used for the subsequent incubation.
The sections were stained with DAB solution and counterstained with
haematoxylin. Images were obtained from a bright-field microscope
(Olympus BX50; Olympus Corporation).
Western blot analysis
Transfected cells were lysed on ice in RIPA buffer
(Thermo Fisher Scientific, Inc.) with proteinase inhibitors.
Bicinchoninic acid protein (BCA) assay kit (Beyotime) was applied
to measure the total protein concentrations. The protein lysates
(30 μg) were separated with 10% SDS-PAGE gel and then
electrotransferred to PVDF which was pretreated with 5% non-fat dry
skim milk in TBST for 2 h at room temperature. Thereafter, the
membranes were incubated with appropriate primary antibodies:
anti-NRP2 (dil, 1:4,000; cat. no. ab185710); anti-GAPDH (dil,
1:1,000; cat. no. ab181603) E-cadherin (dil, 1:2,000; cat. no.
ab15148), N-cadherin (dil, 1:2,000; cat. no. ab18203), Vimentin
(dil, 1:1,000; cat. no. ab137321) (all from Abcam) overnight at
4°C. The membrane was then incubated with anti-rabbit IgG (dil,
1:5,000; cat. no. ab191866; Abcam) at room temperature for 2 h. The
protein bands were detected by chemiluminescent detection system
(Beyotime). GAPDH was the internal reference.
Transwell assays
Cell invasion and migration assays were performed by
Transwell chambers (Coring Costar) with membrane pore size of 8.0
µm. After treated with miR-331-3p mimics, inhibitor or NRP2 siRNA,
CRC cell lines were seeded into the top chamber. For invasion and
migration assays, Transwell chamber was pretreated with or without
Matrigel (BD Biosciences) respectively. The top chamber was added
with serum-free medium when the medium containing 10% FBS was added
into the bottom chambers. After incubation for 48 h at 37°C, the
cells that remained on the top surface were removed with cotton
swabs. At the same time, those that adhered to the bottom surface
were fixed and stained respectively using formaldehyde (4%) and
crystal violet (0.1%) for detecting the images using a microscope
(Olympus Corporation). The values for invasion or migration were
obtained by counting 3 randomly selected fields per membrane and
represented the average of 3 independent experiments.
In silico analysis and luciferase
reporter assay
TargetScan database (http://www.targetscan.org/vert_72/) was utilized to
scan for the potential target gene of miR-331-3p that may
participate in CRC (19).
The amplified NRP2-3′-UTR-WT and corresponding
NRP2-3′-UTR-MUT were respectively inserted into pGL3 luciferase
vectors (Promega). CRC cells were cotransfected with miR-331-3p
mimics and luciferase reporter vectors of the wild-type or
mutant-type 3′-UTR of NRP2 by Lipofectamine™ 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Subsequently, the Dual luciferase
reporter assay kit (Promega) was used to detect the relative
luciferase activities 48 h after the transfections.
Statistical analysis
The above assays were conducted at least three
times. SPSS 17.0 (SPSS Inc.) was used to perform the statistical
analysis with Student's t-test or one-way ANOVA test followed by
post hoc test. Data are indicated as means ± SD. Correlation
between expression levels of miR-331-3p and NRP2 was estimated
using the Pearson's correlation method. The differences were
identified as statistically significant at P<0.05.
Results
miR-331-3p is downregulated and NRP2
is upregulated in CRC
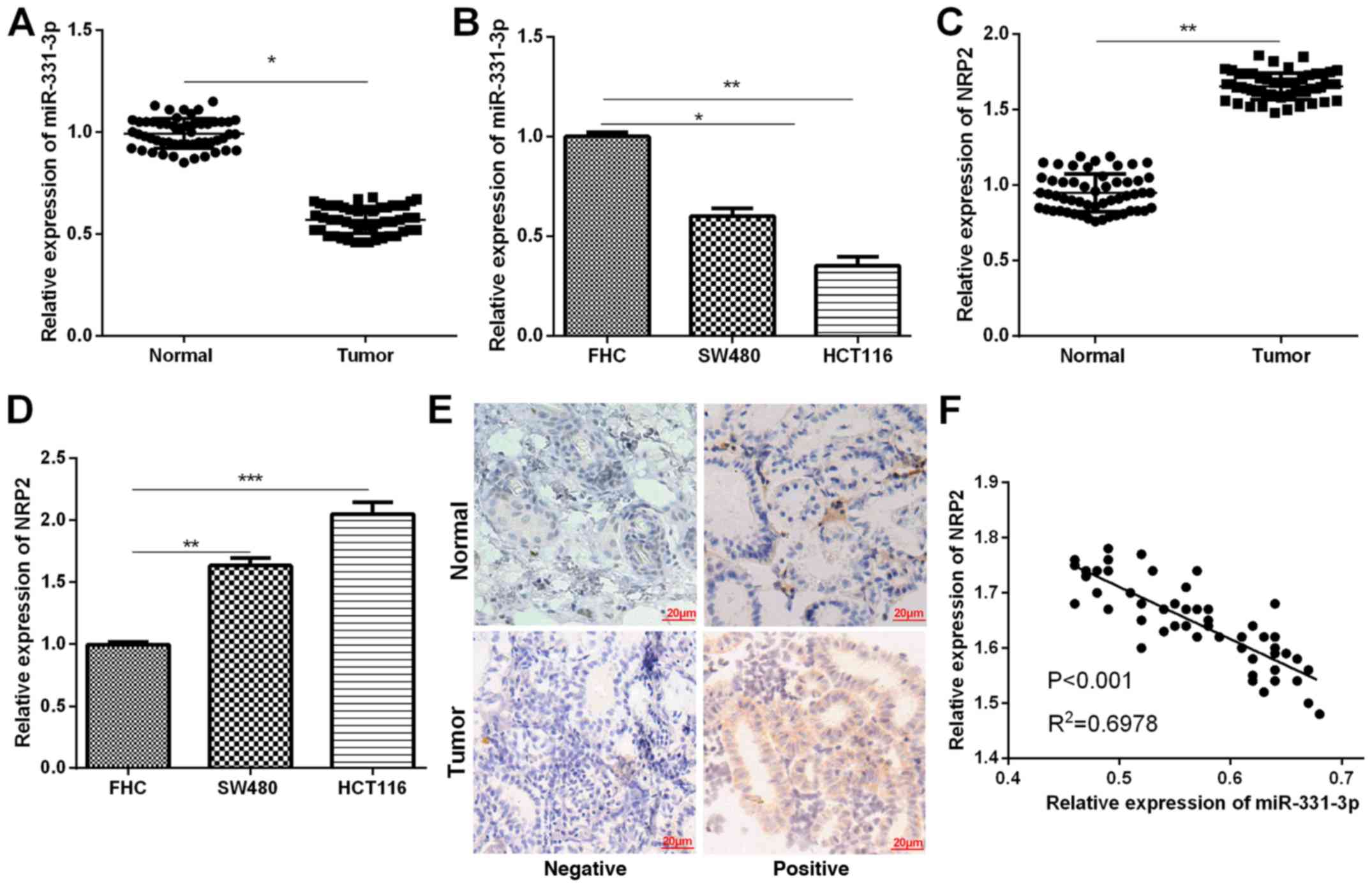
To determine whether miR-331-3p was involved in CRC
carcinogenesis, its expression in 54 pairs of CRC tissue samples
and two cell lines was detected by RT-qPCR. The results indicated
that, when compared to the matched normal tissue samples, the
miR-331-3p expression in CRC tissue samples was significantly
decreased (Fig. 1A). Similarly,
RT-qPCR results also indicated that miR-331-3p expression in CRC
cells was significantly lower than that in normal colonic cells
(Fig. 1B). Furthermore, NRP2
expression levels in CRC tissues and cells were measured. RT-qPCR
analysis demonstrated significantly higher mRNA levels of NRP2 in
both CRC tissues and cells compared to the corresponding controls
(Fig. 1C and D). In addition, we
analyzed the correlation between the expression of NRP2 and
miR-331-3p in CRC tissues to better understand their functions in
CRC progression. The results demonstrated that the miR-331-3p
expression had a negative correlation with the expression of NRP2
in CRC tissues (Fig. 1E). In
addition, all the enrolled CRC patients were assigned into high or
low miR-331-3p expression groups based on the mean miR-331-3p
level. Clinicopathologic analysis demonstrated that CRC patients
with low miR-331-3p expression presented malignant
clinicopathological features (Table
I).
 | Table I.Correlation of miR-331-3p expression
with the clinicopathological characteristics of the colorectal
carcinoma patients. |
Table I.
Correlation of miR-331-3p expression
with the clinicopathological characteristics of the colorectal
carcinoma patients.
|
|
|
miR-331-3pa expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases (n=54) | High (n=18) | Low (n=36) | P-value |
|---|
| Age (years) |
|
|
| 0.563 |
|
>60 | 26 | 10 | 16 |
|
| ≤60 | 28 | 8 | 20 |
|
| Sex |
|
|
| 0.471 |
| Male | 30 | 12 | 18 |
|
|
Female | 24 | 6 | 18 |
|
| Tumor size
(cm) |
|
|
| 0.312 |
|
≥5.0 | 27 | 7 | 20 |
|
|
<5.0 | 27 | 11 | 16 |
|
| TNM stage |
|
|
| 0.015b |
|
I–II | 21 | 15 | 6 |
|
|
III | 33 | 3 | 30 |
|
| Lymph node
metastasis |
|
|
| 0.006b |
|
Yes | 31 | 5 | 26 |
|
| No | 23 | 13 | 10 |
|
| Location |
|
Colon | 27 | 12 | 15 | 0.316 |
|
Rectum | 27 | 6 | 21 |
|
| Distant
metastasis |
|
|
| 0.072 |
|
Yes | 28 | 9 | 19 |
|
| No | 26 | 9 | 17 |
|
miR-331-3p inhibits CRC cell invasion
and migration
To further understand the effects of miR-331-3p on
CRC progression, SW480 and HCT116 cells were trasnfected with
miR-331-3p mimics or inhibitor to overexpress or inhibit miR-331-3p
expression. RT-qPCR analysis was performed to confirm the
successful miR-331-3p overexpression or downregulation in SW480 or
HCT116 cells (Fig. 2A and B).
Subsequently, we explored the functions of miR-331-3p in SW480 and
HCT116 cell invasion and migration through performing Transwell
assays. Fig. 2C shows that
overexpression of miR-331-3p could markedly repress the invasion
and migration capacities of SW480 cells when decreased expression
of miR-331-3p enhanced the SW480 invasion and migration.
Additionally, similar functions of miR-331-3p in HCT116 cell
invasion and migration were confirmed by Transwell assays (Fig. 2D). Results suggested that miR-331-3p
was able to inhibit CRC cell invasion and migration.
miR-331-3p upregulation suppresses CRC
cell EMT
It was reported that EMT is regarded as a crucial
representations in cancer metastasis and invasion. Thus, to address
molecular mechanism of miR-331-3p-induced anti-metastatic effect on
CRC cells, western blot analysis was performed to detect the
protein levels involved in EMT occurrence. It was demonstrated that
in SW480 cells, the expression levels of E-cadherin were
significantly increased while the expression levels of N-cadherin
and Vimentin were significantly decreased by miR-331-3p mimics
(Fig. 3A). On the other hand,
miR-331-3p inhibitor in SW480 cells had the opposite functions in
EMT-related proteins (Fig. 3B).
Moreover, we examined the protein expression in HCT116 cells, and a
similar influence of miR-331-3p on the expression of proteins which
were closely related to EMT was identified (Fig. 3C and D).
miR-331-3p interacts with NRP2 in CRC
cells by directly binding to the NRP2 3′-UTR
The correlation was investigated between NRP2 and
miR-331-3p to fully understand the mechanisms of miR-331-3p in
regulating CRC. Based on Targetscan, miR-331-3p was predicted to
bind to NRP2 3′-UTR (Fig. 4A),
suggesting that NRP2 was a potential target for miR-331-3p. Then,
to confirm whether NRP2 was directly targeted by miR-331-3p, we
performed dual-luciferase reporter assays. The luciferase reporter
vectors which contained NRP2 3′-UTR-WT or NRP2 3′-UTR-MUT were
constructed and cotransfected into CRC cells with miR-331-3p
mimics. The relative luciferase activities of the reporter
containing the NRP2 3′-UTR-WT were significantly reduced by
miR-331-3p mimics; however, the luciferase activity of NRP2
3′-UTR-MUT was not notably affected by miR-331-3p mimics (Fig. 4B). RT-qPCR and western blot analyses
of the NRP2 expression demonstrated that NRP2 expression levels in
CRC cells were significantly decreased by miR-331-3p mimics in
contrast to the controls, whereas NRP2 expressions in cells with
transfection of miR-331-3p inhibitor demonstrated notably increased
NRP2 levels compared with the NC (Fig.
4C and D).
Silencing of NRP2 partially reverses
the miR-331-3p inhibitor-mediated functions in promoting SW480 cell
invasion and migration
To elucidate whether miR-331-3p exerted anti-CRC
functions through regulating NRP2, NRP2 siRNA and miR-331-3p
inhibitor were co-transfected into CRC cell lines. NRP2 siRNA was
transfected into CRC cells to knock down NRP2, RT-qPCR and western
blot results showed that, transfection with NRP2 siRNA resulted in
marked downregulation of NRP2 expression in CRC cells (Fig. 5A and B). Moreover, similar results
were also identified in CRC cells transfected with NRP2 siRNA and
miR-331-3p inhibitor (Fig. 5A and
B). Subsequently, the Transwell assays were carried out to
determine the functions of NRP2 siRNA in SW480 cell migration and
invasion. Results demonstrated that the invasion and migration
abilities of SW480 cell lines cotransfected with miR-331-3p
inhibitor and NRP2 siRNA were markedly suppressed compared to that
of the only miR-331-3p downregulated SW480 cell lines (Fig. 5D and E). The findings suggested that
deletion of NRP2 markedly reversed miR-331-3p inhibitor-mediated
promotion of cell invasion and migration in SW480 cell lines.
Discussion
CRC is a critical challenge both for public health
and clinical practice. In recent decades, although the life
expectancy of CRC patients has been improved due to the advances in
CRC screening and therapy (20), CRC
still remains a leading health problem worldwide. Thus, more
attention should been given to the specific mechanisms of the CRC
initiation and development. Growing evidence has indicated that
miRNAs play important functions in human CRC development (21). Moreover, miRNAs have been determined
to play a crucial role in regulating gene expression, and in other
relevant processes, such as invasion and metastasis (22).
miR-331-3p has been identified as a tumor-associated
miRNA. As an independent prognostic factor, miR-331-3p was reported
to modulate tumor progression. Epis et al (23) found that miR-331-3p inhibited
prostate cancer progression with Aurora Kinase inhibitor II
cotreatment; Chen et al(24)
reported that in hepatocellular carcinoma patients, serum
miR-331-3p and miR-182 functioned as therapic biomarkers; Cao et
al (25) verified that
miR-331-3p suppressed VHL expression in HCC. Given that miRNAs are
widely known as tumor regulators, we provide further evidence in
this study that miR-331-3p plays important roles in human CRC.
miR-331-3p was identified as the downregulated miRNA in CRC by
RT-qPCR. Moreover, we found that decreased miR-331-3p was
associated with the aggressive clinicopathological features of CRC
patients. Over-expression of miR-331-3p was able to inhibit CRC
cell invasion and migration by targeting NRP2 and regulating EMT.
Collectively, the findings of this research revealed that
miR-331-3p played anti-tumor roles in CRC.
Neuropilins (NRPs) are type I transmembrane
receptors that form heterodimeric complexes with two key classes of
signaling transmembrane receptors: Plexins and vascular endothelial
growth factor receptors (VEGFRs) (26). There are two main NRP receptors (NRP1
and NRP2), with multiple extracellular and transmembrane isoforms
observed for each in vivo (27). NRPs are thought primarily to modulate
the affinity and specificity of extracellular ligand binding upon
co-receptor complex formation. Plexin-NRP co-receptor complexes
bind semaphorins (Semas), which are a large class of extracellular,
dimeric ligands that act as either attractive or repulsive cues
during cell migration in a diverse array of processes (28). VEGFR-NRP co-receptor complexes bind
vascular endothelial growth factor (VEGF), which plays a major role
in the induction of endothelial cell proliferation and increase of
the vascular endothelium permeability (29,30). NRP
is now considered a candidate specific receptor for VEGF (31). Given the diversity of biological
processes in which Sema and VEGF modulate cell migration,
dysregulation of NRP-dependent signaling has been linked to a
variety of cancers. The role of NRPs as co-receptors of Semas and
VEGF in tumor angiogenesis and metastases is the basis for current
trials. Various research has reported the effects and mechanisms of
NRP2 on tumor progression. Fung et al (32) indicated that NRP2 promoted
oesophageal squamous cell carcinoma metastasis and tumorigenicity;
Dallas et al (33) further
demonstrated that NRP2 regulated pancreatic adenocarcinoma
angiogenesis and growth; Moriarty et al (34) found that NRP2 promoted melanoma
progression and growth. To our knowledge, there is no previous
report on research investigating the association between NRP2 and
miR-331-3p in CRC. The current study provided preliminary strong
evidence that NRP2 was directly targeted by miR-331-3p and
implicated in CRC invasion and migration. The data also revealed
that knockdown of NRP2 reversed the functions of miR-331-3p
inhibitor in cell invasion and migration of CRC cells. These
results suggest that miR-331-3p exerted cancer suppressive roles in
CRC via targeting NRP2.
In conclusion, miR-331-3p was downregulated in CRC,
which indicates poor outcomes of CRC patients. miR-331-3p
overexpression suppressed migration and invasion through regulating
NRP2 and EMT. In addition, the suppression function of miR-331-3p
in invasion and migration of CRC cells was partially mediated by
direct deregulation of NRP2. Thus, the findings in the current
study may help to better determine the mechanisms of miR-331-3p and
NRP2 implicated in CRC progression, and to discover sensitive
prognostic and therapeutic biomarkers for CRC.
Acknowledgements
Not applicable.
Funding
This study was supported by Shandong Traditional
Chinese Medicine Science and Technology Development Plan
(2017-474).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ as the first author contributed significantly to
statistics analysis and manuscript preparation. RW wrote the
manuscript and helped perform the statistics analysis with
constructive discussions. MW as the corresponding author
contributed to the conception of the study and provided clinical
data of the patients as well as crucial experiment materials. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Linyi Central Hospital (Linyi, China). Written informed consent
from each patient was received before the samples were
collected.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bae JM, Kim JH, Kwak Y, Lee DW, Cha Y, Wen
X, Lee TH, Cho NY, Jeong SY, Park KJ, et al: Distinct clinical
outcomes of two CIMP-positive colorectal cancer subtypes based on a
revised CIMP classification system. Br J Cancer. 116:1012–1020.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hadjipetrou A, Anyfantakis D, Galanakis
CG, Kastanakis M and Kastanakis S: Colorectal cancer, screening and
primary care: A mini literature review. World J Gastroenterol.
23:6049–6058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haraldsdottir S, Einarsdottir HM,
Smaradottir A, Gunnlaugsson A and Halfdanarson TR: Colorectal
cancer - review. Laeknabladid. 100:75–82. 2014.(In Icelandic).
PubMed/NCBI
|
|
4
|
Lieberman D, Ladabaum U, Cruz-Correa M,
Ginsburg C, Inadomi JM, Kim LS, Giardiello FM and Wender RC:
Screening for colorectal cancer and evolving issues for physicians
and patients: A Review. JAMA. 316:2135–2145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu W, Sun Y, Zhang L and Xing BC:
Negative surgical margin improved long-term survival of colorectal
cancer liver metastases after hepatic resection: A systematic
review and meta-analysis. Int J Colorectal Dis. 30:1365–1373. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Massat NJ, Moss SM, Halloran SP and Duffy
SW: Screening and primary prevention of colorectal cancer: A review
of sex-specific and site-specific differences. J Med Screen.
20:125–148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bonfrate L, Altomare DF, Di Lena M,
Travaglio E, Rotelli MT, De Luca A and Portincasa P: MicroRNA in
colorectal cancer: New perspectives for diagnosis, prognosis and
treatment. J Gastrointestin Liver Dis. 22:311–320. 2013.PubMed/NCBI
|
|
8
|
Karatas OF, Suer I, Yuceturk B, Yilmaz M,
Hajiyev Y, Creighton CJ, Ittmann M and Ozen M: The role of miR-145
in stem cell characteristics of human laryngeal squamous cell
carcinoma Hep-2 cells. Tumour Biol. 37:4183–4192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu L, Li Q, Xu D, Wang Q, An Y, Du Q,
Zhang J, Zhu Y and Miao Y: hsa-miR-141 downregulates TM4SF1 to
inhibit pancreatic cancer cell invasion and migration. Int J Oncol.
44:459–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang G, Shen N, Cheng L, Lin J and Li K:
Downregulation of miR-22 acts as an unfavorable prognostic
biomarker in osteosarcoma. Tumour Biol. 36:7891–7895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bai QL, Hu CW, Wang XR, Shang JX and Yin
GF: miR-616 promotes proliferation and inhibits apoptosis in glioma
cells by suppressing expression of SOX7 via the Wnt signaling
pathway. Eur Rev Med Pharmacol Sci. 21:5630–5637. 2017.PubMed/NCBI
|
|
12
|
Wang Z, Liao H, Deng Z, Yang P, Du N,
Zhanng Y and Ren H: miRNA-205 affects infiltration and metastasis
of breast cancer. Biochem Biophys Res Commun. 441:139–143. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Staton CA, Kumar I, Reed MW and Brown NJ:
Neuropilins in physiological and pathological angiogenesis. J
Pathol. 212:237–248. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pellet-Many C, Frankel P, Jia H and
Zachary I: Neuropilins: Structure, function and role in disease.
Biochem J. 411:211–226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Drabkin HA, Starkova J and Gemmill RM: A
triad of NRP2, DLX and p53 proteins in lung cancer metastasis.
Oncotarget. 8:96464–96465. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fujii T, Shimada K, Asano A, Tatsumi Y,
Yamaguchi N, Yamazaki M and Konishi N: MicroRNA-331-3p suppresses
cervical cancer cell proliferation and E6/E7 expression by
targeting NRP2. Int J Mol Sci. 17:172016. View Article : Google Scholar
|
|
17
|
Yasuoka H, Kodama R, Tsujimoto M,
Yoshidome K, Akamatsu H, Nakahara M, Inagaki M, Sanke T and
Nakamura Y: Neuropilin-2 expression in breast cancer: Correlation
with lymph node metastasis, poor prognosis, and regulation of CXCR4
expression. BMC Cancer. 9:2202009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta DeltaC(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Burstein HJ and Schwartz RS: Molecular
origins of cancer. N Engl J Med. 358:5272008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
El Sharawy A, Röder C, Becker T, Habermann
JK, Schreiber S, Rosenstiel P and Kalthoff H: Concentration of
circulating miRNA-containing particles in serum enhances miRNA
detection and reflects CRC tissue-related deregulations.
Oncotarget. 7:75353–75365. 2016.PubMed/NCBI
|
|
22
|
Suzuki HI, Katsura A, Matsuyama H and
Miyazono K: MicroRNA regulons in tumor microenvironment. Oncogene.
34:3085–3094. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Epis MR, Giles KM, Beveridge DJ,
Richardson KL, Candy PA, Stuart LM, Bentel J, Cohen RJ and Leedman
PJ: miR-331-3p and Aurora kinase inhibitor II co-treatment
suppresses prostate cancer tumorigenesis and progression.
Oncotarget. 8:55116–55134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen L, Chu F, Cao Y, Shao J and Wang F:
Serum miR-182 and miR-331-3p as diagnostic and prognostic markers
in patients with hepatocellular carcinoma. Tumour Biol.
36:7439–7447. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao Y, Zhang J, Xiong D, Wang D, Wu T,
Huang A and Tang H: Hsa-miR-331-3p inhibits VHL expression by
directly targeting its mRNA 3-UTR in HCC cell lines. Acta Biochim
Pol. 62:77–82. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zachary IC, Frankel P, Evans IM and
Pellet-Many C: The role of neuropilins in cell signalling. Biochem
Soc Trans. 37:1171–1178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rossignol M, Gagnon ML and Klagsbrun M:
Genomic organization of human neuropilin-1 and neuropilin-2 genes:
Identification and distribution of splice variants and soluble
isoforms. Genomics. 70:211–222. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Niland S and Eble JA: Neuropilins in the
context of tumor vasculature. Int J Mol Sci. 20:202019. View Article : Google Scholar
|
|
29
|
Favier B, Alam A, Barron P, Bonnin J,
Laboudie P, Fons P, Mandron M, Herault JP, Neufeld G, Savi P, et
al: Neuropilin-2 interacts with VEGFR-2 and VEGFR-3 and promotes
human endothelial cell survival and migration. Blood.
108:1243–1250. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ferrara N, Gerber HP and LeCouter J: The
biology of VEGF and its receptors. Nat Med. 9:669–676. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Caunt M, Mak J, Liang WC, Stawicki S, Pan
Q, Tong RK, Kowalski J, Ho C, Reslan HB, Ross J, et al: Blocking
neuropilin-2 function inhibits tumor cell metastasis. Cancer Cell.
13:331–342. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fung TM, Ng KY, Tong M, Chen JN, Chai S,
Chan KT, Law S, Lee NP, Choi MY, Li B, et al: Neuropilin-2 promotes
tumourigenicity and metastasis in oesophageal squamous cell
carcinoma through ERK-MAPK-ETV4-MMP-E-cadherin deregulation. J
Pathol. 239:309–319. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dallas NA, Gray MJ, Xia L, Fan F, van
Buren G II, Gaur P, Samuel S, Lim SJ, Arumugam T, Ramachandran V,
et al: Neuropilin-2-mediated tumor growth and angiogenesis in
pancreatic adenocarcinoma. Clin Cancer Res. 14:8052–8060. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moriarty WF, Kim E, Gerber SA, Hammers H
and Alani RM: Neuropilin-2 promotes melanoma growth and progression
in vivo. Melanoma Res. 26:321–328. 2016. View Article : Google Scholar : PubMed/NCBI
|