Introduction
The increasing incidence and mortality rates of
cancer in humans worldwide have exerted serious effects on the
health and quality of life of affected individuals, particularly in
developed countries (1,2). Hepatocellular carcinoma (HCC), a very
common malignant gastrointestinal tumor that occurs in
geographically heterogeneous regions, in particular Eastern Asia
and sub-Saharan Africa, where medical service is relatively
limited, accounts for 75–85% of primary liver cancer cases. The
major risk factors, including chronic hepatitis B and hepatitis C,
excessive intake of alcohol, aflatoxin exposure and metabolic liver
disease, have increased the threat of HCC worldwide (2). HCC has been revealed to have
sex-specific differences, as the prevalence and mortality rates are
2–3 times higher among men than women (3,4). This
predisposition may be attributed to the frequent development of
chronic hepatitis B virus infection, alcoholism, fatty liver
disease and hepatitis C virus infection in men (5–7).
Although substantial advances have been made in the therapeutic
field of HCC, frequent recurrence, advanced diagnosis and
metastasis remain challenging issues to overcome and are
preventative factors in obtaining a good prognosis (8,9). Thus, a
useful marker for estimating the diagnosis and curative outcome of
HCC is essential.
Mature microRNAs (miRNAs), commonly recognized as a
major family of endogenous and small non-coding RNAs with a length
of ~22 nucleotides, bind to the 3′-untranslated region (UTR) of
corresponding target messenger RNAs (mRNAs), resulting in the
inhibition of protein translation or degradation of mRNAs (10–12).
Importantly, it is generally acknowledged that miRNAs serve
important roles in biological processes, including cell
proliferation, expansion, differentiation, infiltration and
apoptosis, by regulating the expression of related genes in tissues
or blood plasma (13–15). Moreover, miRNA dysregulation is an
important factor contributing to tumorigenesis and progression,
furthermore, certain specific miRNAs, such as miR-92a-2, miR-133b,
miR-34a and miR-96, can act as either oncogenes or tumor
suppressors (16–18).
miR-139-5p, one of the most common subtypes of
miR-139, which is located on chromosome 11q13.4, has become
established as a critical focus of studies examining the initiation
and progression of diverse cancers, including oral squamous
carcinoma (19), colorectal cancer
(20,21), bladder cancer (22), endometrial cancer (23) and HCC (24–27).
Wang et al (27) reported
that miR-139-5p inhibits HCC development, however, the study
included a limited number of HCC specimens. Moreover, a number of
studies have reported a close association between differential
miR-139-5p expression and the prognosis of HCC (25,26,28,29).
Nevertheless, literature reporting the precise molecular mechanisms
and available published studies analyzing miR-139-5p regulation in
HCC are inconclusive, thus additional comprehensive studies are
required.
The current meta-analysis was performed by
integrating miRNA sequencing and microarray data, and
comprehensively analyzing published articles that mentioned a
relationship between miR-139-5p regulation and clinical data from
human HCC samples. In addition, a bioinformatics analysis was
conducted in order to confirm the characteristics of miR-139-5p and
its targeted genes in HCC pathology- and development-related
mechanisms. The present meta-analysis may provide further
suggestions regarding the development of novel molecular
therapeutic strategies for patients with HCC.
Materials and methods
Data mining and literature search
Microarray profiles associated with miRNA expression
in HCC were searched and downloaded from the Gene Expression
Omnibus (GEO) (30), The Cancer
Genome Atlas (TCGA) (31),
ArrayExpress (32) and Oncomine
(33) databases. The related records
were updated on March 28, 2019 and the following search terms were
used: (malignan* OR cancer OR tumor OR tumour OR neoplas* OR
carcinoma) AND (hepatocellular OR liver OR hepatic OR HCC).
Initially, studies were chosen based on the following inclusion
criteria: i) Human HCC confirmed by pathological analysis and ii)
information about miR-139-5p regulation in HCC and nontumorous
samples.
For the published literature search, scientific
databases, including PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), ScienceDirect
(https://www.sciencedirect.com/), Web of
Science (http://wokinfo.com/), Wang Fang
(http://www.wanfangdata.com.cn/index.html), ChongQing
VIP (http://cstj.cqvip.com/) and the China
National Knowledge Infrastructure (https://www.cnki.net/), were comprehensively searched
to determine the most relevant studies published up to March 28,
2019. The key search terms were: (miR-139-5p OR miRNA-139-5p OR
microRNA139-5p OR miR139-5p OR miRNA139-5p OR miR 139-5p OR miRNA
139-5p OR microRNA 139-5p OR microRNA-139-5p) AND (malignan* OR
cancer OR tumor OR tumour OR neoplas* OR carcinoma) AND
(hepatocellular OR liver OR hepatic OR HCC). The inclusion criteria
regarding the published studies were: i) HCC confirmed by an
operation and pathological analysis; ii) information on the
miR-139-5p expression levels (high vs. low) and relative
clinicopathological data, including odds ratios (ORs) with 95%
confidence intervals (CIs) of some clinicopathological parameters
in HCC; and iii) direct clinical prognostic information, including
overall survival (OS), recurrence-free survival (RFS) or
disease-free survival (DFS) with 95% CIs. Furthermore, when the
Kaplan-Meier curves were drawn clearly, we could artificially
extract and calculate the effect sizes, including OS, RFS, DFS with
corresponding 95% CIs statistically.
The following exclusion criteria were applied: i)
Lack of information on miR-139-5p expression in patients with HCC;
ii) the full study was not published in English or Chinese; iii)
reported data were insufficient for the calculation of clinical
pathological parameters and prognostic indicators; and iv) the
articles were reviews, conference abstracts or case reports.
Data extraction
The following information was considered for
microarray data selection: Dataset name, case number (N), mean (M)
and standard deviation (SD) of corresponding datasets among HCC and
nontumorous tissues, as well as area under the curve (AUC) values.
Available parameters from the TCGA clinicopathological analysis
included: Age, sex, race, neoplasm status, tumor grade,
pathological stage, tumor-node-metastasis (TNM) stage, lymph node
metastasis and metastasis status.
The following data were extracted from the
scientific publications: First author, year published, country,
sample sizes, methods, sample sources, follow-up time, outcomes and
hazard ratios (HRs) with corresponding 95% CIs for OS, RFS or DFS.
Kaplan-Meier curves were generated using the officially recommended
method by Tierney et al (34).
Statistical analysis
Statistical analyses were performed using Stata
(version 14.0; StataCorp LP), SPSS software (version 24.0; SPSS,
IBM Corp.) and GraphPad Prism software (version 9; GraphPad
Software, Inc.). Initially, microarray data were separated into two
groups, namely the HCC tissue group and the nontumorous liver
tissue group. miR-139-5p expression levels were calculated from
each included dataset, the N and M ± SD of the two groups were then
recorded and compared using Student's t-test. More than two groups
in the TCGA clinical parameter calculation were analyzed using
one-way ANOVA. In addition, scatter plots presenting miR-139-5p
expression levels were generated using GraphPad Prism software, and
the receiver operating characteristic (ROC) curve analysis of each
dataset was performed using SPSS software. Meta-DiSc (version 1.4;
Clinical Biostatistics Unit Miscellaneous Shareware), which is
capable of testing the precision of meta-analyses, was utilized to
assess diagnostic efficiency, including the summary receiver
operating characteristic (sROC) curve, positive likelihood ratio
(PLR), negative likelihood ratio (NLR), and diagnostic sensitivity
and specificity. Moreover, the merged standard mean differences
(SMDs) and 95% CIs for all appropriate datasets were assessed using
Stata 14.0 software to analyze miR-139-5p expression levels in
patients with HCC. The merged SMD was <0 and the 95% CI did not
include 0, indicating a relatively low expression level of
miR-139-5p in HCC tissues. Furthermore, Kaplan-Meier survival
curves were manually extracted from published studies via Engauge
Digitizer software (version 4.1; http://markummitchell.github.io/engauge-digitizer/),
and outcomes were recorded as HRs and 95% CIs. These values were
ultimately merged using Stata 14.0 software aimed at confirming the
prognostic effect of miR-139-5p. The HR was considered significant
if P<0.05 and the 95% CI did not exceed 1; otherwise, no
significant effect of miR-139-5p regulation was observed in
patients with HCC. Heterogeneity analysis was evaluated by the
Cochrane Q and inconsistency index (I2) tests (35). A random-effects model was employed if
clear heterogeneity was indicated with I2>50% or
P<0.05; if not, a fixed-effects model was selected.
Additionally, publication bias was detected by Begg's funnel test,
and influence analysis was performed to determine whether the
studies or datasets included in the present study exerted excessive
effects on pooled outcomes by independently removing each
study.
Bioinformatics analysis
Twelve online tools, namely miRWalk3.0 (http://mirwalk.umm.uni-heidelberg.de/),
MiRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php),
TarBase6.0 (http://www.microrna.gr/tarbase), TargetMiner
(http://www.mybiosoftware.com/targetminer-microrna-target-prediction.html),
polymiRTS3.0 (http://compbio.uthsc.edu/miRSNP), RNA22 (https://cm.jefferson.edu/rna22/), microRNA.org (http://www.microrna.org/), PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
mirRNAMAP2.0 (http://mirnamap.mbc.nctu.edu.tw/), TargetScan
(http://www.targetscan.org/vert_71/),
miRDB (http://mirdb.org/) and PicTar-vert (http://dorina.mdc-berlin.de) (36–47),
were employed to comprehensively investigate the impact of
miR-139-5p on biological processes. Genes were investigated to
predict possible target genes and differentially expressed genes
(DEGs) in HCC tissues were downloaded from TCGA. Next, a Venn
diagram (http://bioinfogp.cnb.csic.es/tools/venny/) was
generated to obtain intersecting DEGs as final target genes from
the aforementioned DEGs and predicted genes. Furthermore, the final
target genes were subjected to Gene Ontology (GO) enrichment, Kyoto
Encyclopedia of Genes and Genomes (KEGG) and Panther pathway
analyses using the Database for Annotation, Visualization and
Integrated Discovery (DAVID; version 6.8; http://david.ncifcrf.gov) (48). P<0.05 was considered to indicate a
statistically significant difference in all pathways. Moreover,
images of correlation networks were presented in a bubble diagram
that was constructed using the ImageGP online website (http://www.ehbio.com/ImageGP/). The Search Tool for
the Retrieval of Interacting Genes (STRING; http://string-db.org; version 11.0) was employed to
explore the protein-protein interaction (PPI) network and identify
the top downstream target genes with the greatest number of
connections of each node that were most likely regulated by
miR-139-5p in HCC (49,50). Additionally, Gene Expression
Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/; version 1.0) (51) was used to determine the expression
and relative prognostic roles of downstream target genes in the
development of HCC. Intersecting DEGs were identified as hub genes
from the top downstream genes and the target genes that were
prominently enriched in the first pathway, which included the most
statistically significant differences of KEGG. Additionally, the
Human Protein Atlas database (52)
and CBio Cancer Genomics Portal (53) were searched to determine the
deregulation and mutations of hub genes. The expression patterns of
hub genes were validated using immunohistochemical (IHC) staining
images of pathological sections obtained from the Human Protein
Atlas, and comparing normal and HCC tissue samples. The correlation
between hub gene and miR-139-5p expression in TCGA data was
evaluated by the calculation of Pearson's correlation coefficients
using SPSS software.
Results
Characteristics of the included
studies, miRNA-sequencing (miRNA-seq, and microarray data
In the literature search performed using the
aforementioned retrieval strategy, 205 studies were preliminarily
obtained. After thorough examination of all the studies, 201
studies were excluded due to 36 articles being duplicated, and no
available clinical information being explicitly provided in 165
articles. Consequently, only four studies that provided
associations with the survival of 367 patients could be included
(25,28,29,42).
Three studies presented survival curve data that allowed OS
calculation, and one directly provided accurate OS data. However,
only one study separately (28,29)
described DFS and RFS, which were not merged to confirm the
significance of these two effect sizes. Details of the included
terms are presented in Table I, and
a schematic of the literature and microarray data screening
processes is shown in Fig. 1.
 | Table I.Major characteristics of the analyzed
publications reporting associations with miR-139-5p expressed in
HCC. |
Table I.
Major characteristics of the analyzed
publications reporting associations with miR-139-5p expressed in
HCC.
| Study | Year | Country | N | Cut-off value | Method | Sample | Follow-up
(months) | Result | HR (95%
CI)a | Source | (Refs.) |
|---|
| Li et
al | 2014 | China | 31 | NA | RT-qPCR | Plasma | 12 | OS | OS: 1.56
(0.3–8.07) | ED | (54) |
| Wang et
al | 2016 | China | 153 | NA | RT-qPCR | Tissue | – | OS CPP | OS: 2.68
(1.42–5.05) | ED | (25) |
| Wong et
al | 2011 | China | 67 | NA | RT-qPCR | Tissue | 60 | OS DFS CPP | OS: 1.75
(0.36–8.48) | ED | (29) |
|
|
|
|
|
|
|
|
|
| DFS: 1.67
(0.57–4.87) |
|
|
| Wang et
al | 2015 | China | 116 | NA | RT-qPCR | Tissue | – | OS RFS | OS: 1.195
(0.90–1.57) | RD | (28) |
|
|
|
|
|
|
|
|
|
| RFS: 1.20
(0.61–2.37) |
|
|
The miRNA-seq and microarray data were initially
retrieved from 90,880 datasets, and after careful examination and
verification of the data, 18 datasets were identified to provide
miR-139-5p expression data regarding HCC and nontumor liver tissues
that conformed to the inclusion criteria of the present study.
These 18 datasets were retrieved from the TCGA, GEO, ArrayExpress
and Oncomine databases. Specifically, 14 datasets were obtained
from GEO and Oncomine, including GSE12717, GSE21279, GSE21362,
GSE36915, GSE39678, GSE40744, GSE41874, GSE50013, GSE54751,
GSE64632, GSE67882, GSE69580, GSE98269 and GSE115016. Three
datasets were retrieved from the ArrayExpress database: E-MTAB-511,
E-MTAB-3347 and E-MTAB-4809. The remaining TCGA dataset containing
available data of miR-139-5p expression profile was downloaded from
TCGA miRNA-seq profiles. Detailed information is presented in
Table II.
 | Table II.Essential features of datasets
involving miR-139-5p expression founded on GEO, TCGA, Oncomine and
Array Express databases. |
Table II.
Essential features of datasets
involving miR-139-5p expression founded on GEO, TCGA, Oncomine and
Array Express databases.
|
| HCC | Non-tumor |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Dataset | N | M | SD | N | M | SD | t | P-value | AUC |
|---|
| GSE12717 | 10 | 5.9442 | 1.33223 | 6 | 8.2381 | 0.44433 | −4.036 | 0.001 | 0.917 |
| GSE21279 | 4 | 3.3278 | 2.49676 | 12 | 3.3192 | 3.49780 | 0.005 | 0.996 | 0.531 |
| GSE21362 | 73 | 4.8902 | 1.51216 | 73 | 6.3078 | 1.02803 | −6.624 | <0.001 | 0.833 |
| GSE36915 | 68 | 12.0473 | 1.20972 | 21 | 13.5388 | 0.60042 | −7.583 | <0.001 | 0.891 |
| GSE39678 | 16 | 7.9983 | 0.57305 | 8 | 8.7452 | 0.64627 | −2.888 | 0.009 | 0.828 |
| GSE40744 | 26 | 8.5754 | 1.41834 | 50 | 9.4392 | 0.47678 | −3.018 | 0.005 | 0.735 |
| GSE41874 | 3 | 0.8372 | 0.06337 | 4 | 1.1558 | 0.06887 | −6.252 | 0.002 | 1.000 |
| GSE50013 | 14 | 1.7929 | 4.95168 | 13 | 0.9769 | 2.13937 | 0.548 | 0.589 | 0.448 |
| GSE54751 | 10 | 0.0110 | 0.00801 | 10 | 0.0435 | 0.02363 | −4.119 | 0.002 | 0.920 |
| GSE64632 | 3 | 0.3118 | 0.38826 | 3 | 0.1204 | 0.06350 | 0.843 | 0.484 | 0.444 |
| GSE67882 | 3 | 12.1139 | 1.96061 | 8 | 9.9169 | 3.02709 | 1.149 | 0.280 | 0.250 |
| GSE69580 | 5 | 1.3212 | 1.21214 | 5 | 10.8835 | 3.12460 | −6.380 | <0.001 | 1.000 |
| GSE98269 | 3 | 5.4040 | 0.25288 | 3 | 5.9628 | 0.14467 | −3.322 | 0.029 | 1.000 |
| GSE115016 | 12 | 9.5731 | 12.11435 | 12 | 48.9893 | 26.09749 | −4.746 | <0.001 | 0.944 |
| TCGA | 371 | 6.4189 | 1.23762 | 49 | 8.5117 | 0.45390 | −22.926 | <0.001 | 0.953 |
| E-MTAB-511 | 8 | 8.1250 | 10.04899 | 10 | 6.7000 | 9.35771 | 0.311 | 0.760 | 0.450 |
| E-MTAB-3347 | 4 | 6.7444 | 0.34443 | 4 | 6.8057 | 0.38966 | −0.236 | 0.821 | 0.500 |
| E-MTAB-4809 | 12 | 9.2651 | 1.27329 | 12 | 5.1714 | 1.42375 | 7.424 | <0.001 | 0.000 |
Analysis of miRNA-seq and microarray data
acquired from public databases and the scientific literature
miR-139-5p expression data in HCC
tissues obtained from TCGA
As presented in Table
III, lower miR-139-5p expression levels were observed in HCC
tissues (n=371) than in normal tissues (n=49) (6.4189±1.2376 vs.
8.5117±0.4539; t=−22.926; P<0.01). Statistically significant
associations (P<0.05) were observed among miR-139-5p expression
and tumor grade (G1/G2/G3/G4), pathological stage (stage
I/II/III/IV) and T stage (T1-2/3–4). However, no statistically
significant association was observed between miR-139-5p expression
and the remaining clinicopathological characteristics.
 | Table III.Clinicopathological parameters of
miRNA-139-5p regulation in HCC tissues from TCGA. |
Table III.
Clinicopathological parameters of
miRNA-139-5p regulation in HCC tissues from TCGA.
|
|
| MiR-139-5p relevant
expression (log2x) |
|---|
|
|
|
|
|---|
| Clinicopathological
features | n | Mean ± SD | t | P-value |
|---|
| Tissue |
|
| −22.926 | <0.01 |
|
HCC | 371 | 6.4189±1.2376 |
|
|
| Normal
tissue | 49 | 8.5117±0.4539 |
|
|
| Age (years) |
|
| −0.059 | 0.953 |
|
≥60 | 200 | 6.5022±1.4621 |
|
|
|
<60 | 170 | 6.5107±1.3107 |
|
|
| Sex |
|
| 1.196 | 0.233 |
|
Female | 119 | 6.4136±1.4532 |
|
|
|
Male | 251 | 6.5982±1.2365 |
|
|
| Race |
|
| F=0.662 | 0.576 |
|
White | 181 | 6.5850±1.4719 |
|
|
| Black
or African American | 17 | 6.6571±0.9780 |
|
|
|
Asian | 161 | 6.4148±1.2035 |
|
|
|
American Indian or Alaska
native | 2 | 5.9092±2.1116 |
|
|
| Cancer status |
|
| −1.605 | 0.109 |
| Tumor
free | 202 | 6.6343±1.3739 |
|
|
| With
tumor | 150 | 6.3997±1.3318 |
|
|
| Grade |
|
| F=8.305 | <0.001 |
| G1 | 55 | 7.1548±1.1886 |
|
|
| G2 | 173 | 6.6150±1.3285 |
|
|
| G3 | 124 | 6.1845±1.2038 |
|
|
| G4 | 13 | 6.1198±1.1988 |
|
|
| Pathological
stage |
|
| F=7.575 | <0.001 |
| Stage
I | 172 | 6.8524±1.1347 |
|
|
| Stage
II | 85 | 6.2293±1.3337 |
|
|
| Stage
III | 85 | 6.1721±1.4300 |
|
|
| Stage
IV | 5 | 6.3922±1.6006 |
|
|
| T stage |
|
| −2.595 | 0.010 |
|
T1-2 | 275 | 6.6139±1.3039 |
|
|
|
T3-4 | 93 | 6.1970±1.4403 |
|
|
| Node
involvement |
|
| 1.506 | 0.133 |
| No | 232 | 6.3513±1.5065 |
|
|
|
Yes | 4 | 7.4924±1.1574 |
|
|
| Metastasis |
|
| −0.554 | 0.580 |
| No | 267 | 6.4695±1.3007 |
|
|
|
Yes | 4 | 6.1048±1.6927 |
|
|
| Survival
status |
|
| −1.756 | 0.080 |
|
Dead | 129 | 6.3757±1.4070 |
|
|
|
Alive | 241 | 6.6261±1.2509 |
|
|
Meta-analysis of miRNA-seq and
microarray data from four public databases
As illustrated in Fig.
2, miR-139-5p downregulation in HCC groups was clearly observed
in the following datasets: GSE12717, GSE21362, GSE36915, GSE39678,
GSE40744, GSE41874, GSE54751, GSE69580, GSE98269, GSE115016 and
TCGA. However, higher miR-139-5p expression levels in HCC tissues
were identified in the E-MTAB-4809 dataset acquired from
ArrayExpress, and no statistically significant difference was
detected in the remaining datasets. In addition, corresponding ROC
curves were constructed and AUC values were calculated (Fig. 3). Moreover, pooled SMDs with 95% CIs
for all included datasets are presented as a forest plot in
Fig. 4A, suggesting reduced
miR-139-5p expression in HCC tissues, as determined by a
random-effects model (SMD=−0.84; 95% CI: −1.36 to −0.32;
P<0.001; I2=85.7%). Additionally, the Begg's funnel
plot of combined datasets revealed clear symmetry, indicating no
evident publication bias in the current analysis of miRNA-seq and
microarray data (Fig. 4B).
Furthermore, the results from the sensitivity analysis (Fig. 4C) demonstrated no indication of any
individual study significantly affecting the final SMD and its 95%
CI.
Determination of the diagnostic
efficacy of miR-139-5p expression in HCC
As shown in Fig. 5,
the combined diagnostic OR was 14.39 (CI: 5.96–34.75) and a slight
heterogeneity was indicated by the I2, which was found
to be 56.3%. Furthermore, the sensitivity and specificity of the
included datasets were calculated, and the corresponding results
were 0.87 (0.85–0.90) and 0.66 (0.60–0.72), respectively. Moreover,
the pooled PLR and NLR values were 2.50 (CI: 1.53–4.07) and 0.23
(CI: 0.16–0.33), respectively. The AUC result of the sROC curve was
0.8978, indicating that miR-139-5p low-expression has a relatively
high diagnostic efficacy to discriminatee HCC from non-tumor liver
tissue based on the miRNA-seq and microarray data.
Clinical role of miR-139-5p expression
in HCC tissue samples determined from scientific publications
Four publications (25,28,29,54)
offered correlative prognostic information to verify the clinical
role of miR-139-5p in HCC. HRs for OS were combined with 95% CIs
(HR=1.37; 95% CI: 1.07–1.76; P=0.001) and calculated using a
fixed-effects model due to a lack of notable heterogeneity
(I2=43.9%; P=0.148). The results indicated that
decreased miR-139-5p expression was negatively associated with
patient prognosis, as shown in Table
IV and Fig. 6A. Furthermore,
Begg's funnel test was performed to evaluate publication bias in
the four included studies (Fig. 6B).
No marked bias was observed based on the symmetry of the funnel
plot. Moreover, as shown in Fig. 6C,
it was concluded that individual studies did not affect the final
outcomes. Only two studies (25,29)
provided correlations among clinicopathological parameters and
miR-139-5p expression in HCC tissues, while other studies reported
on DFS (28) and RFS (29) rates (DFS: HR=1.67, 95% CI: 0.57–4.87;
RFS: HR=1.20, 95% CI: 0.61–2.37).
 | Table IV.Merged HRs and corresponding 95% CIs
for OS in patients with HCC obtained using a meta-analysis based on
fixed-effects model. |
Table IV.
Merged HRs and corresponding 95% CIs
for OS in patients with HCC obtained using a meta-analysis based on
fixed-effects model.
| Study | Year | HR | LL | UL | p | I2
(%) | P-value | (Refs.) |
|---|
| Li et
al | 2014 | 1.56 | 0.30 | 8.07 |
|
|
| (54) |
| Wang et
al | 2016 | 2.68 | 1.42 | 5.05 |
|
|
| (25) |
| Wong et
al | 2011 | 1.75 | 0.36 | 8.48 |
|
|
| (29) |
| Wang et
al | 2015 | 1.20 | 0.91 | 1.58 |
|
|
| (28) |
| Pooled |
| 1.37 | 1.07 | 1.76 | 0.001 | 43.9 | 0.148 |
|
Bioinformatics analysis
Target genes of miR-139-5p and their
clinical roles
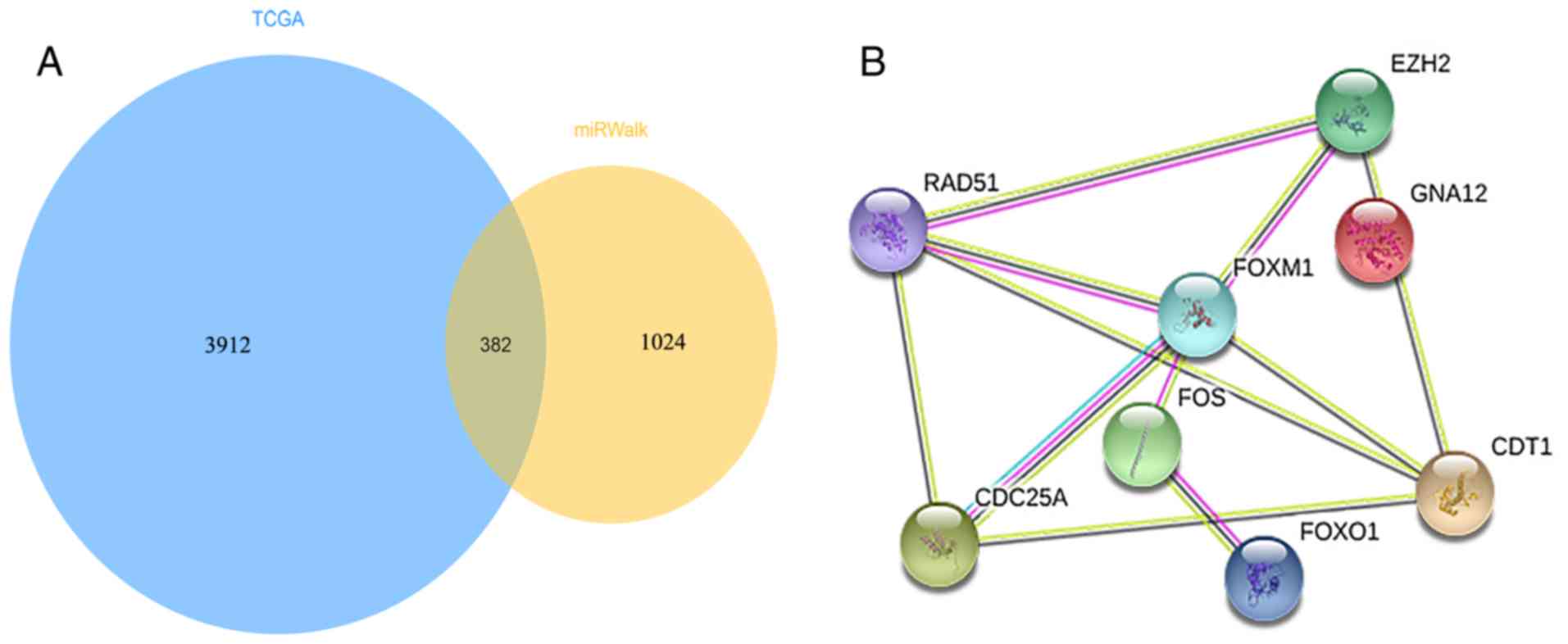
A total of 4,294 differentially expressed genes
obtained from TCGA, 1,406 targeted genes predicted by the miRWalk
website, and a total of 382 intersecting genes were downloaded as
final target genes (Fig. 7A).
Furthermore, DAVID was employed to conduct a GO functional
enrichment analysis, in addition to KEGG and Panther pathway
analyses for the purpose of identifying specific molecular
mechanisms of miR-139-5p related to HCC progression and prognosis.
As shown in Fig. 8 and Table V, the genes were significantly
enriched in ‘response to drug’, ‘signal transduction’, ‘cell
migration involved in sprouting angiogenesis’ and ‘positive
regulation of cell proliferation’ (P<0.01), based on the
biological process of GO. Regarding the cellular component of GO,
the genes were specifically focused on ‘plasma membrane’, ‘integral
component of plasma membrane’ and ‘neuron projection’ (P<0.01).
Concerning the molecular function of GO, the functional genes were
primarily assembled in ‘transcription factor activity,
sequence-specific DNA binding’ and ‘Ras guanyl-nucleotide exchange
factor activity’ (P<0.01). Moreover, as presented in Table VI, KEGG and Panther pathway analyses
were performed on several signaling pathways, including ‘pathways
in cancer’, ‘Ras signaling pathway’ and ‘endogenous cannabinoid
signaling’ (P<0.05).
 | Table V.Enrichment of downstream target genes
regulated by miR-139-5p in Gene Ontology terms in the categories of
biological processes, cellular components and molecular
functionsa. |
Table V.
Enrichment of downstream target genes
regulated by miR-139-5p in Gene Ontology terms in the categories of
biological processes, cellular components and molecular
functionsa.
| Term | Counts | P-value |
|---|
| Biological
process |
|
|
|
Response to drug | 16 |
1.19×10−03 |
| Signal
transduction | 39 |
1.72×10−03 |
| Cell
migration involved in sprouting angiogenesis | 4 |
2.97×10−03 |
|
Positive regulation of cell
proliferation | 19 |
5.81×10−03 |
|
Positive regulation of neuron
projection development | 7 |
8.64×10−03 |
|
Positive regulation of GTPase
activity | 21 |
9.52×10−03 |
|
Regulation of insulin
secretion | 6 |
1.07×10−02 |
|
Response to insulin | 6 |
1.07×10−02 |
|
Activation of protein kinase
activity | 5 |
1.21×10−02 |
|
Positive regulation of
transcription, DNA-templated | 19 |
1.52×10−02 |
| Cellular
component |
|
|
| Plasma
membrane | 111 |
1.30×10−04 |
|
Integral component of plasma
membrane | 48 |
2.12×10−04 |
| Neuron
projection | 15 |
2.35×10−04 |
|
Integral component of
membrane | 125 |
3.30×10−03 |
|
Neuronal cell body | 14 |
9.02×10−03 |
|
External side of plasma
membrane | 11 |
9.07×10−03 |
|
Postsynaptic membrane | 10 |
2.27×10−02 |
|
Anchored component of
membrane | 7 |
2.30×10−02 |
| Z
disc | 7 |
2.77×10−02 |
|
Terminal bouton | 5 |
3.24×10−02 |
| Molecular
function |
|
|
|
Transcription factor activity,
sequence-specific DNA binding | 36 |
4.37×10−04 |
| Ras
guanyl-nucleotide exchange factor activity | 8 |
8.28×10−03 |
| Steroid
binding | 4 |
1.62×10−02 |
| mRNA
3′-UTR binding | 5 |
1.63×10−02 |
|
Sequence-specific DNA
binding | 19 |
1.65×10−02 |
|
Transcriptional activator
activity, RNA polymerase II core promoter proximal region
sequence-specific binding | 11 |
2.05×10−02 |
| Kinase
activity | 11 |
2.33×10−02 |
|
Pyridoxal phosphate
binding | 5 |
2.69×10−02 |
| DNA
binding | 45 |
3.55×10−02 |
|
Calmodulin binding | 9 |
3.62×10−02 |
 | Table VI.Downstream predicted genes of
miR-139-5p are principally concentrated in five KEGG pathways and
five Panther pathwaysa. |
Table VI.
Downstream predicted genes of
miR-139-5p are principally concentrated in five KEGG pathways and
five Panther pathwaysa.
| Term | P-value | Counts | Genes |
|---|
| hsa05200:Pathways
in cancer |
6.01×10−04 | 20 | CKS1B, AR, RET,
E2F3, PDGFB, GNA12, FOXO1, KITLG, FGF13, FGF12, CTNNA3, RAD51,
TPM3, EDNRB, FOS, WNT7B, PAX8, TGFA, GNG4, PLCB1 |
| hsa04720:Long-term
potentiation |
1.27×10−02 | 6 | GRM5, RPS6KA6,
GRIN2B, CAMK4, RAPGEF3, PLCB1 |
| hsa04014:Ras
signaling pathway |
2.16×10−02 | 11 | LAT, GAB2, GRIN2B,
RASGRF2, PDGFB, EFNA3, KITLG, FGF13, FGF12, GNG4, ABL2 |
|
hsa04080:Neuroactive ligand-receptor
interaction |
3.31×10−02 | 12 | GRM5, EDNRB,
GABRG3, PTGIR, ADORA3, GRIN2B, P2RY4, CNR1, ADRA1A, CHRNB2, HTR1D,
GABRQ |
| hsa04015:Rap1
signaling pathway |
3.42×10−02 | 10 | LAT, GRIN2B, PDGFB,
CNR1, EFNA3, KITLG, FGF13, FGF12, RAPGEF3, PLCB1 |
|
P00026:Heterotrimeric G-protein signaling
pathway-Gi alpha and Gs alpha-mediated pathway |
2.26×10−02 | 11 | GRM5, GNAL, RET,
ADORA3, FGB, RGS4, RGS5, GPSM2, ADRA1A, HTR1D, PYGB |
| P05730:Endogenous
cannabinoid signaling |
3.57×10−02 | 4 | GRM5, CNR1, PLCB1,
GNG4 |
|
P00027:Heterotrimeric G-protein signaling
pathway-Gq alpha and Go alpha-mediated pathway |
5.59×10−02 | 9 | GRM5, ADORA3, RGS4,
RGS5, GPSM2, RHOB, PLCB1, GNG4, RHOF |
| P05911:Angiotensin
II-stimulated signaling through G proteins and beta-arrestin |
5.81×10−02 | 5 | EGR1, RHOB, PLCB1,
GNG4, RHOF |
| P00008:Axon
guidance mediated by Slit/Robo |
6.51×10−02 | 4 | ROBO1, RHOB, ABL2,
RHOF |
Construction of target gene networks
and identification of hub genes
The STRING database was employed in order to predict
382 intersected genes that were associated with miR-139-5p
expression by constructing a PPI network. As presented in Fig. 7B, the top eight genes, namely, FBJ
murine osteosarcoma viral oncogene homolog (FOS), chromatin
licensing and DNA replication factor 1 (CDT1), enhancer of zeste
homolog 2 (EZH2), recombination protein (RAD51), forkhead box M1
(FOXM1), forkhead box O1 (FOXO1), cell division cycle 25A (CDC25A),
and guanine nucleotide binding protein alpha 12 (GNA12) with
connection degrees >14 were chosen from the PPI network for
further investigation. Moreover, four genes, FOS, FOXO1, GNA12 and
RAD51, identified in the first signaling pathway termed ‘pathways
in cancer’ were eventually determined as hub genes.
Target gene expression and associated
clinical implications
An analysis was conducted using GEPIA to determine
the top eight genes associated with HCC development and verify
their specific clinical implications. The expression levels of four
genes, CDT1, EZH2, FOXM1 and RAD51, were substantially increased in
HCC tissues, whereas decreased FOS gene expression was observed in
HCC tissues (P<0.05; Fig. 9).
Moreover, the prognostic value of the top eight genes was also
evaluated (Fig. 10). The expression
of six genes, CDT1, EZH2, RAD51, FOXM1, GNA12 and CDC25A, was
negatively associated with the OS of patients with HCC. In
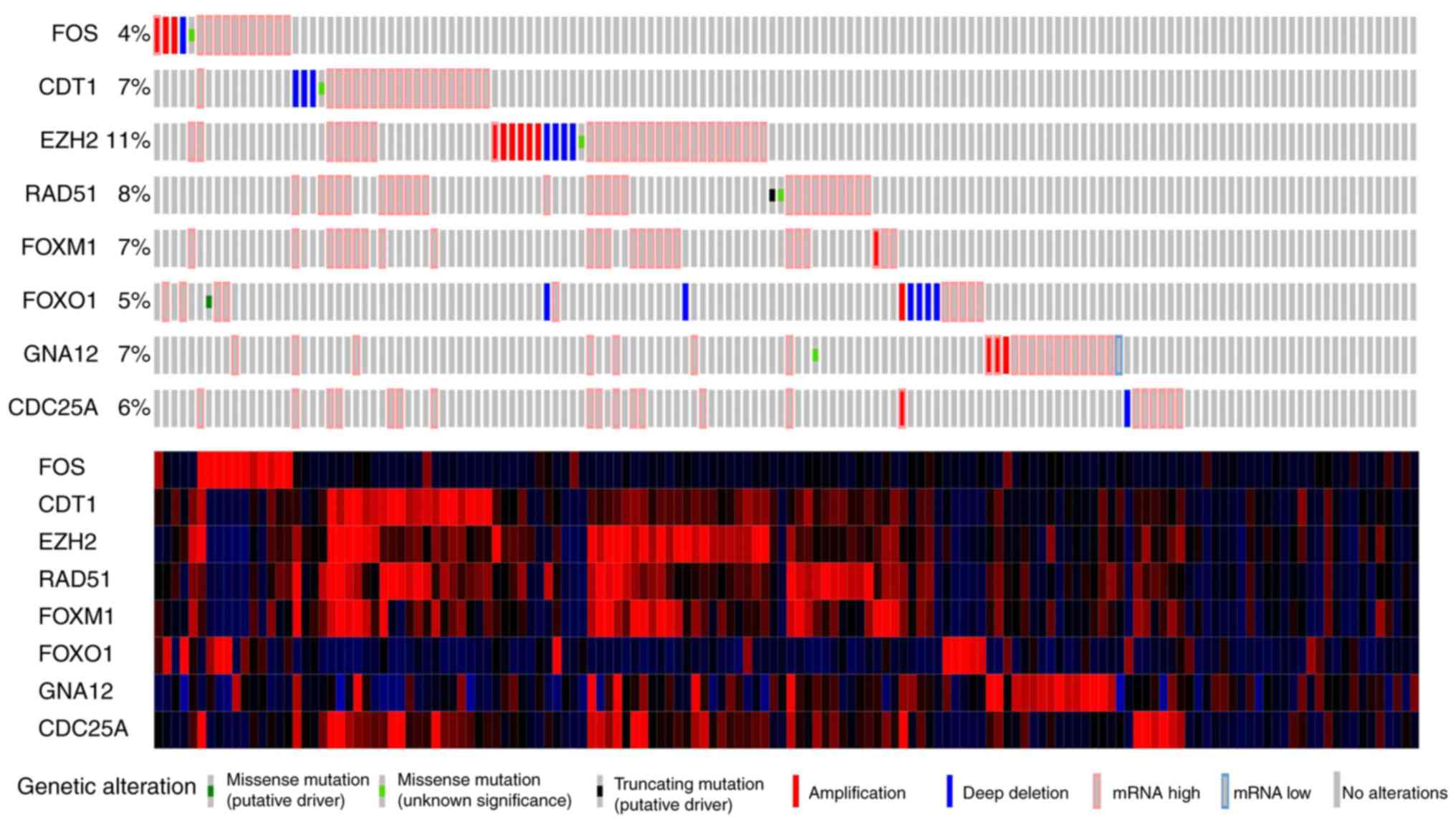
addition, the frequency of mutations in the top eight genes
(Fig. 11), and the expression
patterns of the four hub genes FOS, FOXO1, GNA12 and RAD51 in HCC
and normal liver tissues were explored using the Human Protein
Atlas database (Fig. 12).
Furthermore, based on TCGA data, a positive correlation was
identified between miR-139-5p expression in HCC and the regulation
of FOS and FOXO1 genes, which were also reduced in HCC tissues
(Fig. 13). Consistent with the
aforementioned findings, a negative correlation was detected
between miR-139-5p expression and the regulation of GNA12 and RAD51
in patients with HCC (P<0.001).
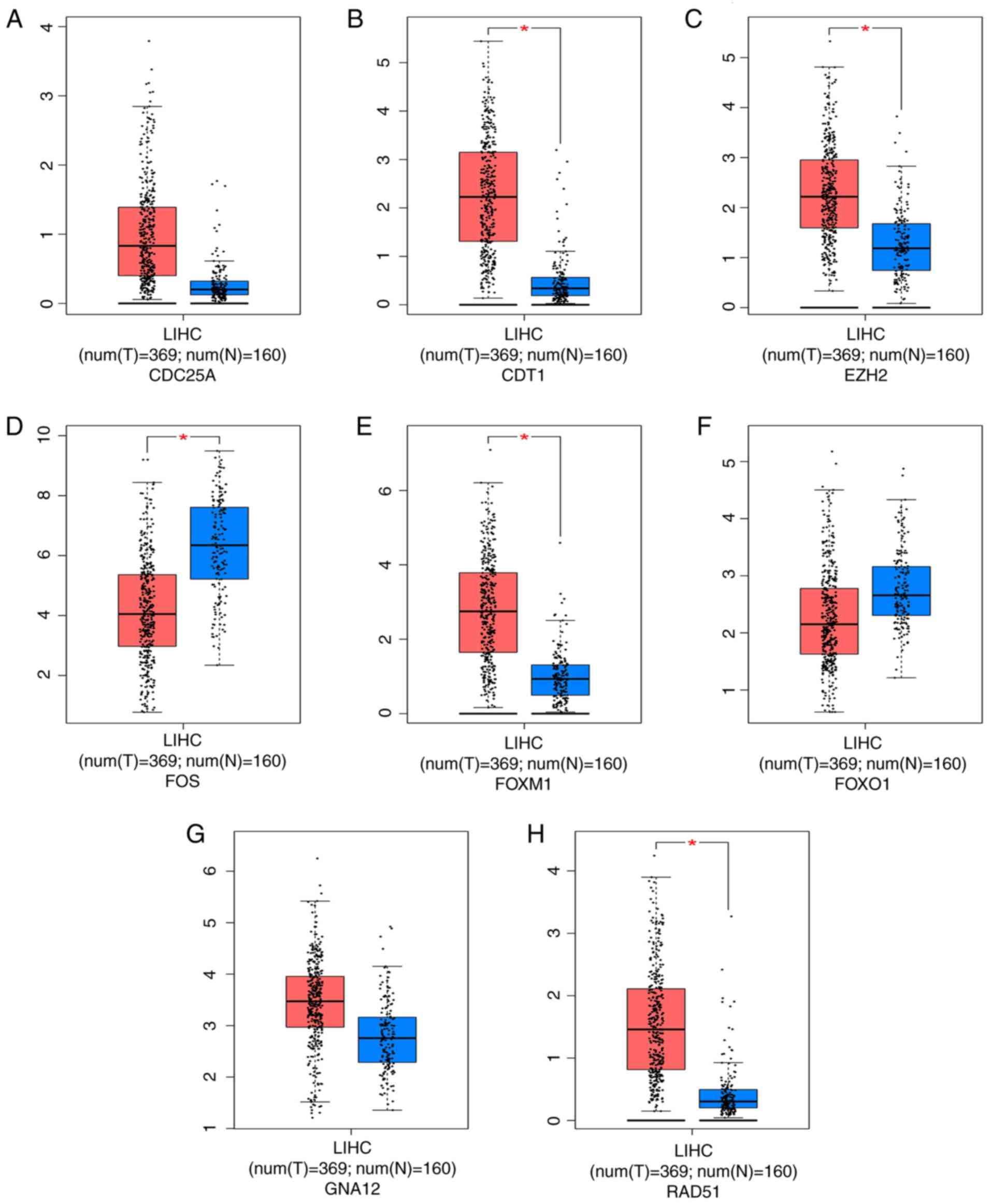 | Figure 9.Expression of the top eight predicted
target genes in HCC and nontumor liver tissues from TCGA. (A)
CDC25A; (B) CDT1; (C) EZH2; (D) FOS; (E) FOXM1; (F) FOXO1; (G)
GNA12; (H) RAD51. Significant upregulation of CDT1, EZH2, FOXM1 and
RAD51 expression and downregulation of FOS in HCC tissues was
observed (*P<0.05). No predominant differences were detected
among the remaining genes, with P>0.05. LIHC, Liver
Hepatocellular Carcinoma; HCC, hepatocellular carcinoma; TCGA, The
Cancer Genome Atlas; CDC25A, cell division cycle 25A; CDT1,
chromatin licensing and DNA replication factor 1; EZH2, enhancer of
zeste homolog 2; FOS, FBJ murine osteosarcoma viral oncogene
homolog; FOXM1, forkhead box M1; FOXO1, forkhead box O1; GNA12,
guanine nucleotide binding protein alpha 12; RAD51, recombination
protein. |
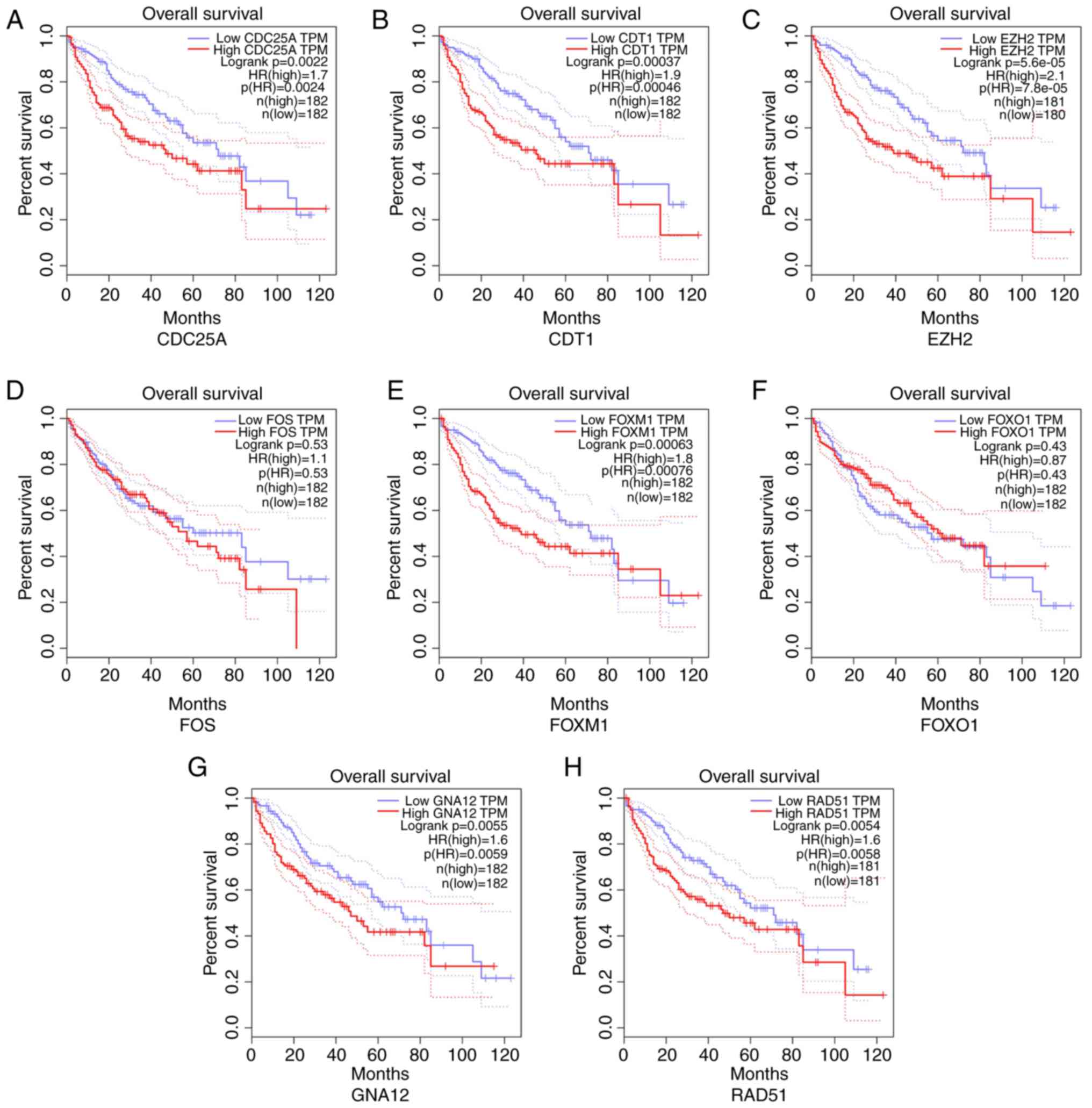 | Figure 10.Kaplan-Meier curves displaying the
associations between the expression of the top target genes and OS
of patients with HCC. (A) CDC25A; (B) CDT1; (C) EZH2; (D) FOS; (E)
FOXM1; (F) FOXO1; (G) GNA12; (H) RAD51. These results indicate that
the high expression of CDT1, EZH2, RAD51, FOXM1, GNA12 and CDC25A
had a significant effect on the OS of patients with HCC (P<0.05)
while no marked effect was detected in the expression of other
genes in HCC (P>0.05). HCC, hepatocellular carcinoma; OS,
overall survival; CDC25A, cell division cycle 25A; CDT1, chromatin
licensing and DNA replication factor 1; EZH2, enhancer of zeste
homolog 2; FOS, FBJ murine osteosarcoma viral oncogene homolog;
FOXM1, forkhead box M1; FOXO1, forkhead box O1; GNA12, guanine
nucleotide binding protein alpha 12; RAD51, recombination protein;
TPM, transcripts per million. |
Discussion
Based on the miRNA-seq and microarray data retrieved
from four public databases and data obtained from published
literature (25,28,29,54),
including human HCC specimens, the primary aim of this study was to
perform additional bioinformatics analyses examining miR-139-5p and
the corresponding target gene expression and molecular mechanism
involved in HCC development. Based on the analytical miRNA-seq and
microarray data, reduced miR-139-5p expression levels were observed
in the HCC group (SMD=−0.84; 95% CI: −1.36 to −0.32; P<0.001).
Based on the data retrieved from the TCGA database, certain
clinicopathological features, including grade, pathological and T
stage, were associated with miR-139-5p expression, indicating that
decreased miR-139-5p expression may facilitate disease progression.
Moreover, the AUC value of the sROC curve for data obtained from
public databases was 0.8978, indicating a relatively high
diagnostic efficacy of miR-139-5p expression. Regarding the
association between clinical characteristics and miR-139-5p
expressed in HCC based on relevant literature, the pooled OS
(HR=1.37; 95% CI: 1.07–1.76) revealed that decreased miR-139-5p
expression led to a poor prognosis in patients with HCC.
Furthermore, bioinformatics software was used to identify 382 genes
regulated by miR-139-5p in HCC, for which GO, KEGG and Panther
pathway analyses were performed. The target genes were
significantly enriched in ‘response to drug’, ‘signal
transduction’, ‘plasma membrane’, ‘transcription factor activity,
sequence-specific DNA binding’, ‘pathways in cancer’, ‘Ras
signaling pathway’ and ‘endogenous cannabinoid signaling’
(P<0.05). In addition, the top eight target genes, FOS, CDT1,
EZH2, RAD51, FOXM1, FOXO1, GNA12 and CDC25A, were selected by PPI
network construction; and their clinical impacts were investigated.
Moreover, FOS, FOXO1, GNA12 and RAD51, which were enriched in
‘pathways in cancer’, were determined as hub genes. The IHC
staining for proteins encoded by these four genes, namely, FOS,
FOXO1, GNA12 and RAD51, was further confirmed using the Human
Protein Atlas database, and the expression atlas of the
pathological sections supported the downregulation of FOS and FOXO1
accompanied by the upregulation of GNA12 and RAD51 in HCC tissues.
Furthermore, a relativity analysis revealed that miR-139-5p
expression was positively correlated with the regulation of FOS and
FOXO1 and negatively correlated with GNA12 and RAD51 regulation,
suggesting that miR-139-5p may mediate the expression of hub genes
involved in the aforementioned signaling pathways, mainly in
‘pathways in cancer’.
Mature miRNAs, which associate with the RNA-induced
silencing complex (RISC), are capable of promoting the interaction
of RISC with the 3′-UTR of the downstream target mRNA, resulting in
abnormal translation of downstream mRNA. In addition, miR-139-5p is
regarded as a tumor suppressor due to its regulatory effects on
multiple malignant tumors. Notably, miR-139-5p controls tumor
progression by regulating target mRNA expression and participates
in downstream signaling pathways in malignancies (55–59),
including HCC. According to a report by Wong et al (29), the progression and invasion of HCC
are inhibited by miR-139-5p via its effects on Rho-kinase 2
expression. In addition, Au et al (60) revealed that miR-139-5p greatly
affects the metastasis-related pathways of HCC cells by increasing
the expression of EZH2, which is involved in sustaining the
transcriptional suppression of genes over sequential cell
generations. The increased expression of EZH2 contributed to
metastasis, and poor prognosis of HCC identified in our analysis
(Figs. 9 and 10). As reported by Hua et al
(24), miR-139-5p regulates aerobic
glycolysis, cell proliferation and motility by interacting with
ETS1 in HCC cells. Moreover, a meta-analysis based on published
articles that explored miR-139-5p expression in digestive system
tumors, including one HCC case that was analyzed prospectively,
indicated an unfavorable effect on the life expectancy of patients
with gastrointestinal tumors (27).
Additionally, downregulation of miR-139-5p has been demonstrated to
result in poor outcome and disease progression (25,26,28).
This finding was confirmed in the present study as indicated by the
merged results with an SMD of −0.84 and HR for OS of 1.37 (95% CI:
1.07–1.76).
To further investigate the oncogenic molecular
mechanism of miR-139-5p expression, bioinformatics analysis was
performed with biological software and the top eight target genes
were ultimately evaluated. Regarding the GO enrichment analysis,
target genes were notably enriched in ‘response to drug’ and
‘positive regulation of cell proliferation’. Certain studies have
reported that miR-139-5p expression has a major impact on pesticide
effects in cancer therapy (58,61).
Yoon et al (62) selected
certain serum miRNAs, including miR-139-5p, associated with
positive radiological responses or improved survival to assess the
efficacy of sorafenib in patients with HCC. However, no
statistically significant difference was reported between
miR-139-5p expression and sorafenib treatment in patients with HCC.
Additional correlative studies focused on drug efficacy and
miR-139-5p expression should be further developed.
In terms of the pathway analysis, ‘pathways in
cancer’, ‘Ras signaling pathway’ and ‘endogenous cannabinoid
signaling’ were strongly enriched by predictive target genes.
Importantly, miR-139-5p regulation serves an important role in
tumor development and progression (63). Results from the present study
revealed an association of miR-139-5p expression with progression
and poor outcome in patients with HCC. Mitogenic signaling cascades
associated with proliferation and translation are facilitated by
activated Ras (63), and Ras
regulates programmed cell death (64). Thus, an association exists between
Ras alterations and carcinogenesis in various malignancies
(65–67). Moreover, miRNAs have the ability to
initiate the Ras-mitogen activated protein kinase pathway related
to cell proliferation and survival, while no publication has
reported an association between the Ras signaling pathway and
miR-139 expression. Endogenous cannabinoid signaling, which is
mainly determined by the activity of endocannabinoids
N-arachidonoylethanolamine and 2-arachidonoylglycerol, as well as
cannabinoid receptor type receptors 1 and 2 has attracted
increasing interest due to its antitumor effects (68,69).
According to Vago et al (70), the suppression of
N-acylethanolamine-hydrolyzing acid amidase activity regulates
tumor cell death and migration, and controls the progression of
bladder cancer, indicating a new therapeutic target for patients
with bladder cancer. Furthermore, Martínez-Martínez et al
(71) verified that activated CB2
accelerates the development of colon cancer via a mechanism
dependent on the AKT/GSK3β signaling pathway. Nevertheless, a
relationship between endogenous cannabinoid signaling and miR-139
regulation has not been experimentally confirmed, therefore further
research is required.
To better understand the downstream mechanisms of
the top eight target genes, biofunctional investigations of FOS,
CDT1, EZH2, RAD51, FOXM1, FOXO1, GNA12 and CDC25A were conducted.
CDT1, which is involved in the formation of the prereplication
complex required for DNA fragment replication, functions as an
oncogene in multiple tumor types, including HCC (72–75).
Karavias et al (75)
confirmed that increased CDT1 expression has a negative influence
on the survival of patients with HCC and observed correlations
between the upregulation of CDT1 and tumor grade and TNM stage by
performing IHC staining in HCC tissues, supporting the results of
the present study (Figs. 9 and
11). Moreover, Yu et al
(76) discovered a combination of
new diagnostic indicators, including CDT1, MCM7, NUDT1, CENPM and
HDAC11, with favorable diagnostic efficiency in HCC. Accordingly,
CDT1 expression is crucial for the development and prognostic
evaluation of HCC, thus further research is required to verify the
association between miR-139-5p and CDT1 expression in HCC. RAD51
recombinase (RAD51), which participates in reestablishing and
repairing homologous DNA, binds to the BRCA1 and BRCA2 genes, which
are involved in tumorigenesis (77).
Previous studies have reported that certain pharmaceutical
molecules, including corylin, melatonin and gefitinib in
combination with irinotecan, were able to enhance cancer cell
sensitivity to chemotherapy by inhibiting RAD51-induced DNA repair
in HCC (78–80). Luo et al (81) observed increased expression of RAD51
in HCC tissues, consistent with the findings of the present study,
and revealed that miRNA-146a-5p enhanced the radio-sensitivity of
HCC cells via the DNA repair pathway. However, studies have not yet
reported an association between miR-139-5p and RAD51 regulation in
patients with HCC. FOXM1 and FOXO1, members of the forkhead family,
serve vital roles in cell proliferation and gene transcription.
According to certain studies (82–85),
FOXM1 overexpression results in poor survival and undesirable
development of HCC, consistent with the present study results
(Figs. 9 and 10). Lin et al (86) confirmed the reduced expression of
FOXO1 in HCC tissues using IHC, and reported that FOXO1 served an
important role in the development of HCC by regulating miRNA
expression (87–89). CDC25A, as a member of the CDC25
family of phosphatases, mediates the G1 to S phase transition. A
strong correlation between CDC25 expression and poor prognosis of
patients with HCC was demonstrated by Xu et al (90) and a relevant study reported that the
suppression of CDC25 expression halts the proliferation and
progression of HCC (91). GNA12 has
been found to possess carcinogenic potential and accelerate the
progression of certain tumors (92–95). No
studies have focused on the relevancy between miR-139-5p expression
and FOXM1, FOXO1, GNA12 and CDC25A in HCC. The present study may
provide novel insights for subsequent studies. FOS has multiple
functions in cell proliferation, differentiation, transformation
and apoptosis. Moreover, an association between FOS expression and
HCC development has been reported (96–98). Fan
et al (98) indicated that
decreased miRNA-139 expression potentially facilitates the
progression and metastasis of HCC via inhibiting FOS expression.
Further research is necessary.
In short, it should be noted that miR-139-5p
expression has a strong impact on the progression and prognosis of
HCC mainly via targeted regulation of EZH2 and FOS expression.
Moreover, the results of the present study clearly demonstrated
that the regulation of FOS and FOXO1 was positively associated with
miR-139-5p expressed in HCC. However, GNA12 and RAD51 expression
levels were negatively associated with miR-139-5p expression based
on the TCGA data. However, no studies have yet demonstrated the
involvement of miR-139-5p in HCC development via targeted
modulation of CDT1, RAD51, FOXM1, FOXO1, GNA12 and CDC25A
expression.
The present study had certain limitations. Firstly,
only four publications described a relationship among miR-139-5p
regulation and prognostic effect size, including OS, DFS and RFS in
HCC. Secondly, the HRs and corresponding 95% CIs were directly
obtained from Kaplan-Meier curves, which generated biased outcomes
due to limited data. Thirdly, the included miRNA-seq and microarray
data were obtained from diverse platforms and channels, suggesting
clear heterogeneity and poor diagnostic efficiency. Accordingly, a
uniform method for detecting miR-139-5p expression in HCC should be
formulated. Moreover, evidence for signaling pathways that are
regulated by miR-139-5p via the targeted modulation of hub gene
expression is lacking, and further in vitro and in
vivo studies are required to determine the precise biological
effects of the target genes on HCC development.
In summary, the findings of the present study
revealed that miR-139-5p expression was lower in HCC tissues than
in nontumorous tissues and strong associations were observed for
miR-139-5p expression with tumor grade, pathological stage and T
stage. Additionally, reduced miR-139-5p expression generally led to
an undesirable prognosis in patients with HCC. Moreover, the top
eight target genes, namely FOS, CDT1, EZH2, RAD51, FOXM1, FOXO1,
GNA12 and CDC25A induced by miR-139-5p expression were revealed to
be involved in several pathways, including ‘response to drug’,
‘pathways in cancer’, ‘Ras signaling pathway’ and ‘endogenous
cannabinoid signaling’. These genes may have a major impact on
carcinogenesis and the development of HCC. The current study
provided a comprehensive investigation of the role of miR-139-5p in
HCC progression. Nevertheless, additional research of the
underlying molecular mechanisms related to HCC, including in
vitro and in vivo experiments, should be further
conducted to confirm the outcomes of the present study.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Guangxi
Science and Technology Program (grant no. GuiKeAB17195020) and the
National Natural Science Foundation of China (grant no.
NSFC81860319).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HQ, BML, XDW, DYW, HY and YH made substantial
contributions to the conception and design of the present study.
HQ, QQ, YTP and CYZ assisted in the acquisition, analysis and
interpretation of data. HQ, DYW, BML, YH and HY were involved in
drafting the manuscript and BML, YH and HY critically revising it
for important intellectual content. All authors gave final approval
for the version of the manuscript to be published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F and Soerjomataram I: The changing
global burden of cancer: Transitions in human development and
implications for cancer prevention and control. View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yeh SH and Chen PJ: Gender disparity of
hepatocellular carcinoma: The roles of sex hormones. Oncology. (78
Suppl 1):S172–S179. 2010. View Article : Google Scholar
|
|
5
|
Aghemo A: Update on HCC management and
review of the new EASL guidelines. Gastroenterol Hepatol (NY).
14:384–386. 2018.
|
|
6
|
Singal AG and El-Serag HB: Hepatocellular
carcinoma from epidemiology to prevention: Translating knowledge
into practice. Clin Gastroenterol Hepatol. 13:2140–2151. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Otedo A, Simbiri KO, Were V, Ongati O and
Estambale BA: Risk factors for liver Cancer in HIV endemic areas of
Western Kenya. Infect Agent Cancer. 13:412018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oweira H, Petrausch U, Helbling D, Schmidt
J, Mehrabi A, Schöb O, Giryes A and Abdel-Rahman O: Prognostic
value of site-specific extra-hepatic disease in hepatocellular
carcinoma: A SEER database analysis. Expert Rev Gastroenterol
Hepatol. 11:695–701. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Intaraprasong P, Siramolpiwat S and
Vilaichone RK: Advances in management of hepatocellular carcinoma.
Asian Pac J Cancer Prev. 17:3697–3703. 2016.PubMed/NCBI
|
|
10
|
Valinezhad Orang A, Safaralizadeh R and
Kazemzadeh-Bavili M: Mechanisms of miRNA-mediated gene regulation
from common downregulation to mRNA-specific upregulation. Int J
Genomics. 2014:9706072014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fanini F and Fabbri M: MicroRNAs and
cancer resistance: A new molecular plot. Clin Pharmacol Ther.
99:485–493. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Manikandan J, Aarthi JJ, Kumar SD and
Pushparaj PN: Oncomirs: The potential role of non-coding microRNAs
in understanding cancer. Bioinformation. 2:330–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Beca F and Schmitt F: MicroRNA signatures
in cytopathology: Are they ready for prime time? Cancer Cytopathol.
124:613–615. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang J, Chen J and Sen S: MicroRNA as
biomarkers and diagnostics. J Cell Physiol. 231:25–30. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Keller A, Rounge T, Backes C, Ludwig N,
Gislefoss R, Leidinger P, Langseth H and Meese E: Sources to
variability in circulating human miRNA signatures. RNA Biol.
14:1791–1798. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Z, Yu T, Cabay RJ, Jin Y, Mahjabeen
I, Luan X, Huang L, Dai Y and Zhou X: miR-486-3p, miR-139-5p, and
miR-21 as biomarkers for the detection of oral tongue squamous cell
carcinoma. Biomark Cancer. 9:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zou F, Mao R, Yang L, Lin S, Lei K, Zheng
Y, Ding Y, Zhang P, Cai G, Liang X and Liu J: Targeted deletion of
miR-139-5p activates MAPK, NF-κB and STAT3 signaling and promotes
intestinal inflammation and colorectal cancer. FEBS J.
283:1438–1452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S,
Li M, You Q and Huang Z: miR-139-5p sensitizes colorectal cancer
cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract.
212:643–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu Y, Deng C, Zhang H, Zhang J, Peng B and
Hu C: Long non-coding RNA XIST promotes cell growth and metastasis
through regulating miR-139-5p mediated Wnt/β-catenin signaling
pathway in bladder cancer. Oncotarget. 8:94554–94568. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu J, Li C, Jiang Y, Wan Y, Zhou S and
Cheng W: Tumor-suppressor role of miR-139-5p in endometrial cancer.
Cancer Cell Int. 18:512018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hua S, Lei L, Deng L, Weng X, Liu C, Qi X,
Wang S, Zhang D, Zou X, Cao C, et al: miR-139-5p inhibits aerobic
glycolysis, cell proliferation, migration, and invasion in
hepatocellular carcinoma via a reciprocal regulatory interaction
with ETS1. Oncogene. 37:1624–1636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Ding Q, Li Y, Liu Q, Wu W, Wu L
and Yu H: Reanalysis of microRNA expression profiles identifies
novel biomarkers for hepatocellular carcinoma prognosis. Tumour
Biol. 37:14779–14787. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ni H, Dai X, Leng X, Deng M, Qin Y, Ji Q,
Xu C, Li J and Liu Y: Higher variety and quantity of
microRNA-139-5p isoforms confer suppressive role in hepatocellular
carcinoma. J Cell Biochem. 119:6806–6813. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang YH, Ji J, Weng H, Wang BC and Wang
FB: MiR-139 in digestive system tumor diagnosis and detection:
Bioinformatics and meta-analysis. Clin Chim Acta. 485:33–41. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang L, Liu M, Zhu H, Rong W, Wu F, An S,
Liu F, Feng L, Wu J and Xu N: Identification of recurrence-related
serum microRNAs in hepatocellular carcinoma following hepatectomy.
Cancer Biol Ther. 16:1445–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wong CC, Wong CM, Tung EK, Au SL, Lee JM,
Poon RT, Man K and Ng IO: The microRNA miR-139 suppresses
metastasis and progression of hepatocellular carcinoma by
down-regulating Rho-kinase 2. Gastroenterology. 140:322–331. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
32
|
Parkinson H, Sarkans U, Kolesnikov N,
Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E,
Holloway E, et al: ArrayExpress update-an archive of microarray and
high-throughput sequencing-based functional genomics experiments.
Nucleic Acids Res. 39:D1002–D1004. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tierney JF, Stewart LA, Ghersi D, Burdett
S and Sydes MR: Practical methods for incorporating summary
time-to-event data into meta-analysis. Trials. 8:162007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vamvakas EC: Meta-analyses of studies of
the diagnostic accuracy of laboratory tests: A review of the
concepts and methods. Arch Pathol Lab Med. 122:675–686.
1998.PubMed/NCBI
|
|
36
|
Sticht C, De La Torre C, Parveen A and
Gretz N: miRWalk: An online resource for prediction of microRNA
binding sites. PLoS One. 13:e02062392018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vergoulis T, Vlachos IS, Alexiou P,
Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N,
Dalamagas T and Hatzigeorgiou AG: TarBase 6.0: Capturing the
exponential growth of miRNA targets with experimental support.
Nucleic Acids Res. 40:D222–D229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bandyopadhyay S and Mitra R: TargetMiner:
MicroRNA target prediction with systematic identification of
tissue-specific negative examples. Bioinformatics. 25:2625–2631.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bhattacharya A, Ziebarth JD and Cui Y:
PolymiRTS Database 3.0: Linking polymorphisms in microRNAs and
their target sites with human diseases and biological pathways.
Nucleic Acids Res. 42:D86–D91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Loher P and Rigoutsos I: Interactive
exploration of RNA22 microRNA target predictions. Bioinformatics.
28:3322–3323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC,
Hsu PW, Wong YH, Chen YH, Chen GH and Huang HD: miRNAMap 2.0:
Genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res.
36:D165–D169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
46
|
Liu W and Wang X: Prediction of functional
microRNA targets by integrative modeling of microRNA binding and
target expression data. Genome Biol. 20:182019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Krek A, Grun D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pontén F, Schwenk JM, Asplund A and
Edqvist PH: The human protein atlas as a proteomic resource for
biomarker discovery. J Intern Med. 270:428–446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li T, Yin J, Yuan L, Wang S, Yang L, Du X
and Lu J: Downregulation of microRNA-139 is associated with
hepatocellular carcinoma risk and short-term survival. Oncol Rep.
31:1699–1706. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jiao W, Zhang J, Wei Y, Feng J, Ma M, Zhao
H, Wang L and Jiao W: MiR-139-5p regulates VEGFR and downstream
signaling pathways to inhibit the development of esophageal cancer.
Dig Liver Dis. 51:149–156. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Qin L, Deng HY, Chen SJ, Wei W and Zhang
YT: miR-139 acts as a tumor suppressor in T-cell acute
lymphoblastic leukemia by targeting CX chemokine receptor 4. Am J
Transl Res. 9:4059–4070. 2017.PubMed/NCBI
|
|
57
|
Chen J, Yu Y, Chen X, He Y, Hu Q, Li H,
Han Q, Ren F, Li J, Li C, et al: MiR-139-5p is associated with poor
prognosis and regulates glycolysis by repressing PKM2 in
gallbladder carcinoma. Cell Prolif. 51:e125102018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang HD, Sun DW, Mao L, Zhang J, Jiang
LH, Li J, Wu Y, Ji H, Chen W, Wang J, et al: MiR-139-5p inhibits
the biological function of breast cancer cells by targeting Notch1
and mediates chemosensitivity to docetaxel. Biochem Biophys Res
Commun. 465:702–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ren Y, Zhu H, Chi C, Yang F and Xu X:
MiRNA-139 regulates oral cancer Tca8113 cells apoptosis through Akt
signaling pathway. Int J Clin Exp Pathol. 8:4588–4594.
2015.PubMed/NCBI
|
|
60
|
Au SL, Wong CC, Lee JM, Fan DN, Tsang FH,
Ng IO and Wong CM: Enhancer of zeste homolog 2 epigenetically
silences multiple tumor suppressor microRNAs to promote liver
cancer metastasis. Hepatology. 56:622–631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xu K, Shen K, Liang X, Li Y, Nagao N, Li
J, Liu J and Yin P: MiR-139-5p reverses CD44+/CD133+-associated
multidrug resistance by downregulating NOTCH1 in colorectal
carcinoma cells. Oncotarget. 7:75118–75129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yoon EL, Yeon JE, Ko E, Lee HJ, Je JH, Yoo
YJ, Kang SH, Suh SJ, Kim JH, Seo YS, et al: An explorative analysis
for the role of serum miR-10b-3p levels in predicting response to
sorafenib in patients with advanced hepatocellular carcinoma. J
Korean Med Sci. 32:212–220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huang LL, Huang LW, Wang L, Tong BD, Wei Q
and Ding XS: Potential role of miR-139-5p in cancer diagnosis,
prognosis and therapy. Oncol Lett. 14:1215–1222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cox AD and Der CJ: The dark side of Ras:
Regulation of apoptosis. Oncogene. 22:8999–9006. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bos JL: Ras oncogenes in human cancer: A
review. Cancer Res. 49:4682–4689. 1989.PubMed/NCBI
|
|
66
|
Bos JL: The ras gene family and human
carcinogenesis. Mutat Res. 195:255–271. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Prior IA, Lewis PD and Mattos C: A
comprehensive survey of Ras mutations in cancer. Cancer Res.
72:2457–2467. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Schwarz R, Ramer R and Hinz B: Targeting
the endocannabinoid system as a potential anticancer approach. Drug
Metab Rev. 50:26–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lu Y and Anderson HD: Cannabinoid
signaling in health and disease. Can J Physiol Pharmacol.
95:311–327. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Vago R, Bettiga A, Salonia A, Ciuffreda P
and Ottria R: Development of new inhibitors for
N-acylethanolamine-hydrolyzing acid amidase as promising tool
against bladder cancer. Bioorg Med Chem. 25:1242–1249. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Martínez-Martínez E, Martín-Ruiz A, Martín
P, Calvo V, Provencio M and García JM: CB2 cannabinoid receptor
activation promotes colon cancer progression via AKT/GSK3β
signaling pathway. Oncotarget. 7:68781–68791. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liontos M, Koutsami M, Sideridou M,
Evangelou K, Kletsas D, Levy B, Kotsinas A, Nahum O, Zoumpourlis V,
Kouloukoussa M, et al: Deregulated overexpression of hCdt1 and
hCdc6 promotes malignant behavior. Cancer Res. 67:10899–10909.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bravou V, Nishitani H, Song SY, Taraviras
S and Varakis J: Expression of the licensing factors, Cdt1 and
Geminin, in human colon cancer. Int J Oncol. 27:1511–1518.
2005.PubMed/NCBI
|
|
74
|
Arentson E, Faloon P, Seo J, Moon E,
Studts JM, Fremont DH and Choi K: Oncogenic potential of the DNA
replication licensing protein CDT1. Oncogene. 21:1150–1158. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Karavias D, Maroulis I, Papadaki H, Gogos
C, Kakkos S, Karavias D and Bravou V: Overexpression of CDT1 is a
predictor of poor survival in patients with hepatocellular
carcinoma. J Gastrointest Surg. 20:568–579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yu Z, Wang R, Chen F, Wang J and Huang X:
Five novel oncogenic signatures could be utilized as AFP-related
diagnostic biomarkers for hepatocellular carcinoma based on
next-generation sequencing. Dig Dis Sci. 63:945–957. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhong Q, Chen CF, Li S, Chen Y, Wang CC,
Xiao J, Chen PL, Sharp ZD and Lee WH: Association of BRCA1 with the
hRad50-hMre11-p95 complex and the DNA damage response. Science.
285:747–750. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Chen CC, Chen CY, Ueng SH, Hsueh C, Yeh
CT, Ho JY, Chou LF and Wang TH: Corylin increases the sensitivity
of hepatocellular carcinoma cells to chemotherapy through long
noncoding RNA RAD51-AS1-mediated inhibition of DNA repair. Cell
Death Dis. 9:5432018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chen CC, Chen CY, Wang SH, Yeh CT, Su SC,
Ueng SH, Chuang WY, Hsueh C and Wang TH: Melatonin sensitizes
hepatocellular carcinoma cells to chemotherapy through long
non-coding RNA RAD51-AS1-mediated suppression of DNA repair.
Cancers (Basel). 10:2018. View Article : Google Scholar
|
|
80
|
Shao J, Xu Z, Peng X, Chen M, Zhu Y, Xu L,
Zhu H, Yang B, Luo P and He Q: Gefitinib synergizes with irinotecan
to suppress hepatocellular carcinoma via antagonizing
Rad51-mediated DNA-repair. PLoS One. 11:e01469682016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Luo J, Si ZZ, Li T, Li JQ, Zhang ZQ, Chen
GS, Qi HZ and Yao HL: MicroRNA-146a-5p enhances radiosensitivity in
hepatocellular carcinoma through replication protein A3-induced
activation of the DNA repair pathway. Am J Physiol Cell Physiol.
316:C299–C311. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chand V, Pandey A, Kopanja D, Guzman G and
Raychaudhuri P: Opposing roles of the forkhead box factors FoxM1
and FoxA2 in liver cancer. Mol Cancer Res. 17:1063–1074. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Liang C, Zhao J, Ge H, Li G and Wu J:
Clinicopathological and prognostic significance of FoxM1 in
hepatocellular carcinoma patients: A meta-analysis. Onco Targets
Ther. 11:3561–3571. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tian C, Wu H, Li C, Tian X, Sun Y, Liu E,
Liao X and Song W: Downreguation of FoxM1 by miR-214 inhibits
proliferation and migration in hepatocellular carcinoma. Gene Ther.
25:312–319. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Song BN and Chu IS: A gene expression
signature of FOXM1 predicts the prognosis of hepatocellular
carcinoma. Exp Mol Med. 50:e4182018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lin P, He RQ, Dang YW, Wen DY, Ma J, He Y,
Chen G and Yang H: An autophagy-related gene expression signature
for survival prediction in multiple cohorts of hepatocellular
carcinoma patients. Oncotarget. 9:17368–17395. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chang Y, Zhou C, Fan L, Qiu G, Wang G, Wei
G, Chang X and Li X: Upregulation of microRNA-300 induces the
proliferation of liver cancer by downregulating transcription
factor FOXO1. Oncol Rep. 40:3561–3572. 2018.PubMed/NCBI
|
|
88
|
Xu H, Li G, Yue Z and Li C: HCV core
protein-induced upregulation of microRNA-196a promotes aberrant
proliferation in hepatocellular carcinoma by targeting FOXO1. Mol
Med Rep. 13:5223–5229. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yang XW, Shen GZ, Cao LQ, Jiang XF, Peng
HP, Shen G, Chen D and Xue P: MicroRNA-1269 promotes proliferation
in human hepatocellular carcinoma via downregulation of FOXO1. BMC
Cancer. 14:9092014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Xu X, Yamamoto H, Sakon M, Yasui M, Ngan
CY, Fukunaga H, Morita T, Ogawa M, Nagano H, Nakamori S, et al:
Overexpression of CDC25A phosphatase is associated with hypergrowth
activity and poor prognosis of human hepatocellular carcinomas.
Clin Cancer Res. 9:1764–1772. 2003.PubMed/NCBI
|
|
91
|
Xu X, Yamamoto H, Liu G, Ito Y, Ngan CY,
Kondo M, Nagano H, Dono K, Sekimoto M and Monden M: CDC25A
inhibition suppresses the growth and invasion of human
hepatocellular carcinoma cells. Int J Mol Med. 21:145–152.
2008.PubMed/NCBI
|
|
92
|
Kelly P, Moeller BJ, Juneja J, Booden MA,
Der CJ, Daaka Y, Dewhirst MW, Fields TA and Casey PJ: The G12
family of heterotrimeric G proteins promotes breast cancer invasion
and metastasis. Proc Natl Acad Sci USA. 103:8173–8178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kelly P, Stemmle LN, Madden JF, Fields TA,
Daaka Y and Casey PJ: A role for the G12 family of heterotrimeric G
proteins in prostate cancer invasion. J Biol Chem. 281:26483–26490.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chia CY, Kumari U and Casey PJ: Breast
cancer cell invasion mediated by Gα12 signaling involves expression
of interleukins-6 and −8, and matrix metalloproteinase-2. J Mol
Signal. 9:62014. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Udayappan UK and Casey PJ: c-Jun
contributes to transcriptional control of GNA12 expression in
prostate cancer cells. Molecules. 22:2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Bakiri L, Hamacher R, Graña O,
Guío-Carrión A, Campos-Olivas R, Martinez L, Dienes HP, Thomsen MK,
Hasenfuss SC and Wagner EF: Liver carcinogenesis by FOS-dependent
inflammation and cholesterol dysregulation. J Exp Med.
214:1387–1409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Watanabe T, Hiasa Y, Tokumoto Y, Hirooka
M, Abe M, Ikeda Y, Matsuura B, Chung RT and Onji M: Protein kinase
R modulates c-Fos and c-Jun signaling to promote proliferation of
hepatocellular carcinoma with hepatitis C virus infection. PLoS
One. 8:e677502013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Fan Q, He M, Deng X, Wu WK, Zhao L, Tang
J, Wen G, Sun X and Liu Y: Derepression of c-Fos caused by
microRNA-139 down-regulation contributes to the metastasis of human
hepatocellular carcinoma. Cell Biochem Funct. 31:319–324. 2013.
View Article : Google Scholar : PubMed/NCBI
|