Introduction
Primary liver cancer is a malignant tumor with the
third highest mortality rate in the world (1). The long-term survival remains
unsatisfactory due to high incidence of recurrence and metastasis
following hepatic resection (2). Due
to the large number of genes and proteins involved, the
pathogenesis of liver cancer is particularly complicated (3–5).
Therefore, revealing the molecular mechanism of liver cancer
pathogenesis is key to the development of effective treatment.
MicroRNAs (miRNAs) are a class of endogenous highly
evolutionarily conserved single-stranded non-coding RNAs with a
total length of 18–25 nucleotides. As regulators of negative
regulatory genes, miRNAs are completely or incompletely
complementary to the 3′untranslated regions (3′-UTRs) of their
target genes, which results in direct degradation or translational
disruption of these genes (6). Ji
et al (7) have demonstrated
that all members of the miRNA (miR)-181 family were upregulated in
epithelial cell adhesion molecule (EpCAM/CD326)-positive hepatic
cancer stem cells isolated from α-fetoprotein (AFP)-positive liver
cancer samples. Tomimaru et al (8) compared serum miR-21 levels in 126 cases
of liver cancer, 30 cases of chronic hepatitis and 50 healthy
volunteers; the results revealed the level of miR-21 expression to
be significantly higher in patients with liver cancer compared with
those with chronic hepatitis and healthy volunteers. Abnormal
expression of miR-221 has been observed in multiple types of
cancer, such as liver (9), gastric
(10) and breast (11) cancer, suggesting that it may be
closely associated with tumorigenesis and may provide a novel
target for tumor diagnosis and treatment. miR-15b dysregulation has
also been reported in various types of cancer. For example, the
miR-15b/HOTAIR/p53 regulatory loop affects glioma cell
proliferation (12). miR-15b
promotes prostate cancer cell proliferation by targeting
reversion-inducing cysteine-rich protein with Kazal motifs and may
also be used as a clinical diagnostic marker in patients with
prostate cancer (13).
The present study aimed to analyze miR-221-3p and
miR-15b-5p expression in liver cancer to establish their potential
roles and target genes.
Materials and methods
Patients and samples
A total of 69 patients with liver cancer (40 men and
29 women; mean age 58.3±5.9 years) who underwent surgical resection
at Affiliated Hospital of Nantong University between October 2010
and May 2012 were recruited for the present study. Tumor and
adjacent non-cancerous tissues (>2 cm from the lesion) were
collected and stored at −80°C until further use. The patients did
not receive chemotherapy, radiotherapy, biological therapy or a
similar treatment regimen prior to surgery. The pathological
classification and staging of liver cancer were consistent with the
7th edition of the American Joint Committee on Cancer
Tumor-Node-Metastasis (TNM) Staging System published in 2010
(14). This study was approved by
the Ethics Committee of The Affiliated Hospital of Nantong
University and written informed consent was obtained from all
patients.
Cell culture and transfection
Human liver cancer cell lines HepG2 and Huh7 were
purchased from the American Type Culture Collection, and Hep3B and
HCCLM3 cell lines were purchased from the type Culture Collection
of the Chinese Academy of Sciences. All cells were authenticated by
short tandem repeat profiling. Cells were cultured with DMEM
(Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS
(Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA) and placed
at 37°C in a humidified incubator containing 5% CO2.
Vectors expressing AXIN2, AXIN2 small interfering RNA (siRNA)
(5′-3′) and scrambled control-sense (5′-3′) were designed and
synthesized by Chang Jing Bio-Tech, Ltd. Lipofectamine®
3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) was used for cell transfections according to the
manufacturer's instructions. The miRNA mimics and inhibitor and
AXIN2 siRNA were purchased from Sangon Biotech Co., Ltd. The
pcDNA3.1A(−) vector and pcDNA3.1A(−)-AXIN2 overexpression plasmid
were obtained from Shanghai GeneChem Co., Ltd. The sequences were
as follows: miR-221-3p mimics, 5′-AGCUACAUUGUCUGCUGGGUUUC-3′,
5′-AACCCAGCAGACAAUGUAGCUUU-3′; miR-221-3p inhibitor,
5′-GAAACCCAGCAGACAAUGUAGCU-3; miR-15b-5p inhibitor,
5′-UGUAAACCAUGAUGUGCUGCUA-3′; miR-15b-5p mimics:
5′-UAGCAGCACAUCAUGGUUUACA-3′, 5′-UAAACCAUGAUGUGCUGCUAUU-3′; and
negative control, 5′-UUCUCCGAACGUGUCACGUTT-3′ and
5′-ACGUGACACGUUCGGAGAATT-3′. The sequences of siRNA and its
negative control were as follows: Axin2, 5′-GCAGAGGGACAGGAATCAT-3′,
and the negative control, 5′-GCAGGGACAAGGTAGACAT-3′.
Transfection of liver cancer cells was performed
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Transfection efficiency was verified by western
blotting 48 h post-transfection. For functional analysis of
miR-221-3p and miR-15b-5p, liver cancer cells were transfected with
miR-221-3p and miR-15b-5p mimics and the control (miR-con) or
miR-221-3p and miR-15b-5p inhibitor and negative control
(scramble).
RNA extraction and quantitative
(q)PCR
According to manufacturer's instructions, total RNA
was extracted by TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) from tissues or liver cancer cells, including
Hep3B, HepG2, HCCLM3 and Huh7 cell lines. Reverse transcription
reaction was performed using SYBR Premix Ex Taq (Takara Bio, Inc.)
at 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. qPCR was
performed using SYBR Premix Ex Taq with 1 µl cDNA from the RT
reaction on an ABI 7500 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The PCR thermocycling conditions were as
follows: 94°C for 2 min followed by 30 cycles of 94°C for 30 sec,
58°C for 30 sec, and 72°C for 30 sec. U6 small RNA was used as an
internal reference. Primers were synthesized by Sangon Biotech Co.,
Ltd. as follows: miR-221-3p, forward, 5′-CGGCTACATTGTCTGCCTG-3′ and
reverse, 5′-CAGTGCGTGTCGTGGAGT-3′; miR-15b-5p forward,
5′-ATGAACTTTCTCTGTCTTGG-3′ and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′;
and U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The reverse universal miR qPCR primers
were included in the PrimeScript™ miRNA RT-PCR kit (cat. no. RR716;
Takara Biotechnology Co., Ltd.). The relative expression levels of
miR-221-3p and miR-15b-5p were calculated using the
2−ΔΔCq method as previously described (15). All experiments were conducted in
triplicate.
Cell viability assay
Cell viability assay was performed using the
3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide
(MTT) method with DMSO as solvent (3,000 cells per well in 96-well
plates). Absorbance was detected at 570 nm in three parallel
samples, and each sample was tested three times.
Colony formation assay
At 24 h post-transfection, cells were seeded into
6-well plates and cultured for two weeks in DMEM containing 12% FBS
at 37°C in a humiliated atmosphere of 5% CO2 (600 cells
per well in 6-well plates). The colonies were fixed and stained
with 1 mg/ml crystal violet, and colonies containing >50 cells
were counted using Leica microscopy (Leica DMR 3000; Leica
Microsystems GmbH).
Transwell assay
Cell invasive ability was determined using a
Transwell assay with Matrigel (BD Biosciences). To assess cell
invasion, Transwell chambers were coated with 30 µl Matrigel,
incubated at 37°C for 40 min and placed in 24-well plates. At 24 h
post-transfection, HepG2 and HCCLM3 cells were seeded in the upper
chamber at a density of 5×104 cells/well in DMEM with 2%
FBS. A total of 500 µl DMEM containing 10% FBS was added to the
lower chamber. After 24 h at 37°C, invaded cells were fixed with 4%
paraformaldehyde for 10 min at 37°C. Non-invaded cells were removed
with a cotton swab. Invaded cells were stained with 1 mg/ml crystal
violet at room temperature for 10 min and counted in five randomly
selected fields using Leica microscope (Leica DMR 3000; Leica
Microsystems GmbH).
Dual luciferase reporter assay
The 3′ UTR of Axin2 was cloned into a pGL3 control
vector (Promega Corporation) containing luciferase genes to
generate pGL3-AXIN2-wild-type plasmids. The primers are as follows:
Axin2 forward, 5′-GCTCTAGAGCCCTGGGGTCTGGCTTTG-3′ and
reverse, 5′-GCTCTAGATTTTGAAAAATATAAAATT-3′. The
pGL3-AXIN2-mutant plasmids were constructed using a Takara
MutanBEST Kit (Takara Biotechnology Co., Ltd.). HCCLM3 and HepG2
cells were seeded into 24-well plates (1×105
cells/well), cultured for 24 h at 37°C and transfected with the
miRNA mimic or inhibitor and the corresponding luciferase reporter
plasmids using Lipofectamine® 3000. pRL-TK
Renilla plasmids were co-transfected into the cells as an
internal reference. At 48 h, the reporter activity was measured
using a Dual Luciferase Reporter Assay Kit (Promega Corporation),
and relative to Renilla luciferase activities were
determined. The online target gene prediction software packages
TargetScan 7.2 (http://www.targetscan.org) and miRanda 3.2 (http://www.mirdb.org) were used.
Western blotting
Total protein of HepG2 cells was extracted according
to the manufacturer's instructions using RIPA lysis buffer
containing protease inhibitors (Promega Corporation). Protein
concentration was determined using bicinchoninic acid assay kit
(Bio-Rad Laboratories, Inc.). Proteins (30 µg per lane) were
separated using 12% SDS-PAGE and transferred onto a polyvinylidene
fluoride membrane. Following blocking with 5% skimmed milk in TBS
at room temperature for 2 h, the membrane was incubated with rabbit
anti-Axin2 polyclonal antibody (1:1,000; cat. no. ab32197; Abcam),
rabbit anti-β-actin polyclonal antibody (1:5,000; cat. no. ab8227;
Abcam) and rabbit anti-GAPDH polyclonal antibody (1:5,000; cat. no.
ab37168; Abcam) for 12 h at 4°C. The membrane was washed three
times in TBS + Tween-20 (0.1% V/V) and incubated with a horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000;
Sigma-Aldrich; Merck KGaA) at room temperature for 2 h bands were
detected using enhanced chemiluminescence substrate, and relative
expression of proteins was normalized to GAPDH using Scion Image v.
4.0.2 software (Scion Corporation).
Statistical analysis
SPSS 17.0 (SPSS, Inc.) was used for statistical
analysis. Data are expressed as the mean ± standard deviation.
Multiple groups were compared using one-way ANOVA followed by the
Student-Newman-Keuls test. The Kaplan-Meier method was used to
evaluate the survival rate. Spearman's rank analysis was used to
identify the correlation between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-221-3p and miR-15b-5p are
upregulated in liver cancer tissues and cell lines
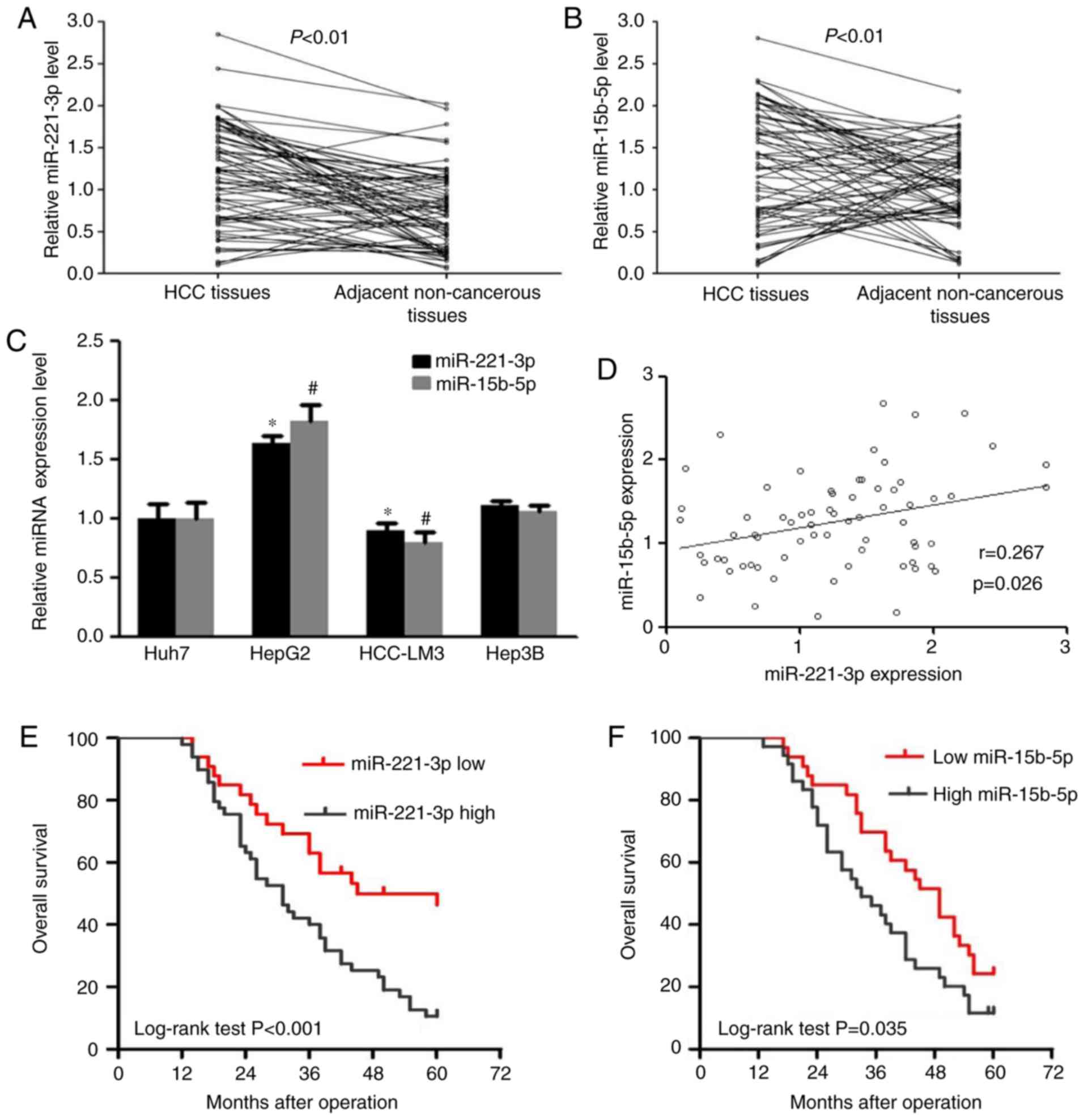
RT-qPCR was used to determine the relative
expression levels of miR-221-3p and miR-15b-5p in liver cancer and
adjacent non-cancerous tissues. The results demonstrated that the
relative expression level of miR-221-3p was 1.85±1.19 in liver
cancer tissues, which was significantly higher compared with that
in adjacent non-cancerous tissues, in which an expression level of
1.02±0.39 was observed (P<0.01; Fig.
1A). miR-15b-5p levels in liver cancer tissues was also
significantly higher compared within the adjacent tissues
(P<0.01; Fig. 1B). The expression
levels of miR-221-3p and miR-15b-5p were also determined in liver
cancer cell lines Hep3B, HepG2, HCCLM3 and Huh7 using RT-qPCR. The
results indicated that miR-221-3p and miR-15b-5p were expressed at
a higher level in HepG2 cells and at a lower level in HCCLM3 cells
compared with Huh7 cells (P<0.05; Fig. 1C). Spearman's rank analysis was used
to identify the correlation between miR-221-3p and miR-15b-5p
expression levels in liver cancer tissues; the results demonstrated
that their expression in tumor tissues exhibited a weak correlation
(r=0.267, P=0.026; Fig. 1D).
miR-221-3p and miR-15b-5p levels are
associated with clinicopathological characteristics and prognosis
of patients with liver cancer
The clinicopathological characteristics of 69
patients with liver cancer are presented in Table I. The results of statistical
Student-Newman-Keuls test demonstrated that miR-221-3p and
miR-15b-5p levels were associated with the TNM stage and tumor
capsular infiltration (P<0.05), but not with sex, age, tumor
size, AFP, hepatitis B virus surface antigen, cirrhosis or tumor
differentiation (P>0.05). Kaplan-Meier analysis revealed that
patients with liver cancer with high expression levels of
miR-221-3p or miR-15b-5p exhibited lower overall survival rates
compared with those with low levels of expression of the respective
miRNA (P<0.001 and P=0.035, respectively; Fig. 1E and F).
 | Table I.Association between miR-221
expression and clinicopathological parameters of patients with
liver cancer. |
Table I.
Association between miR-221
expression and clinicopathological parameters of patients with
liver cancer.
| Clinicopathological
characteristic | N | Relative miR-221-3p
expression | P-value | Relative miR-15b-5p
expression | P-value |
|---|
| Sex |
|
| 0.464 |
| 0.431 |
|
Male | 40 | 1.99±1.68 |
| 1.85±1.48 |
|
|
Female | 29 | 1.73±1.29 |
| 1.64±1.33 |
|
| Age, years |
|
| 0.231 |
| 0.325 |
|
≥60 | 35 | 1.96±1.71 |
| 1.68±1.51 |
|
|
<60 | 34 | 1.37±0.85 |
| 1.48±0.92 |
|
| Tumor size, cm |
|
| 0.169 |
| 0.201 |
| ≥5 | 36 | 2.32±1.41 |
| 1.98±1.68 |
|
|
<5 | 33 | 1.73±0.58 |
| 1.69±0.98 |
|
| AFP, ng/ml |
|
| 0.651 |
| 0.520 |
|
<400 | 27 | 2.11±0.83 |
| 2.01±1.02 |
|
|
≥400 | 42 | 1.96±1.15 |
| 1.99±1.25 |
|
| HBsAg |
|
| 0.523 |
| 0.488 |
|
(+) | 41 | 1.71±1.02 |
| 1.88±1.23 |
|
|
(−) | 28 | 1.68±0.89 |
| 1.72±0.96 |
|
| Clinical TNM
stage |
|
| 0.007a |
| 0.005a |
|
I–II | 29 | 0.75±0.83 |
| 0.82±0.69 |
|
|
III–IV | 40 | 2.14±0.47 |
| 2.25±0.85 |
|
| Tumor capsular
infiltration |
|
| 0.011a |
| 0.016a |
| No | 29 | 0.88±1.11 |
| 0.91±0.90 |
|
|
Yes | 40 | 2.77±1.38 |
| 2.52±1.42 |
|
| Cirrhosis |
|
| 0.722 |
| 0.689 |
|
Yes | 37 | 1.82±1.41 |
| 1.96±1.52 |
|
| No | 32 | 1.62±1.39 |
| 1.79±1.47 |
|
|
Differentiation |
|
| 0.332 |
| 0.433 |
| Poorly
differentiated | 31 | 1.91±1.28 |
| 1.87±1.48 |
|
|
Moderately/well
differentiated | 38 | 2.09±1.56 |
| 2.15±1.34 |
|
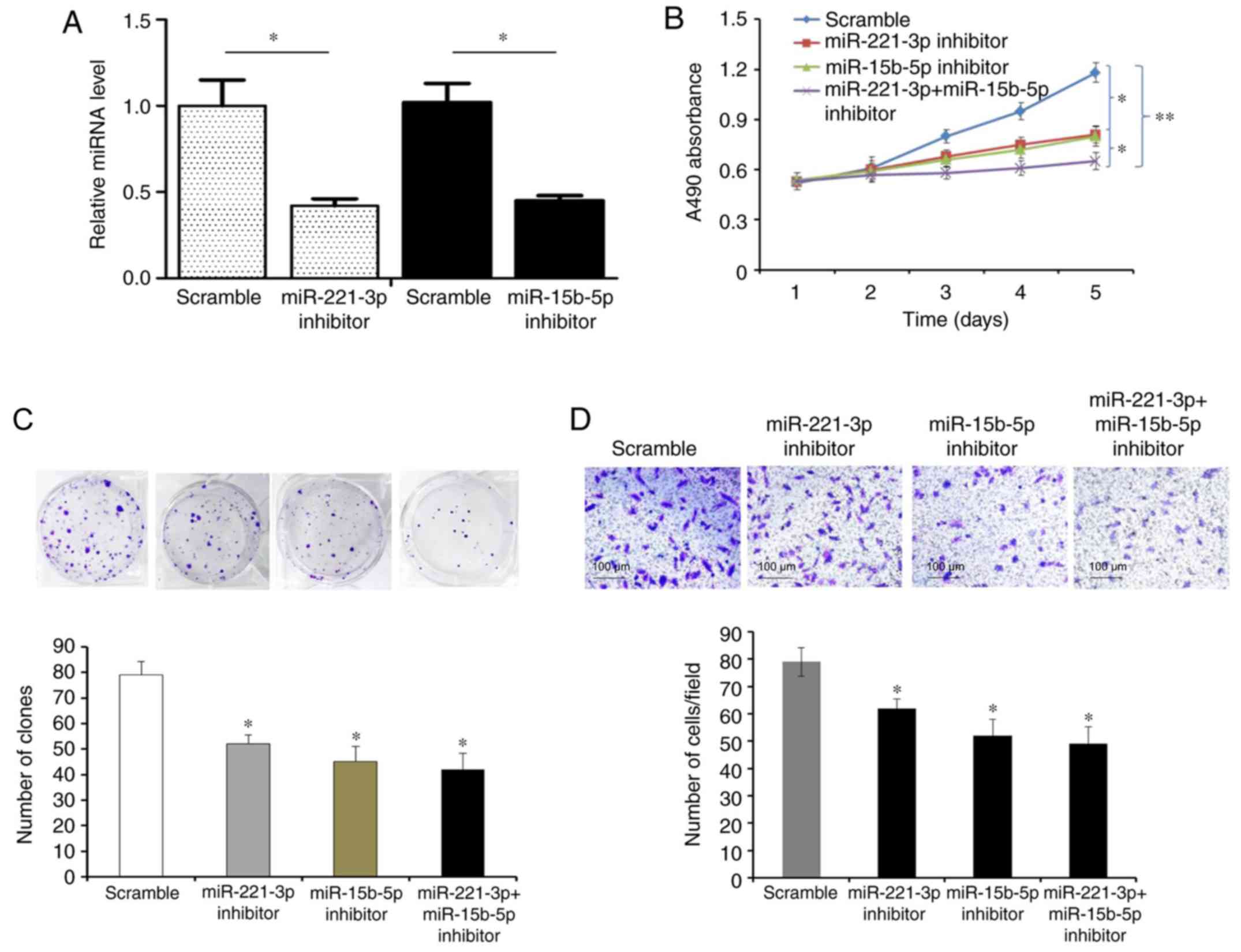
Promotive effects of miR-221-3p and
miR-15b-5p on liver cancer cell proliferation and invasion in
vitro
Based on the analysis of the patient
clinicopathological data, it was hypothesized that miR-221-3p and
miR-15b-5p may promote liver cancer cell proliferation and
invasion. miR-221-3p and miR-15b-5p expression levels were the
lowest in HCCLM3 cells, which were selected for overexpression
experiments by transfection with miR-221-3p and miR-15b-5p mimics
(P<0.05; Fig. 2A). Overexpression
of miR-221-3p and/or miR-15b-5p significantly promoted the
proliferation of HCCLM3 cells, as indicated by the results of MTT
(P<0.05 and P<0.01; Fig. 2B)
and colony formation (P<0.05; Fig.
2C) assays. Overexpression of miR-221-3p and/or miR-15b-5p also
stimulated cell invasion (P<0.05; Fig. 2D).
Compared with the other liver cancer cell lines, the
relative levels of miR-221-3p and miR-15b-5p were the highest in
HepG2 cells, which were selected for transfection with miR-221-3p
and miR-15b-5p inhibitors to knock down endogenous miR-221-3p and
miR-15b-5p expression (P<0.05, Fig.
3A); knockdown of miR-221-3p and/or miR-15b-5p significantly
inhibited cell proliferation, colony formation and invasion
(P<0.05; Fig. 3B-D).
Axin2 is a common target of miR-221-3p
and miR-15b-5p
To establish the specific mechanism by which
miR-221-3p and miR-15b-5p may promote the proliferation of liver
cancer cells, the common target gene of miR-221-3p and miR-15b-5p
was predicted through bioinformatic analysis using online target
gene prediction software packages TargetScan 7.2 and miRanda 3.2 to
determine whether the candidate target gene 3′UTR contained
miR-221-3p and/or miR-15b-5p binding sites (Fig. 4A and B). The results of a dual
luciferase activity assay revealed that overexpression of
miR-221-3p or miR-15b-5p inhibited the luciferase activity of the
pGL3-Axin2-3′-UTR reporter, but not the pGL3-mut-Axin2-3′-UTR
reporter (P<0.05; Fig. 4C and D).
Western blotting results demonstrated that Axin2 protein levels
were significantly lower in liver cancer cells transfected with
miR-221-3p or miR-15b-5p mimics (P<0.01; Fig. 4E). Western blotting experiments were
also performed to assess Axin2 expression in liver cancer tissues
and corresponding adjacent non-cancerous tissues, and the results
revealed that the protein expression level of Axin2 in HCC tissues
was significantly lower compared with that in adjacent
non-cancerous tissues (P<0.05; Fig.
4F).
 | Figure 4.Axin2 is a direct common target of
miR-221-3p and miR-15b-5p in liver cancer. (A) The schematic map of
the pGL3-AXIN2-3′UTR and pGL3-AXIN2-mut-3′UTR binding site for
miR-221-3p. (B) The schematic map of pGL3-AXIN2-3′UTR and
pGL3-AXIN2-mut-3′UTR binding site for miR-15b-5p. (C) miR-221-3p
targeted the AXIN2 wild type 3′UTR, but not the mutant. (D)
miR-15b-5p targeted the wild type 3′UTR, but not the mutant. (E)
Western blotting results demonstrated that overexpression of
miR-221-3p or miR-15b-5p inhibited Axin2 protein expression in
HepG2 cells. (F) Western blotting experiments were performed to
detect Axin2 expression in liver cancer tissues and corresponding
adjacent non-cancerous tissues; the results indicated the Axin2
protein expression in liver cancer tissues was significantly lower
compared with that in adjacent non-cancerous tissues. *P<0.05
vs. miR-con or adjacent non-cancerous tissues. Axin2, axis
inhibition protein 2; miR, microRNA; UTR, untranslated region; mut,
mutant; miR-con, mimic control; T, tumor; N, adjacent non-cancerous
tissue; HCC, hepatocellular carcinoma. |
Axin2 is a common functional target of
miR-221-3p and miR-15b-5p in liver cancer cells
To establish the roles of miR-221-3p and miR-15b-5p
in the regulation of Axin2, HepG2 cells were transfected with
si-Axin2 to knock down the endogenous expression of Axin2. The
reduction and overexpression in Axin2 levels was determined by
western blotting (Fig. 5A and B).
Axin2 knockdown had a significant promotive effect on the
proliferation and invasion of HepG2 cells, similar to the promotion
of liver cancer cells by overexpression of miR-221-3p and
miR-15b-5p (Fig. 5C and D). In HepG2
cells stably overexpressing miR-221-3p and miR-15b-5p, the
restoration of Axin2 level reversed the promotive effects of
miR-221-3p and miR-15b-5p in liver cancer cells (Fig. 5E and F). In addition, the results
revealed that when knockdown of Axin2 was followed by transfection
with miR-221-3p and miR-15b-5p inhibitor, Axin2 levels and invasive
and colony formation abilities in liver cancer cells were partially
restored (Fig. 6).
Discussion
Liver cancer is a primary liver tumor that ranks
third in incidence among all gastrointestinal tumors globally
(1,16). Numerous studies have reported that
miRNAs perform important functions in the development of liver
cancer (17–21). Previous studies have demonstrated
abnormal levels of miR-221 and miR-15b in liver cancer tissues,
which affect the progression of liver cancer and exhibit oncogenic
characteristics (9,22–25).
Although the mechanisms of action of these miRNAs in tumors are
unclear, their importance is evident; miR-221 and miR-15b may be
breakthrough targets for tumor prevention, diagnosis and
treatment.
Recent studies have indicated that miR-221-3p and
miR-15b-5p are involved in a number of types of cancer. For
example, miR-221-3p exerts a tumor-suppressive role in epithelial
ovarian cancer and directly targets ADP ribosylation factor 4,
which suggests that miR-221-3p may be a suitable candidate for the
clinical prognosis and therapeutics for epithelial ovarian cancer
(26); additionally, miRNA-221-3p
serves an oncogenic role in gastric carcinoma by inhibiting PTEN
expression (27), promotes the
metastasis of gastric cancer through progestin and adipoQ receptor
family member 3 and may be a biomarker of gastric cancer (28). The results of the present study
indicated that miR-221-3p and miR-15b-5p were frequently
upregulated in liver cancer tissues and cells and that high
expression levels of miR-221-3p and miR-15b-5p were associated with
TNM stage, infiltration and poor prognosis. In addition,
overexpression of miR-221-3p and miR-15b-5p promoted liver cancer
cell proliferation and invasion in vitro. These results
indicated that miR-221-3p and miR-15b-5p may serve crucial roles in
the occurrence of liver cancer and may promote its progression.
Axin2 is an anti-oncogene that negatively regulates
the canonical Wnt/β-catenin signaling pathway, which includes a
series of proteins such as the extracellular factor Wnt, T-cell
factor and the frizzled transmembrane receptor. Axin2 is a scaffold
protein involved in the degradation β-catenin complex when
dephosphorylated (29). Mutations in
the Axin2 gene have been identified in multiple types of human
cancer, including tumors of the digestive tract and melanoma
(30–33). The results of the present study
predicted that Axin2 may be one of the target genes of miR-221
using bioinformatics tools, and luciferase and western blot assay
results revealed that Axin2 was a direct target of miR-221.
Knockdown of Axin2 promoted the proliferation of liver cancer
cells, which further suggested that Axin2 may be a target of
miR-221 in liver cancer.
In conclusion, the results of the present study
demonstrated that miR-221-3p and miR-15b-5p were upregulated in
liver cancer cells and were associated with TNM stage, tumor
capsular infiltration and prognosis of patients with liver cancer.
Overexpression of miR-221-3p and miR-15b-5p promoted liver cancer
cell proliferation and invasion in vitro. In addition, Axin2
was identified as a direct and functional target of miR-221 in
liver cancer cells. These results indicated that miR-221-3p and
miR-15b-5p may be used as prognostic indicators for liver cancer.
The miR-221-3p/miR-15b-5p-Axin2 axis may potentially serve as a
therapeutic target for patients with liver cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by Nantong Science
and Technology Project (grant nos. MS12018066 and MSZ18114).
Availability of data and materials
All data analyzed during the present study are
included in this published article.
Authors' contributions
YD and XT conceived and designed the experiments.
YD, NZ, SZ, XC, FL and XT performed all the experiments. NZ and XT
wrote and revised the manuscript. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Linyi Central Hospital (Lin Yi, China). Written
informed consent was obtained from all patients prior to
enrolment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hernandez-Gea V, Turon F, Berzigotti A and
Villanueva A: Management of small hepatocellular carcinoma in
cirrhosis: Focus on portal hypertension. World J Gastroenterol.
19:1193–1199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang LY, Fang F, Ou DP, Wu W, Zeng ZJ and
Wu F: Solitary large hepatocellular carcinoma: A specific subtype
of hepatocellular carcinoma with good outcome after hepatic
resection. Ann Surg. 249:118–123. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu X, Liao W, Yuan Q, Ou Y and Huang J:
TTK activates Akt and promotes proliferation and migration of
hepatocellular carcinoma cells. Oncotarget. 6:34309–34320.
2015.PubMed/NCBI
|
|
4
|
Whittaker S, Marais R and Zhu AX: The role
of signaling pathways in the development and treatment of
hepatocellular carcinoma. Oncogene. 29:4989–5005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karavias D, Maroulis I, Papadaki H, Gogos
C, Kakkos S, Karavias D and Bravou V: Overexpression of CDT1 is a
predictor of poor survival in patients with hepatocellular
carcinoma. J Gastrointest Surg. 20:568–579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ji J, Yamashita T and Wang XW:
Wnt/beta-catenin signaling activates microRNA-181 expression in
hepatocellular carcinoma. Cell Biosci. 1:42011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tomimaru Y, Eguchi H, Nagano H, Wada H,
Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I,
Umeshita K, et al: Circulating microRNA-21 as a novel biomarker for
hepatocellular carcinoma. J Hepatol. 56:167–175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rong M, Chen G and Dang Y: Increased
miR-221 expression in hepatocellular carcinoma tissues and its role
in enhancing cell growth and inhibiting apoptosis in vitro. BMC
Cancer. 13:212013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK and Kim VN: Functional links
between clustered microRNAs: Suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang
L, Yang A, Zhao J and Jia L: MiR-221 promotes
trastuzumab-resistance and metastasis in HER2-positive breast
cancers by targeting PTEN. BMB Rep. 47:268–273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen LP, Zhang NN, Ren XQ, He J and Li Y:
miR-103/miR-195/miR-15b regulate SALL4 and inhibit proliferation
and migration in glioma. Molecules. 23(pii): E29382018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen R, Sheng L, Zhang HJ, Ji M and Qian
WQ: miR-15b-5p facilitates the tumorigenicity by targeting RECK and
predicts tumour recurrence in prostate cancer. J Cell Mol Med.
22:1855–1863. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsu PK, Wu YC, Chou TY, Huang CS and Hsu
WH: Comparison of the 6th and 7th editions of the American Joint
Committee on Cancer tumor-node-metastasis staging system in
patients with resected esophageal carcinoma. Ann Thorac Surg.
89:1024–1031. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kapodistrias N, Bobori C and
Theocharopoulou G: MiR-140-3p downregulation in association with
PDL-1 overexpression in many cancers: A review from the literature
using predictive bioinformatics tools. Adv Exp Med Biol.
988:225–233. 2017.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang G, Dong F, Xu Z, Sharma S, Hu X, Chen
D, Zhang L, Zhang J and Dong Q: MicroRNA profile in HBV-induced
infection and hepatocellular carcinoma. BMC Cancer. 17:8052017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wen Y, Han J, Chen J, Dong J, Xia Y, Liu
J, Jiang Y, Dai J, Lu J, Jin G, et al: Plasma miRNAs as early
biomarkers for detecting hepatocellular carcinoma. Int J Cancer.
137:1679–1690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morgul MH, Klunk S, Anastasiadou Z, Gauger
U, Dietel C, Reutzel-Selke A, Felgendref P, Hau HM, Tautenhahn HM,
Schmuck RB, et al: Diagnosis of HCC for patients with cirrhosis
using miRNA profiles of the tumor-surrounding tissue-A statistical
model based on stepwise penalized logistic regression. Exp Mol
Pathol. 101:165–171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu L, Guo D, Chen X, Xiong W, Jie S and Li
H: Abnormal miRNAs targeting chromosome open reading frame genes
were enriched in microvesicles derived from the circulation of HCC.
Biochem Genet. 54:120–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fornari F, Gramantieri L, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM,
Bolondi L and Negrini M: MiR-221 controls CDKN1C/p57 and CDKN1B/p27
expression in human hepatocellular carcinoma. Oncogene.
27:5651–5661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park JK, Kogure T, Nuovo GJ, Jiang J, He
L, Kim JH, Phelps MA, Papenfuss TL, Croce CM, Patel T and
Schmittgen TD: miR-221 silencing blocks hepatocellular carcinoma
and promotes survival. Cancer Res. 71:7608–7616. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Duan Z, Gao Y, Shen J, Choy E, Cote G,
Harmon D, Bernstein K, Lozano-Calderon S, Mankin H and Hornicek FJ:
miR-15b modulates multidrug resistance in human osteosarcoma in
vitro and in vivo. Mol Oncol. 11:151–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahmad P, Sana J, Slavik M, Gurin D, Radova
L, Gablo NA, Kazda T, Smilek P, Horakova Z, Gal B, et al:
MicroRNA-15b-5p predicts locoregional relapse in head and neck
carcinoma patients treated with intensity-modulated radiotherapy.
Cancer Genomics Proteomics. 16:139–146. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Q, Ren X, Zhang Y, Fu X, Li Y, Peng Y,
Xiao Q, Li T, Ouyang C, Hu Y, et al: MiR-221-3p targets ARF4 and
inhibits the proliferation and migration of epithelial ovarian
cancer cells. Biochem Biophys Res Commun. 497:1162–1170. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi J, Zhang Y, Jin N, Li Y, Wu S and Xu
L: MicroRNA-221-3p plays an oncogenic role in gastric carcinoma by
inhibiting PTEN expression. Oncol Res. 25:523–536. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao C, Li Y, Chen G, Wang F, Shen Z and
Zhou R: Overexpression of miR-15b-5p promotes gastric cancer
metastasis by regulating PAQR3. Oncol Rep. 38:352–358. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lammi L, Arte S, Somer M, Jarvinen H,
Lahermo P, Thesleff I, Pirinen S and Nieminen P: Mutations in AXIN2
cause familial tooth agenesis and predispose to colorectal cancer.
Am J Hum Genet. 74:1043–1050. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rivera B, Perea J, Sanchez E, Villapún M,
Sánchez-Tomé E, Mercadillo F, Robledo M, Benítez J and Urioste M: A
novel AXIN2 germline variant associated with attenuated FAP without
signs of oligondontia or ectodermal dysplasia. Eur J Hum Genet.
22:423–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Castiglia D, Bernardini S, Alvino E,
Pagani E, De Luca N, Falcinelli S, Pacchiarotti A, Bonmassar E,
Zambruno G and D'Atri S: Concomitant activation of Wnt pathway and
loss of mismatch repair function in human melanoma. Genes
Chromosomes Cancer. 47:614–624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim MS, Kim SS, Ahn CH, Yoo NJ and Lee SH:
Frameshift mutations of Wnt pathway genes AXIN2 and TCF7L2 in
gastric carcinomas with high microsatellite instability. Hum
Pathol. 40:58–64. 2009. View Article : Google Scholar : PubMed/NCBI
|