MicroRNAs (miRNAs/miRs) are a family of non-coding
RNAs that were discovered in 1993 (7). miRNAs consist of 22–26 nucleotides but
have a complex structure that confers the ability induce cleavage
or translational repression of target mRNAs (7). miRNAs are therefore important
regulators of gene expression (8).
Breast cancer arises through the accumulation of genetic mutations
and epigenetic modifications; therefore, miRNAs are important
factors in breast carcinogenesis (9). A number of miRNAs are expressed at
higher or lower levels in tumor tissues compared with normal
tissues and may serve as tumor markers in breast cancer (10,11). In
fact, miRNAs have attracted great interest as cancer biomarkers in
the last decade (12–14).
Certain miRNAs have been identified in previous
research as breast cancer biomarkers, including miR-30a,
miR-361-5p, miR-27a and miR-193b (12,15–18).
However, research based on large sample sizes is lacking,
particularly research employing clinical data (19–21). The
Cancer Genome Atlas (TCGA) project, which was launched in 2006 by
the National Cancer Institute and the National Human Genome
Research Institute, contains data on 33 types of cancer from
>10,000 patients (22,23). TNBC data available from TCGA consist
of tissue miRNA-sequencing (seq) data and clinical information. The
present study analyzed the aforementioned data in four steps: i)
Identification of differentially expressed miRNAs between breast
cancer and adjacent normal tissues; ii) screening of the miRNAs
obtained in the first step using the Kaplan-Meier survival method,
which yielded a total of six specific miRNAs; iii) identification
of three of these six miRNAs that predicted patient survival rate
based on statistical analysis; and iv) identification of the
biological pathways regulated by these three miRNAs.
The data analyzed in the present study were
downloaded from TCGA (up to January 28, 2016). The clinical data
for each patient were derived from a variety of methods used to
detect HER-2 levels; therefore, ‘patient HER2 immunohistochemistry
receptor status’ was used as a standard to determine patient HER-2
status (24). When confirming ER, PR
and HER status, only patients with ‘positive’ and ‘negative’ data
were enrolled, whereas patients with data deemed ‘close’ or ‘not
available’ were excluded. The TNBC miRNA-seq data for miRNA
differential expression consisted of data on tumor tissues (n=187)
and matched normal tissues (n=12). All of the miRNA expression data
were reported as ‘reads-per-million-miRNA-mapped’ and were
normalized by log2 transformation. The inclusion criteria regarding
clinical information for survival analysis were as follows: i)
Complete follow-up data were available for 1–60 months (30-1,825
days); ii) the clinical data were complete (patients with uncertain
or missing clinical data, with the exception of metastasis state,
which contains various Mx stages, were excluded) and iii) miRNA-seq
data integrity was validated (patients without individual miRNA
values were exluded). Finally, a total of 151 patients who met
these criteria were enrolled in the present study. In order to
incorporate as much data as possible, miRNA differential expression
and survival analyses were performed independently.
Two online miRNA databases were employed to predict
the target genes of the three prognostic miRNAs: TargetScan
(version 7.2) (26) and miRDB
(27,28). The overlapping set of target genes
was depicted in Venn diagrams for subsequent analysis. The
overlapping genes were analyzed using the Database for Annotation,
Visualization and Integrated Discovery (DAVID; version 6.8;
david.ncifcrf.gov). DAVID is a bioinformatics
database that integrates biological data and analysis tools,
provides systematic and comprehensive bioinformatics annotations
for large-scale gene or protein lists and aids users to extract
bioinformatics data. The tools in DAVID perform numerous functions,
including gene function enrichment, which improves the
understanding of the biological roles of specific gene sets
(29,30). GO (http://geneontology.org/) and KEGG (https://www.genome.jp/kegg; Release 88.0) pathway
enrichment analyses were performed to identify the biological
processes and pathways involving target genes. The cut-off criteria
were P<0.05 and gene count ≥3 (31).
The TNBC miRNA-seq data in the present study
included data on 187 tumor tissues and 12 matched normal tissues. A
total of 194 differentially expressed miRNAs were identified using
the cut-off criteria of P<0.05 and |log2FC|>2. Of these
miRNAs, 65 were downregulated and 129 were upregulated (Fig. 1; Table
SI).
A total of 151 patients with validated data were
enrolled in the present study. A total of 77 miRNAs significantly
associated with OS time were identified from the survival analysis
(Table SII). The intersection of
the 77 miRNAs and 194 differentially expressed miRNAs revealed six
miRNAs, including miR-4742-3p, miR-21-3p, miR-659-5p, miR-500b-3p,
miR-429 and miR-200b-5p. Log-rank tests were performed to evaluate
the survival curves. miR-4742-3p and miR-500b-3p were eliminated
based on the cut-off criteria (P<0.05; Table SIII). The prognostic value of each
of the remaining four miRNAs was evaluated using the Cox
proportional hazards model, and miR-21-3p, miR-659-5p and
miR-200b-5p were identified as independent risk factors associated
with OS time (Fig. 2A-C). Among
these miRNAs, miR-200b-5p was identified as a risk factor, whereas
miR-21-3p and miR-659-5p were identified as protective factors.
Additionally, the association between the aforementioned three
miRNAs and clinical features was investigated (Table I). The results revealed that
miR-218-1 was associated with metastasis state (P=0.031). A risk
score based on the expression levels of the three miRNAs determined
using the Cox regression coefficient for each miRNA was calculated
as follows: Risk score=(0.627 × expression of miR-200b-5p)-(1.181 ×
expression of miR-659-5p)-(0.889 × expression of miR-21-3p). The
risk score was used to divide the 151 patients into a high-risk
group (n=76; risk score=−9.05~-5.99) and a low-risk group (n=75;
risk score=−12.96~-9.06). Compared with the low-risk group, the
high-risk group exhibited poor survival rate outcomes as assessed
using the Kaplan-Meier method (Fig.
2D).
Univariate and multivariate Cox regression analyses
were performed to test the efficacy of the three-miRNA signature
(high vs. low risk) in predicting OS time by considering the
following clinical features: Age at initial pathological diagnosis
(<60 years vs. ≥60 years), histological type [invasive ductile
carcinoma (IDC) vs. non-IDC], tumor size [T in the American Joint
Committee on Cancer (AJCC; http://cancerstaging.org/) Tumor, Node, Metastasis
(TNM) staging system (T1-2 vs. T3-4)], lymph node status (N in the
AJCC TNM staging system, N0-1 vs. N2-3), metastasis (M in AJCC TNM
staging system, M0 vs. Mx) and AJCC pathological stage (G1-2 vs.
G3-4). In the univariate analysis, the three-miRNA signature
[hazard ratio (HR)=6.893; P=0.012; 95% confidence interval (CI)
1.52–30.69], pathological stage (HR=5.953; P=0.001; 95% CI
2.04–17.34) and lymph node status (HR=7.850; P=0.001; 95% CI
2.39–25.76) were associated with OS time in patients with TNBC
(Table II). In the multivariate
analysis, only the three-miRNA signature was identified as an
independent prognostic factor (HR=7.396; P=0.011; 95% CI
1.59–34.41; Table II).
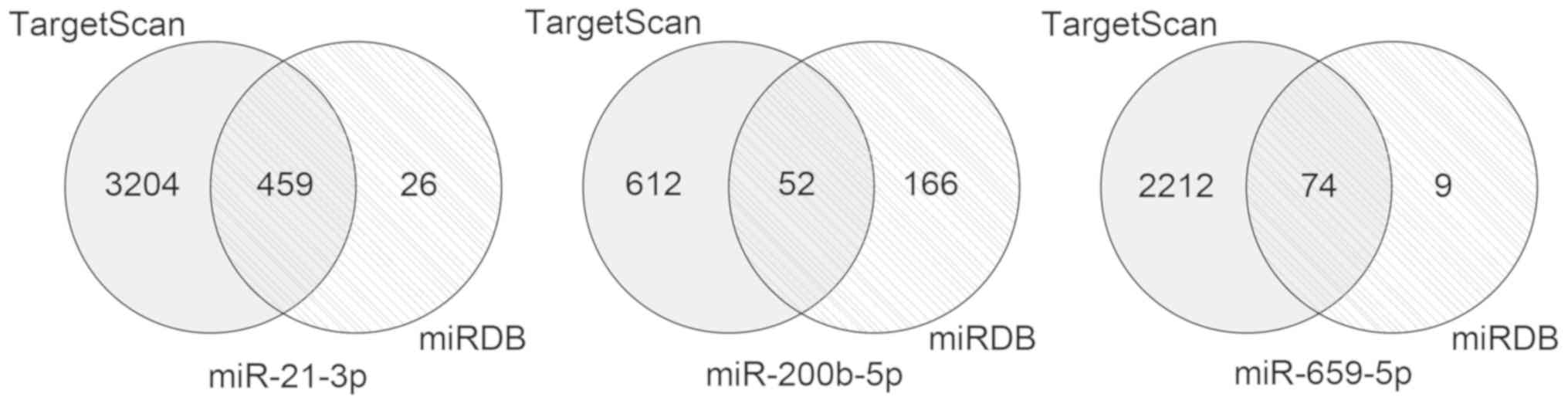
A total of 459 overlapping target genes for
miR-21-3p, 52 overlapping target genes for miR-200b-5p and 74
overlapping target genes for miR-659-5p were identified (Fig. 3). The GO results revealed that these
overlapping genes were enriched in 79 GO accessions and were mainly
involved in ‘cell protein metabolism’, ‘RNA transcriptional
regulation’ and ‘cell migration’ (Fig.
4). KEGG pathway analysis revealed enrichment in six pathways,
including ‘ubiquitin-mediated proteolysis’, ‘long-term
potentiation’, ‘MAPK signaling pathway’, ‘ErbB signaling pathway’,
‘prolactin signaling pathway’ and ‘adherens junction’ (Fig. 5).
The present study identified a three-miRNA signature
associated with the survival rate of patients with TNBC. Patients
were divided into two groups according to their risk score based on
the expression pattern of the three miRNAs (miR-21-3p, miR-659-5p
and miR-200b-5p). Significant differences in OS time were observed
between the two groups. In the multivariate Cox model, the
three-miRNA signature was identified as an independent prognostic
factor of the OS time of patients with TNBC [HR=7.396; 95% CI,
1.59–34.41]. KEGG and GO analyses of the target genes of the three
miRNAs indicated that they serve important roles in cell protein
metabolism, RNA transcriptional regulation and cell migration,
suggesting the aforementioned three miRNAs are implicated in the
pathogenesis, progression and prognosis of TNBC. The 95% CI
exhibited a wide range, possibly due to the small sample size
(n=151). In a similar study, a wide 95% CI interval was also
obtained (20). Future studies
investigating larger sample sizes are required to assess the
accuracy of the three-miRNA signature established in the present
study.
The present study investigated the associations
between clinical factors, such as age, tumor size and clinical
stage, and miRNA levels. However, factors such as co-morbidities,
smoking status and treatment received were not taken into account.
Databases are unlikely to have records of all of the clinical
factors affecting miRNA levels, therefore it is not possible to
consider the impact of these clinical factors on patients. In order
to circumvent this problem, the limma package in R was used to
analyze differentially expressed miRNAs in the present study. The
comparisons between tumor and normal tissues performed in limma
were not conducted using a 1:1 ratio of tumor to healthy tissue, as
healthy tissue specimens were not obtained from the majority of
patients. Therefore, the tumor tissue from each patient was
compared with all healthy tissue specimens. This approach
eliminates the impacts of individual patient treatment options or
other diseases (32).
miR-21 is one of the most studied miRNAs in breast
cancer, but research focusing on the association between miR-21-3p
and TNBC is lacking (33–35). In the present study, miR-21-3p
expression was increased in tumor tissues compared with healthy
tissues, which is consistent with previous studies (36,37).
However, there is conflicting evidence regarding the association
between miR-21-3p expression in tumor tissues and OS time. Previous
studies revealed that miR-21-3p is an oncogenic factor in breast
cancer, which upregulates the expression levels of phosphorylated
protein kinase B (AKT) and L1 cell adhesion molecule (37,38).
However, another in vitro study suggested that miR-21-3p
reduces cancer cell growth (39).
Furthermore, Jiao et al (40)
reported that miR-21-3p functions as a tumor suppressor; however,
further studies are required to investigate the effect of miR-21-3p
on the OS time. Jiao et al (40) proposed that miR-21-3p causes
transcript decay of miR-21-5p, a typical oncogene. The present
study demonstrated that miR-21-3p was a protective factor in TNBC.
However, this result requires further corroboration using a larger
sample size.
Aberrant expression of miR-200 family members,
including miR-200-5p, has been observed in several cancer studies
(13,41). The present study revealed that
miR-200-5p expression was increased in TNBC tissues compared with
healthy tissues, consistent with previous studies (19,21), and
that miR-200-5p is a risk factor for OS time. However, previous
studies have demonstrated that miR-200b/c activates the focal
adhesion kinase/phosphoinositide 3-kinase/AKT/nuclear factor-κB
signaling pathway and zinc-finger E-box-binding homeobox factor to
inhibit cancer cell invasion and migration (42–44). The
aforementioned studies all involved in vitro cell
experiments, and one study reported a different conclusion using
TCGA data (21). The aforementioned
study reported no statistically significant difference in OS time
between patients with low vs. high expression of miR-200b/c,
highlighting the complex biology of cancer (21). There may be two possibilities to
explain the discrepancy between the present study and previously
published studies: i) The present study did not analyze a
sufficient number of samples to generalize the entire population of
patients with TNBC, and ii) in vitro experiments do not
reflect the complex microenvironment in vivo, and some
unknown factor may influence the function of miR-200-5p in
vivo. These key issues require further investigation in future
studies.
The present study demonstrated that miR-659-5p is an
independent protective factor associated with OS time in patients
with TNBC. To the best of our knowledge, the present study was the
first to describe an association between miR-659 and breast cancer.
Due to the lack of systematic research, little is known about the
role of miR-659. One study demonstrated that miR-659 targets the
mitogen-activated protein kinase 9 (MAPK9) gene. In patients with
nasopharyngeal carcinoma, MAPK9 expression levels influence
polymorphic lactotransferrin(LTF) haplotypes, which are positively
associated with the incidence of cancer (45). Other studies have focused on the role
of miR-659 in muscle development and nervous system and metabolic
diseases, such as frontotemporal dementia and diabetes mellitus
(46–48). Thus, the mechanism of miR-659-5p in
breast cancer requires further investigation.
The present study investigated the biological
activities of the selected three miRNAs using KEGG and GO analyses
and identified six enriched pathways. Enhanced ubiquitin-mediated
proteolysis causes the rapid hydrolysis of proteins, including
tumor protein p53 and interferon α inducible protein 27, which are
closely associated with invasiveness, malignancy, clinical stage
and poor prognosis in breast cancer (49). Ubiquitin-mediated proteolysis has
been suggested to be a marker of the risk of breast cancer
(50). Zhang et al (51) reported that mitogen-activated protein
kinase (MAPK) expression levels in TNBC tissues were significantly
increased compared with normal adjacent tissues, indicating an
association between MAPK expression and TNBC. In addition, studies
have indicated that MAPK protein expression is associated with
clinical stage and lymph node metastasis in TNBC (52,53). The
epidermal growth factor receptor (EGFR) serves important roles in
TNBC (54). Previous studies have
revealed the upregulation of EGFR, which affects carcinogenesis,
progression and metastasis of breast cancer, in 70% of patients
with TNBC (55,56). A previous study reported enrichment
of the prolactin (PRL) signaling pathway in TNBC (57). López-Ozuna et al (58) reported that the PRL signaling pathway
may be used to predict the response to prodifferentiation therapy
in TNBC. The adherens junction pathway is another pathway that was
reported to be enriched in TNBC-associated processes (59). Vascular endothelial-cadherin, heparan
and the probe for protein kinase C α have previously been
implicated in the adherens junction pathway (60–62). The
main component of the cell-cell adherens junction is E-cadherin,
which plays a major role in maintaining normal breast epithelial
cell morphology (63). Breast cancer
has been found to downregulate or interfere with the expression of
E-cadherin, which is closely associated with the
epithelial-to-mesenchymal transition (EMT) (63), a key step in the invasion and
metastasis of breast cancer (64,65). The
induction of EMT requires the synergy of the MAPK signaling pathway
(66,67). The aforementioned observations
indicate an association between the MAPK signaling and adherens
junction pathways, which requires further investigation.
The present study had a number of limitations. These
included a relatively small sample size and a lack of validation
experiments, such as reverse-transcription quantitative polymerase
chain reaction analysis, for the selected three miRNA transcripts
at the tissue and plasma levels. Further research based on a larger
cohort of patients with TNBC is required to validate the findings
obtained.
In conclusion, the present study identified a
three-miRNA signature as a potential prognostic predictor of
patients with TNBC. Due to the complex biology of cancer, the
molecular mechanisms involving miRNA signatures warrant further
investigation.
Not applicable.
The present study was supported by The Science and
Technology Program of Quanzhou, China (grant no. 2018Z119).
The datasets analyzed during the current study are
available in The Cancer Genome Atlas repository, cancergenome.nih.gov.
XW wrote the manuscript, designed the study and
interpreted the data. MD performed the statistical analysis and
interpreted the results. JL was involved in designing the study and
critically revising the manuscript. All authors read and approved
the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wen YY, Liu WT, Sun HR, Ge X, Shi ZM, Wang
M, Li W, Zhang JY, Liu LZ and Jiang BH: IGF-1-mediated PKM2/β-
catenin/miR-152 regulatory circuit in breast cancer. Sci Rep.
7:158972017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shimelis H, LaDuca H, Hu C, Hart SN, Na J,
Thomas A, Akinhanmi M, Moore RM, Brauch H, Cox A, et al:
Triple-negative breast cancer risk genes identified by multigene
hereditary cancer panel testing. J Natl Cancer Inst. 110:855–862.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu Q, Yang F, Xiang T, Huai G, Yang X, Wei
L, Yang H and Deng S: A novel microRNA signature predicts survival
in liver hepatocellular carcinoma after hepatectomy. Sci Rep.
8:79332018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karsli-Ceppioglu S, Dagdemir A, Judes G,
Ngollo M, Penault-Llorca F, Pajon A, Bignon YJ and Bernard-Gallon
D: Epigenetic mechanisms of breast cancer: An update of the current
knowledge. Epigenomics. 6:651–664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hafez MM, Hassan ZK, Zekri AR, Gaber AA,
Al Rejaie SS, Sayed-Ahmed MM and Al Shabanah O: MicroRNAs and
metastasis-related gene expression in Egyptian breast cancer
patients. Asian Pac J Cancer Prev. 13:591–598. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang YY, Lai LC, Tsai MH and Chuang EY:
Deep sequencing reveals a MicroRNA expression signature in
triple-negative breast cancer. Methods Mol Biol. 1699:99–111. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: Opportunities and challenges.
Biomed Res Int. 2015:1250942015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zuberi M, Mir R, Das J, Ahmad I, Javid J,
Yadav P, Masroor M, Ahmad S, Ray PC and Saxena A: Expression of
serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in
epithelial ovarian cancer and their association with
clinicopathological features. Clin Transl Oncol. 17:779–787. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alunni-Fabbroni M, Majunke L, Trapp EK,
Tzschaschel M, Mahner S, Fasching PA, Fehm T, Schneeweiss A, Beck
T, Lorenz R, et al: Whole blood microRNAs as potential biomarkers
in post-operative early breast cancer patients. BMC Cancer.
18:1412018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Garcia-Vazquez R, Ruiz-García E, Meneses
García A, Astudillo-de la Vega H, Lara-Medina F, Alvarado-Miranda
A, Maldonado-Martínez H, González-Barrios JA, Campos-Parra AD,
Rodríguez Cuevas S, et al: A microRNA signature associated with
pathological complete response to novel neoadjuvant therapy regimen
in triple-negative breast cancer. Tumour Biol.
39:10104283177028992017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han J, Yu J, Dai Y, Li J, Guo M, Song J
and Zhou X: Overexpression of miR-361-5p in triple-negative breast
cancer (TNBC) inhibits migration and invasion by targeting RQCD1
and inhibiting the EGFR/ PI3K/Akt pathway. Bosn J Basic Med Sci.
19:52–59. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ren YQ, Fu F and Han J: MiR-27a modulates
radiosensitivity of triple-negative breast cancer (TNBC) cells by
targeting CDC27. Med Sci Monit. 21:1297–1303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wahdan-Alaswad RS, Cochrane DR, Spoelstra
NS, Howe EN, Edgerton SM, Anderson SM, Thor AD and Richer JK:
Metformin-induced killing of triple-negative breast cancer cells is
mediated by reduction in fatty acid synthase via miRNA-193b. Horm
Cancer. 5:374–389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cascione L, Gasparini P, Lovat F, Carasi
S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, et
al: Integrated microRNA and mRNA signatures associated with
survival in triple negative breast cancer. PLoS One. 8:e559102013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kleivi Sahlberg K, Bottai G, Naume B,
Burwinkel B, Calin GA, Børresen-Dale AL and Santarpia L: A serum
microRNA signature predicts tumor relapse and survival in
triple-negative breast cancer patients. Clin Cancer Res.
21:1207–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Braicu C, Raduly L, Morar-Bolba G,
Cojocneanu R, Jurj A, Pop LA, Pileczki V, Ciocan C, Moldovan A,
Irimie A, et al: Aberrant miRNAs expressed in HER-2 negative breast
cancers patient. J Exp Clin Cancer Res. 37:2572018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
23
|
No authors listed: The future of cancer
genomics. Nat Med. 21:992015. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dorman SN, Viner C and Rogan PK: Splicing
mutation analysis reveals previously unrecognized pathways in lymph
node-invasive breast cancer. Sci Rep. 4:70632014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lossos IS, Czerwinski DK, Alizadeh AA,
Wechser MA, Tibshirani R, Botstein D and Levy R: Prediction of
survival in diffuse large-B-cell lymphoma based on the expression
of six genes. N Engl J Med. 350:1828–1837. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43((Database Issue)): D146–D152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X: Improving microRNA target
prediction by modeling with unambiguously identified
microRNA-target pairs from CLIP-ligation studies. Bioinformatics.
32:1316–1322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang B, Li Y and Wang T: A three miRNAs
signature predicts survival in cervical cancer using bioinformatics
analysis. Sci Rep. 7:56242017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Phipson B, Lee S, Majewski IJ, Alexander
WS and Smyth GK: Robust hyperparameter estimation protects against
hypervariable genes and improves power to detect differential
expression. Ann Appl Stat. 10:946–963. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W,
Chang G, Li X, Li Q, Wang S and Wang W: MicroRNA profiling implies
new markers of chemoresistance of triple-negative breast cancer.
PLoS One. 9:e962282014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mavrogiannis AV, Kokkinopoulou I, Kontos
CK and Sideris DC: Effect of vinca alkaloids on the expression
levels of microRNAs targeting apoptosis-related genes in breast
cancer cell lines. Curr Pharm Biotechnol. 19:1076–1086. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Calvano Filho CM, Calvano-Mendes DC,
Carvalho KC, Maciel GA, Ricci MD, Torres AP, Filassi JR and Baracat
EC: Triple-negative and luminal A breast tumors: Differential
expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol.
35:7733–7741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Farazi TA, Horlings HM, Ten Hoeve JJ,
Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F,
van Kouwenhove M, et al: MicroRNA sequence and expression analysis
in breast tumors by deep sequencing. Cancer Res. 71:4443–4453.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Aure MR, Leivonen SK, Fleischer T, Zhu Q,
Overgaard J, Alsner J, Tramm T, Louhimo R, Alnæs GI, Perälä M, et
al: Individual and combined effects of DNA methylation and copy
number alterations on miRNA expression in breast tumors. Genome
Biol. 14:R1262013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Doberstein K, Bretz NP, Schirmer U, Fiegl
H, Blaheta R, Breunig C, Müller-Holzner E, Reimer D, Zeimet AG and
Altevogt P: miR-21-3p is a positive regulator of L1CAM in several
human carcinomas. Cancer Lett. 354:455–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lo TF, Tsai WC and Chen ST:
MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates
human methionine adenosyltransferases 2A and 2B and inhibits
hepatoma cell growth. PLoS One. 8:e756282013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiao W, Leng X, Zhou Q, Wu Y, Sun L, Tan
Y, Ni H, Dong X, Shen T, Liu Y and Li J: Different miR-21-3p
isoforms and their different features in colorectal cancer. Int J
Cancer. 141:2103–2111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yeh TS, Wang F, Chen TC, Yeh CN, Yu MC,
Jan YY and Chen MF: Expression profile of microRNA-200 family in
hepatocellular carcinoma with bile duct tumor thrombus. Ann Surg.
259:346–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun Y, Yang D, Xi L, Chen Y, Fu L, Sun K,
Yin J, Li X, Liu S, Qin Y, et al: Primed atypical ductal
hyperplasia-associated fibroblasts promote cell growth and polarity
changes of transformed epithelium-like breast cancer MCF-7 cells
via miR-200b/c-IKKβ signaling. Cell Death Dis. 9:1222018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pang Y, Liu J, Li X, Xiao G, Wang H, Yang
G, Li Y, Tang SC, Qin S, Du N, et al: MYC and DNMT3A-mediated DNA
methylation represses microRNA-200b in triple negative breast
cancer. J Cell Mol Med. 22:6262–6274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Choi SK, Kim HS, Jin T, Hwang EH, Jung M
and Moon WK: Overexpression of the miR-141/200c cluster promotes
the migratory and invasive ability of triple-negative breast cancer
cells through the activation of the FAK and PI3K/AKT signaling
pathways by secreting VEGF-A. BMC Cancer. 16:5702016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo G, Zhou Y, Yi W and Yi H: Expression
levels of JNK associated with polymorphic lactotransferrin
haplotypes in human nasopharyngeal carcinoma. Oncol Lett.
12:1085–1094. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Piscopo P, Albani D, Castellano AE,
Forloni G and Confaloni A: Frontotemporal lobar degeneration and
MicroRNAs. Front Aging Neurosci. 8:172016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zitman-Gal T, Green J, Pasmanik-Chor M,
Golan E, Bernheim J and Benchetrit S: Vitamin D manipulates
miR-181c, miR-20b and miR-15a in human umbilical vein endothelial
cells exposed to a diabetic-like environment. Cardiovasc Diabetol.
13:82014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dmitriev P, Barat A, Polesskaya A,
O'Connell MJ, Robert T, Dessen P, Walsh TA, Lazar V, Turki A,
Carnac G, et al: Simultaneous miRNA and mRNA transcriptome
profiling of human myoblasts reveals a novel set of myogenic
differentiation-associated miRNAs and their target genes. BMC
Genomics. 14:2652013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Asher G, Tsvetkov P, Kahana C and Shaul Y:
A mechanism of ubiquitin-independent proteasomal degradation of the
tumor suppressors p53 and p73. Genes Dev. 19:316–321. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao Q, Song W, He DY and Li Y:
Identification of key gene modules and pathways of human breast
cancer by co-expression analysis. Breast Cancer. 25:213–223. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Wei L, Yu J, Li G, Zhang X, Wang
A, He Y, Li H and Yin D: Targeting of the β6 gene to suppress
degradation of ECM via inactivation of the MAPK pathway in breast
adenocarcinoma cells. Oncol Rep. 32:1787–1795. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shah SP, Roth A, Goya R, Oloumi A, Ha G,
Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al: The clonal
and mutational evolution spectrum of primary triple-negative breast
cancers. Nature. 486:395–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bartholomeusz C, Gonzalez-Angulo AM, Liu
P, Hayashi N, Lluch A, Ferrer-Lozano J and Hortobágyi GN: High ERK
protein expression levels correlate with shorter survival in
triple-negative breast cancer patients. Oncologist. 17:766–774.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Masuda H, Zhang D, Bartholomeusz C,
Doihara H, Hortobagyi GN and Ueno NT: Role of epidermal growth
factor receptor in breast cancer. Breast Cancer Res Treat.
136:331–345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tilch E, Seidens T, Cocciardi S, Reid LE,
Byrne D, Simpson PT, Vargas AC, Cummings MC, Fox SB, Lakhani SR, et
al: Mutations in EGFR, BRAF and RAS are rare in triple-negative and
basal-like breast cancers from Caucasian women. Breast Cancer Res
Treat. 143:385–392. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ,
Kim YJ, Kim JH, Kang E, Kim SW, Kim IA and Park SY: High EGFR gene
copy number predicts poor outcome in triple-negative breast cancer.
Mod Pathol. 27:1212–1222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Burstein MD, Tsimelzon A, Poage GM,
Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK,
Hilsenbeck SG, Chang JC, et al: Comprehensive genomic analysis
identifies novel subtypes and targets of triple-negative breast
cancer. Clin Cancer Res. 21:1688–1698. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
López-Ozuna VM, Hachim IY, Hachim MY,
Lebrun JJ and Ali S: Prolactin Pro-differentiation pathway in
triple negative breast cancer: Impact on prognosis and potential
therapy. Sci Rep. 6:309342016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Martignetti L, Tesson B, Almeida A,
Zinovyev A, Tucker GC, Dubois T and Barillot E: Detection of miRNA
regulatory effect on triple negative breast cancer transcriptome.
BMC Genomics. 16 (Suppl):S42015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pham TND, Perez White BE, Zhao H,
Mortazavi F and Tonetti DA: Protein kinase C α enhances migration
of breast cancer cells through FOXC2-mediated repression of
p120-catenin. BMC Cancer. 17:8322017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Di Modica M, Regondi V, Sandri M, Iorio
MV, Zanetti A, Tagliabue E, Casalini P and Triulzi T: Breast
cancer-secreted miR-939 downregulates VE-cadherin and destroys the
barrier function of endothelial monolayers. Cancer Lett.
384:94–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lim HC, Multhaupt HA and Couchman JR: Cell
surface heparan sulfate proteoglycans control adhesion and invasion
of breast carcinoma cells. Mol Cancer. 14:152015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li Q and Mattingly RR: Restoration of
E-cadherin cell-cell junctions requires both expression of
E-cadherin and suppression of ERK MAP kinase activation in
Ras-transformed breast epithelial cells. Neoplasia. 10:1444–1458.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wendt MK, Taylor MA, Schiemann BJ and
Schiemann WP: Down-regulation of epithelial cadherin is required to
initiate metastatic outgrowth of breast cancer. Mol Biol Cell.
22:2423–2435. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mali AV, Joshi AA, Hegde MV and Kadam SS:
Enterolactone modulates the ERK/NF-κB/Snail signaling pathway in
triple-negative breast cancer cell line MDA-MB-231 to revert the
TGF-β-induced epithelial-mesenchymal transition. Cancer Biol Med.
15:137–156. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Javle MM, Gibbs JF, Iwata KK, Pak Y,
Rutledge P, Yu J, Black JD, Tan D and Khoury T:
Epithelial-mesenchymal transition (EMT) and activated extracellular
signal-regulated kinase (p-Erk) in surgically resected pancreatic
cancer. Ann Surg Oncol. 14:3527–3533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wen S, Hou Y, Fu L, Xi L, Yang D, Zhao M,
Qin Y, Sun K, Teng Y and Liu M: Cancer-associated fibroblast
(CAF)-derived IL32 promotes breast cancer cell invasion and
metastasis via integrin β3-p38 MAPK signalling. Cancer Lett.
442:320–332. 2019. View Article : Google Scholar : PubMed/NCBI
|