Introduction
Bladder cancer (BC), one of the most common
malignancies worldwide, is classified into two subtypes based on
cancer cell infiltration into the muscle layer of the bladder.
Non-muscle invasive BC (NMIBC) is less aggressive but has a high
recurrence rate, whereas muscle invasive BC (MIBC) tends to
metastasize and has a relatively poor prognosis (1–3). High
throughput techniques such as microarray analysis and next
generation sequencing, which are used commonly in the fields of
genetics and epigenetics, have identified several genes involved in
cancer pathogenesis, and have led to identification of cancer
biomarkers and to development of novel effective gene targeted
therapies (4). In a previous study,
we used next generation sequencing and miRNA microarray assays to
identify several miRNAs and their target genes that are
differentially expressed in BC (5).
We found that a novel gene, Rho GTPase-activating protein 9
(ARHGAP9), is down-regulated in BC. In addition,
hsa-miR-3620, which interacts with ARHGAP9, is
up-regulated.
Rho GTPases are key regulators of the actin
cytoskeleton, which plays an important role in cell adhesion and
migration. The switch mechanism of Rho GTPases is controlled by
binding to GTP or GDP (6–8). ARHGAP9 contains a diverse combination
of functional protein domains, including the RhoGAP, SH3, WW, and
PH domains (9). Binding of the
RhoGAP domain to GTP-bound Rho proteins accelerates GTPase
activity, and defective Rho GTPase signaling is implicated in
tumorigenesis and metastasis (10,11).
Silencing ARHGAP9 inhibits proliferation, migration, and invasion
of breast cancer cells (12).
Activated ARHGAP9 inhibits adhesion of a human leukemia cell line,
KG-1, to fibronectin and collagen through activation of cdc42 and
Rac1 but not RhoA (6).
Here, we asked whether ARHGAP9 is a novel
prognostic biomarker for BC. We used real-time polymerase chain
reaction (PCR) to compare expression of ARHGAP9 mRNA in
human BC and control tissues (the latter comprised normal tissue
surrounding BC and normal bladder mucosa); and analyzed its ability
to predict prognosis of NMIBC and MIBC. ARHGAP9, known as a MAP
kinase docking protein, was encoded by ARHGAP9 gene, which
shares 16 bases with Gli1 in their 3′ ends (9,13).
Accordingly, we asked whether ARHGAP9 plays a role in the
MAPK and Hedgehog signaling pathways.
Materials and methods
Patients and tissue samples
The biospecimens used in the present study were
provided by the Chungbuk National University Hospital, a member of
the National Biobank of Korea, which is supported by the Ministry
of Health, Welfare, and Family Affairs. The study was approved by
the Institutional Review Board at Chungbuk National University
(GR2010-12-010), and the experiments were undertaken with the
informed written consents of all participants. The study
methodologies conformed with the standards set by the Declaration
of Helsinki. The baseline characteristics of the case subjects
(n=237 bladder tissue samples) are shown in Table I. Among these, 140 samples were from
primary BC patients and were histologically verified as
transitional cell carcinomas; the remaining 97 samples used as the
control set comprised normal bladder mucosa or normal tissues from
the area surrounding BC. To reduce the chances of confounding
factors affecting the analyses, patients diagnosed with concomitant
carcinoma in situ or carcinoma in situ lesions alone were excluded.
Voided urine cytology was tested before surgical treatment to
assist BC diagnosis and/or prognosis. Fresh-frozen specimens were
obtained during surgical resection of transitional cell carcinoma
at Chungbuk National University Hospital. All tumors were
macro-dissected, typically within 15 min of surgical resection.
Each specimen was confirmed by pathological analysis of a part of
fresh-frozen specimens obtained from radical cystectomy and
transurethral resection of bladder tumor (TURBT). Tumors were
staged (2002 TNM Classification) and graded (2004 WHO
Classification), according to standard criteria (14). Clinically metastatic disease and
non-cystectomy cases were not excluded from the study. Each patient
was followed and managed suggested management according to standard
recommendations (15–17). Surveillance was performed by
cystoscopic examination and upper urinary tract imaging in
accordance with European Association of Urology guidelines
(16). Recurrence was defined as
relapse of primary NMIBC of the same pathologic stage, and
progression of NMIBC and MIBC was defined as TNM stage progression
after disease recurrence. The mean follow-up period for NMIBC
patients was 72.95 months (range, 3.2–172.2). The mean follow-up
period for MIBC patients was 36.18 months (range, 3.0–141.4).
 | Table I.Clinicopathological features of
primary BC patient and control tissues (surrounding normal tissues
and normal bladder mucosae). |
Table I.
Clinicopathological features of
primary BC patient and control tissues (surrounding normal tissues
and normal bladder mucosae).
|
| BC (140) |
|
|
|---|
|
|
|
|
|
|---|
| Variables | NMIBC | MIBC | Control | P-value |
|---|
| No. | 97 | 43 | 97 |
|
| Mean age ± SD | 63.45±13.79 | 67.60±9.84 | 61.98±14.32 | 0.083a |
| Sex (%) |
|
|
| 0.975a |
|
Male | 80 (82.5%) | 36 (83.7%) | 81 (83.5%) |
|
|
Female | 17 (17.5%) | 7 (16.3%) | 16 (16.5%) |
|
| Operation (%) |
|
|
|
<0.001b |
|
TUR-BT | 97 (100.0%) | 17 (39.5%) |
|
|
| Radical
cystectomy | 0 | 26 (60.5%) |
|
|
| Tumor size (%) |
|
|
| 0.003b |
| ≤1
cm | 16 (16.5%) | 2 (4.7%) |
|
|
| 2–3
cm | 37 (38.1%) | 11 (25.6%) |
|
|
| >3
cm | 37 (38.1%) | 28 (65.1%) |
|
|
| Multiplicity
(%) |
|
|
| 0.108b |
|
Single | 52 (53.6%) | 30 (69.8%) |
|
|
|
2–7 | 28 (28.9%) | 7 (16.3%) |
|
|
|
>7 | 11 (11.3%) | 4 (9.3%) |
|
|
| Grade, 2004 WHO
grading system (%) |
|
|
|
<0.001b |
|
Low | 72 (74.2%) | 8 (18.6%) |
|
|
|
High | 25 (25.8%) | 35 (81.4%) |
|
|
| Stage (%) |
|
|
|
<0.001b |
|
TaN0M0 | 26 (26.8%) |
|
|
|
|
T1N0M0 | 71 (73.2%) |
|
|
|
|
T2N0M0 |
| 13 (30.2%) |
|
|
|
T3N0M0 |
| 6 (14.0%) |
|
|
| T≥4 or N≥1 or
M1 |
| 24 (55.8%) |
|
|
| Chemotherapy
(%) |
|
|
|
<0.001b |
| No | 97 (100.0%) | 23 (53.5%) |
|
|
|
Yes | 0 | 20 (46.5%) |
|
|
| BCG therapy
(%) |
|
|
|
<0.001b |
| No | 56 (57.7%) | 38 (88.4%) |
|
|
|
Yes | 40 (41.2%) | 5 (11.6%) |
|
|
| Recurrence, no. of
patients (%) |
| No | 59 (60.8%) | – |
|
|
|
Yes | 38 (39.2%) | – |
|
|
| Progression, no. of
patients (%) |
|
|
| 0.126b |
| No | 79 (81.4%) | 30 (69.8%) |
|
|
|
Yes | 18 (18.6%) | 13 (30.2%) |
|
|
| Survival, no. of
patients (%) |
|
|
| 0.009b |
|
Alive | 64 (66.0%) | 21 (48.8%) |
|
|
|
Non-cancer-specific death | 18 (18.6%) | 3 (7.0%) |
|
|
|
Cancer-specific death | 15 (15.5%) | 19 (44.2%) |
|
|
| Mean
follow-up, months (range) | 72.95
(3.20–172.20) | 36.18
(3.00–141.40) |
|
|
RNA extraction
Total RNA was extracted from tissues using TRIzol
reagent (Invitrogen), as described previously (18), and stored at −80°C. Next, cDNA was
synthesized from 1 µg of total RNA using a First Strand cDNA
Synthesis kit (Clontech, TAKARA), according to the manufacturer's
protocol.
Microarray analysis
Five hundred nanograms of total RNA was used for
labeling and hybridization prior to analysis, according to the
manufacturer's protocols (Illumina). After the bead chips were
scanned with an Illumina Bead Array Reader, the Robust Multiarray
Average in R package was used to perform global correction,
quantile normalization, and median polish summarization of the
microarray data. P-values (t test) were calculated from bead
mRNA signal intensities (19–21). The
full set of microarray data set are available online at http://www.ncbi.nlm.nih.gov/geo/under
data series accession number GSE13507 (21).
mRNA sequencing
Total sequencing reads were subjected to
preprocessing as follows: Adapter trimming was performed using
cutadapt with default parameters, and quality trimming (Q30) was
performed using FastQC with default parameters. Processed reads
were mapped to the human reference genome [Ensembl 72 (GRCh37:
hg19)] using tophat and cufflink with default parameters (22). Fragments Per Kilobase of exon per
million fragments Mapped (FPKM) values were normalized and
quantitated using R package Tag Count Comparison (TCC) (23) to determine statistical significance
(e.g., P and Q values) and differential expression
(e.g., -fold changes).
Quantitative PCR analysis
Tissue mRNAs were amplified by quantitative PCR
performed using a Rotor Gene 6000 instrument (Qiagen) and
quantified using the 2−∆∆cq method (24). QuantitativePCR reactions were carried
out using the SYBR Premix Ex Taq II (Clontech, TAKARA). The
following primers were used to amplify candidate genes:
ARHGAP9 (Gene ID: ENSG00000123329), sense,
5′-CAGAGCAGTGCCTCTCTC-3′ (18 bp, Tm 58°C); antisense,
5′-CTGCTGGGTCAGATGTCTC-3′ (19 bp, Tm 58°C) and the amplicon size
was 179 bp. The control GAPDH (Gene ID: ENSG00000111640)
primers were as follows: sense, 5′-CATGTTCGTCATGGGTGTGA-3′ (20 bp,
Tm 60°C); antisense, 5′-ATGGCATGGACTGTGGTCAT-3′ (20 bp, Tm 60°C)
and the amplicon size was 156 bp. The PCR reaction was performed in
a final volume of 10 µl, comprising 5 µl of 2× SYBR Premix EX Taq
buffer, 0.5 µl of each 5′and 3′ primer (10 pM/µl), and 2 µl, of
sample cDNA. A known concentration of the PCR product was then
10-fold serially diluted from 100 pg/µl to 0.1 pg/µl and used to
establish a standard curve. The real-time PCR conditions were as
follows: 1 cycle at 96°C for 20 sec, followed by 40 cycles of 3 sec
at 96°C for denaturation, 15 sec at 60°C for annealing, and 15 sec
at 72°C for extension. The melting program was performed at 72–95°C
at a heating rate of 1°C per 45 sec. Rotor-Gene Q software 2.3.1.49
was used for capturing and analyzing spectral data. All samples
were run in triplicate. Gene expression was normalized to the
expression of GAPDH.
Statistical analysis
To reduce variation among microarrays, the intensity
values for each microarray were rescaled using a quantile
normalization method (19). Gene
expression values were loge-transformed and median-centered across
samples. The significance of various clinicopathological variables
was evaluated using univariate and multivariate Cox proportional
hazard regression models. Hazard ratios (HRs) and 95% confidence
intervals (CIs) were calculated to investigate relative risk.
Survival curves to determine the prognostic value of the genetic
biomarker were plotted using the Kaplan-Meier method and compared
using the log-rank test. The Kruskal-Wallis H test and
Mann-Whitney U test were used to examine expression of
ARHGAP9 in BC tissues versus control tissues. Correlations
between ARHGAP9 and genes involved in the MAPK and Hedgehog
signaling pathways were examined by calculating non-parametric
Spearman's correlation coefficients. Statistical analyses were
performed using IBM SPSS Statistics ver. 20.0 (IBM) and GraphPad
Prism 7 (GraphPad Software). P-values <0.05 were considered
significant.
Results
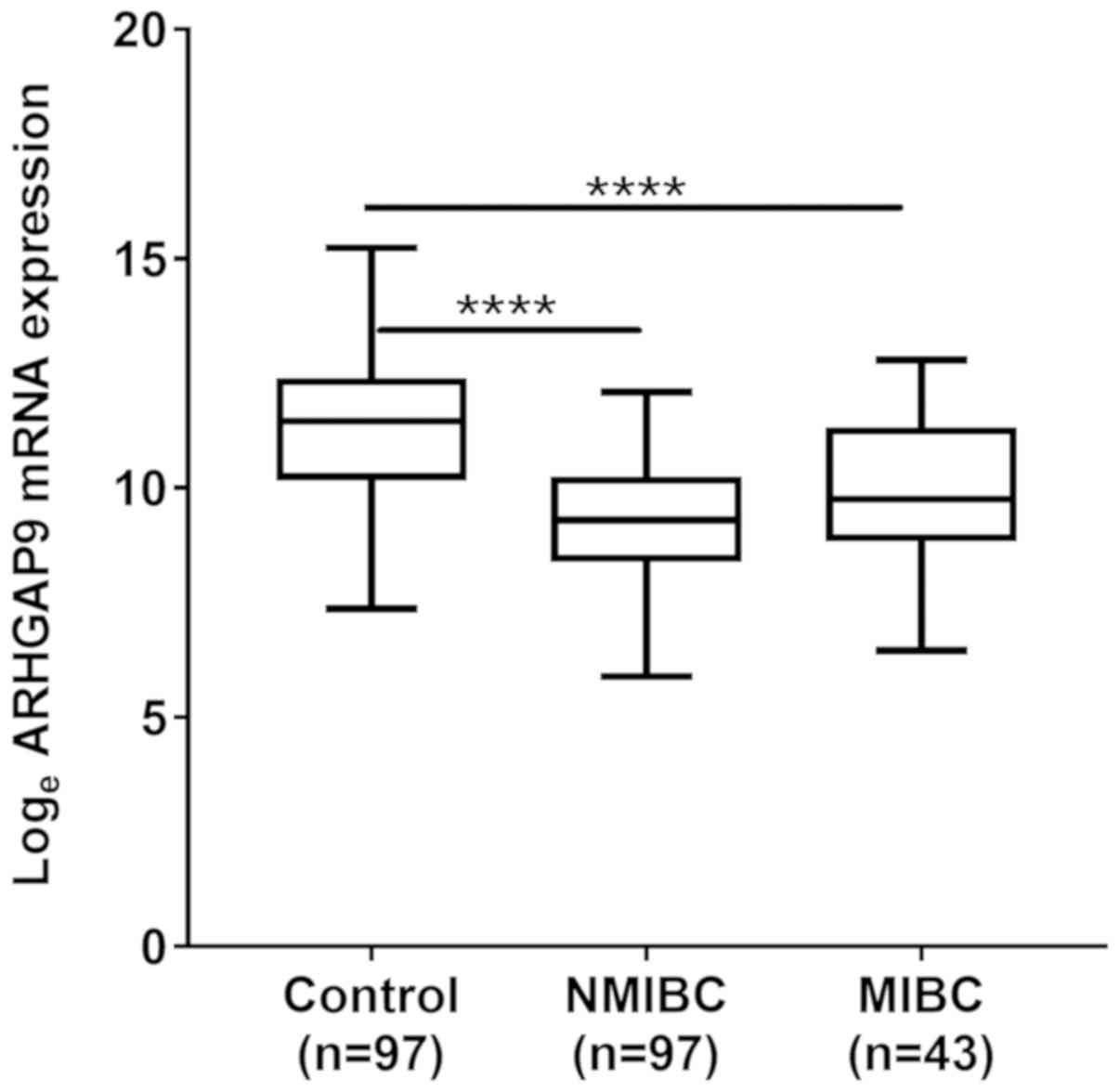
Expression of ARHGAP9 mRNA in BC
tissue
Microarray analysis revealed that expression of mRNA
encoding ARHGAP9 in BC tissues was lower than that in
control samples. The validation test showed that the real-time PCR
results were identical to those of the microarray, i.e., expression
of mRNA encoding ARHGAP9 was significantly lower in NMIBC
and MIBC tissues than in normal control tissues (P<0.001;
Fig. 1).
Expression of ARHGAP9 correlates with
NMIBC prognosis
Univariate and multivariate Cox regression analyses
revealed that expression of ARHGAP9 in NMIBC patients was an
independent predictor of recurrence-free survival (RFS) (HR, 2.436;
95% CI, 1.132–5.243; P=0.023; Table
II). Kaplan-Meier analysis demonstrated that NMIBC patients
with ARHGAP9 expression levels in the upper 50th percentile
experienced less recurrence than those with expression levels in
the lower 50th percentile (log-rank test, P=0.043; Fig. 2A). Particularly, for T1 high
grade(HG) BC patients, univariate and multivariate Cox regression
analysis identified ARHGAP9 expression as an independent
risk factor for T1HG BC recurrence (HR, 7.264; 95% CI,
1.291–45.091; P=0.025) and progression (HR, 14.987; 95% CI,
1.093–205.567; P=0.043; Table
III). The RFS and progression-free survival (PFS) of T1HG BC
patients with ARHGAP9 expression levels in the upper 50th
percentile experienced less recurrence and progression than those
with expression levels in the lower 50th percentile (log-rank test,
P=0.013 and 0.026 respectively; Fig. 2B
and C).
 | Table II.Univariate and multivariate Cox
regression analysis to predict NMIBC recurrence. |
Table II.
Univariate and multivariate Cox
regression analysis to predict NMIBC recurrence.
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age |
| ≤70 (Ref.) vs.
>70 | 2.994
(1.579–5.680) | 0.001a | 1.727
(0.820–3.637) | 0.151 |
| Sex |
| Male (Ref.) vs.
female | 1.314
(0.577–2.993) | 0.516 |
|
|
| Tumor size |
|
|
|
|
| ≤1
cm | Ref. | 0.028a | Ref. | 0.574 |
| 2–3
cm | 1.700
(0.474–6.100) | 0.416 | 1.251
(0.341–4.593) | 0.736 |
| >3
cm | 3.686
(1.093–12.425) | 0.035a | 1.779
(0.484–6.547) | 0.386 |
| Multiplicity |
|
|
|
|
|
Single | Ref. | 0.141 |
|
|
|
2–7 | 1.071
(0.479–2.395) | 0.867 |
|
|
|
>7 | 2.383
(0.985–5.767) | 0.054 |
|
|
| 2004 WHO Grade Low
(Ref.) vs. high | 2.450
(1.275–4.708) | 0.007a | 1.823
(0.809–3.568) | 0.147 |
| Stage |
| Ta (Ref.) vs.
T1 | 2.938
(1.144–7.540) | 0.025a | 2.347
(0.803–6.857) | 0.119 |
| BCG |
| No (Ref.) vs.
yes | 1.918
(1.009–3.647) | 0.047a | 1.744
(0.852–3.568) | 0.128 |
| ARHGAP9
expression High expression (Ref.) vs. Low expression | 1.939
(1.009–3.726) | 0.047a | 2.436
(1.132–5.243) | 0.023a |
 | Table III.Univariate and multivariate Cox
regression analysis to predict T1 high grade NMIBC recurrence and
progression. |
Table III.
Univariate and multivariate Cox
regression analysis to predict T1 high grade NMIBC recurrence and
progression.
|
| Recurrence | Progression |
|---|
|
|
|
|
|---|
|
| Univariate Cox
analysis | Multivariate Cox
analysis | Univariate Cox
analysis | Multivariate Cox
analysis |
|---|
|
|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age |
| ≤70 (Ref.) vs.
>70 | 2.342
(0.625–8.776) | 0.207 |
|
| 1.567
(0.390–6.297) | 0.527 |
|
|
| Sex |
| Male (Ref.) vs.
female | 1.327
(0.342–5.154) | 0.682 |
|
| 2.748
(0.548–13.781) | 0.219 |
|
|
| Tumor size |
| ≤1
cm | Ref. | 0.976 |
|
| Ref. | 0.468 |
|
|
| 2–3
cm | 29604.104 | 0.948 |
|
| 9687.884 | 0.968 |
|
|
|
|
(0.000–2.839×10138) |
|
|
|
(0.000–3.269×10201) |
|
|
|
| >3
cm | 25622.270 | 0.949 |
|
| 36480.741 | 0.964 |
|
|
|
|
(0.000–2.454×10138) |
|
|
|
(0.000–1.226×10202) |
|
|
|
| Multiplicity |
|
Single | Ref. | 0.618 |
|
| Ref. | 0.850 |
|
|
|
2–7 | 1.450
(0.417–5.040) | 0.559 |
|
| 1.548
(0.345–6.943) | 0.568 |
|
|
|
>7 | 2.933
(0.296–29.074) | 0.358 |
|
| 0.000
(0.000–0.000) | 0.991 |
|
|
| BCG |
| No (Ref.) vs.
yes | 1.247
(0.336–4.624) | 0.741 |
|
| 0.459
(0.119–1.766) | 0.257 |
|
|
| ARHGAP9
expression High (Ref.) vs. low expression | 5.126
(1.247–21.066) | 0.023a | 7.264 | 0.025a | 6.041
(1.026–35.571) | 0.047a | 14.987 | 0.043a |
|
|
|
| (1.291–45.019) |
|
|
|
(1.093–205.567) |
|
Expression of ARHGAP9 correlates with
MIBC prognosis
For MIBC patients, univariate and multivariate Cox
regression analysis identified ARHGAP9 expression as an
independent risk factor for disease progression (HR, 5.241; 95% CI,
1.456–18.870; P=0.011) and cancer-specific death (HR, 2.923; 95%
CI, 1.192–7.163; P=0.019) (Tables
IV and V). PFS and cancer
specific survival (CSS) of patients with ARHGAP9 expression
in the upper 50th percentile were significantly higher than those
of patients in the lower 50th percentile (log-rank test, P=0.020
and 0.031, respectively; Fig. 3A and
B).
 | Table IV.Univariate and multivariate Cox
regression analysis to predict MIBC progression. |
Table IV.
Univariate and multivariate Cox
regression analysis to predict MIBC progression.
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age |
| ≤70 (Ref.) vs.
>70 | 1.302
(0.432–3.926) | 0.639 |
|
|
| Sex |
| Male (Ref.) vs.
female | 5.625
(1.766–17.912) | 0.003a | 7.255
(2.062–25.528) | 0.002a |
| Operation |
| TURBT (Ref.) vs.
Radical cystectomy | 0.948
(0.309–2.905) | 0.926 |
|
|
| Tumor size |
| ≤1
cm | Ref. | 0.406 |
|
|
| 2–3
cm | 12417.036
(0.000–2.033×10143) | 0.954 |
|
|
| >3
cm | 35009.555
(0.000–5.718×10143) | 0.949 |
|
|
| Multiplicity |
|
Single | Ref. | 0.507 |
|
|
|
2–7 | 0.358
(0.046–2.800) | 0.328 |
|
|
|
>7 | 1.483
(0.324–6.787) | 0.611 |
|
|
| 2004 WHO Grade |
| Low (Ref.) vs.
high | 31.010
(0.132–7298.224) | 0.218 |
|
|
| Stage |
| T2 | Ref. | 0.851 |
|
|
| T3 | 1.630
(0.297–8.958) | 0.574 |
|
|
| T4 or N1 or M1 | 1.229
(0.358–4.222) | 0.744 |
|
|
| Chemotherapy |
| No (Ref.) vs.
yes | 3.912
(1.076–14.218) | 0.038a | 2.859
(0.752–10.868) | 0.123 |
| ARHGAP9
expression High expression (Ref.) vs. Low expression | 3.818
(1.145–12.733) | 0.029a | 5.241
(1.456–18.870) | 0.011a |
 | Table V.Univariate and multivariate Cox
regression analysis for predicting the cancer-specific survival of
patients with MIBC. |
Table V.
Univariate and multivariate Cox
regression analysis for predicting the cancer-specific survival of
patients with MIBC.
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age |
| ≤70 (Ref.) vs.
>70 | 1.860
(0.791–4.371) | 0.155 |
|
|
| Gender |
| Male (Ref.) vs.
female | 3.379
(1.273–8.967) | 0.014a | 4.046
(1.491–10.976) | 0.006a |
| Operation |
| TURBT (Ref.) vs.
Radical cystectomy | 1.026
(0.435–2.417) | 0.954 |
|
|
| Tumor size |
| ≤1
cm | Ref. | 0.386 |
|
|
| 2–3
cm | 14923.217
(0.000–1.565E+115) | 0.941 |
|
|
| >3
cm | 32178.497
(0.000–3.369E+115) | 0.937 |
|
|
| Multiplicity |
|
Single | Ref. | 0.730 |
|
|
|
2–7 | 0.709
(0.206–2.438) | 0.585 |
|
|
|
>7 | 0.611
(0.137–2.725) | 0.519 |
|
|
| 2004 WHO Grade |
| Low (Ref.) vs.
high | 3.009
(0.699–12.950) | 0.139 |
|
|
| Stage |
| T2 | Ref. | 0.480 |
|
|
| T3 | 0.909
(0.181–4.563) | 0.908 |
|
|
| T4 or
N1 or M1 | 1.671
(0.641–4.358) | 0.294 |
|
|
| Chemotherapy |
| No (Ref.) vs.
yes | 1.482
(0.633–3.472) | 0.365 |
|
|
| ARHGAP9
expression High expression (Ref.) vs. Low expression | 2.554
(1.058–6.163) | 0.037a | 2.923
(1.192–7.163) | 0.019a |
Relationship between ARHGAP9 and genes
regulating the MAPK and Hedgehog signaling pathways in BC
To identify whether expression of ARHGAP9
correlates with that of genes regulating the MAPK and Hedgehog
signaling pathways, we undertook gene network depiction and
analysis using the GeneMANIA (http://www.genemania.org) web tool. We selected seven
genes (ARHGAP9, epidermal growth factor receptor
(EGFR), mitogen-activated protein kinase 1 (MAPK1,
also known as ERK2), mitogen-activated protein kinase 14
(MAPK14, also known as p38α), mitogen-activated
protein kinase kinase 3 (MKK3), mitogen-activated protein
kinase kinase 6 (MKK6), and glioma-associated oncogene
homolog 1 (Gli1)) showing potential inter-correlations
(Supplementary Fig. S1).
Non-parametric Spearman's correlation coefficients (based on
microarray data) identified interactions among ARHGAP9, EGFR,
MAPK1 (ERK2), MAPK14 (p38α), MKK3,
MKK6, and Gli1. Table VI
shows that expression of ARHGAP9 correlated positively with
that of Gli1, which regulates the Hedgehog signaling
pathway. In addition, ARHGAP9 interacted with MKK6
and MAPK1 (ERK2), both of which are essential
components of the MAPK signal transduction pathway (P<0.05 for
both).
 | Table VI.Spearman correlation coefficients of
Gli1, ARHGAP9, EGFR, MKK3, MKK6, MAPK1 (ERK2) and
MAPK14 (p38α) in BC. |
Table VI.
Spearman correlation coefficients of
Gli1, ARHGAP9, EGFR, MKK3, MKK6, MAPK1 (ERK2) and
MAPK14 (p38α) in BC.
|
| Gli1 | ARHGAP9 | EGFR | MKK3 | MKK6 | MAPK1 (ERK2) | MAPK14
(p38α) |
|---|
| Gli1 |
|
Spearman's Rho | 1.000 | 0.518b | −0.009 | 0.099 | −0.042 | 0.178a | −0.202b |
|
P-value | . | 0.000 | 0.911 | 0.205 | 0.589 | 0.022 | 0.009 |
| ARHGAP9 |
|
Spearman's Rho | 0.518b | 1.000 | 0.084 | 0.125 | −0.168a | 0.233b | −0.138 |
|
P-value | 0.000 | . | 0.283 | 0.109 | 0.031 | 0.003 | 0.076 |
| EGFR |
|
Spearman's Rho | −0.009 | 0.084 | 1.000 | 0.194a | −0.118 | 0.301b | 0.192b |
|
P-value | 0.911 | 0.283 | . | 0.012 | 0.130 | 0.000 | 0.013 |
| MKK3 |
|
Spearman's Rho | 0.099 | 0.125 | 0.194a | 1.000 | 0.101 | 0.327b | 0.315b |
|
P-value | 0.205 | 0.109 | 0.012 | . | 0.195 | 0.000 | 0.000 |
| MKK6 |
|
Spearman's Rho | −0.042 | −0.168a | −0.118 | 0.101 | 1.000 | −0.093 | −0.056 |
|
P-value | 0.589 | 0.031 | 0.130 | 0.195 | . | 0.233 | 0.472 |
|
MAPK1(ERK2) |
|
Spearman's Rho | 0.178a | 0.233b | 0.301b | 0.327b | −0.093 | 1.000 | 0.167a |
|
P-value | 0.022 | 0.003 | 0.000 | 0.000 | 0.233 | . | 0.032 |
| MAPK14
(p38α) |
|
Spearman's Rho | −0.202b | −0.138 | 0.192a | 0.315b | −0.056 | 0.167a | 1.000 |
|
P-value | 0.009 | 0.076 | 0.013 | 0.000 | 0.472 | 0.032 | . |
Discussion
ARHGAP9 sits adjacent to Gli1 on human
chromosome 12q13.3; two genes have overlapping 16 bases in their
3′-ends (13), suggesting that
Gli1 and ARHGAP9 may regulate each other. Studies
suggest that Gli1 is down-regulated in BC (25); indeed, Gli1 is considered to
be the most reliable biomarker of Hedgehog pathway activity
(25–27). The microarray data presented herein
shows that mRNA expression of Gli1 and ARHGAP9 were
down-regulated in BC tissues, and that there was a positive
correlation between the two (Table
VI); this indicates that ARHGAP9, which lies adjacent to
Gli1, might be a novel regulator of Gli1.
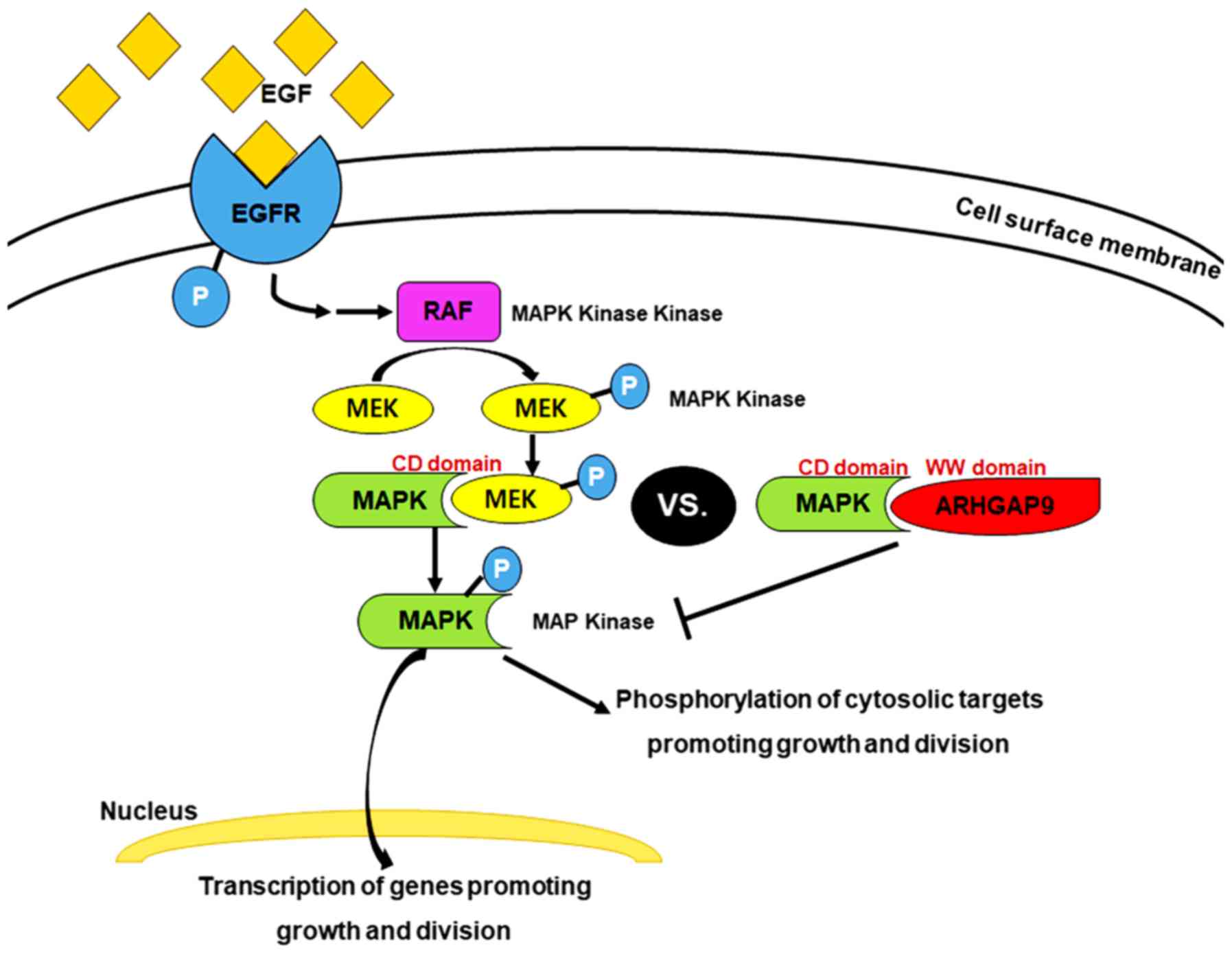
As a novel MAP kinase docking protein, ARHGAP9
associates specifically with ERK2 and p38α via complementarily
charged residues within the WW domain of ARHGAP9 and the CD domains
of ERK2 and p38α. This interaction suppresses MAP kinase
activation; but does not affect that of RhoGAP (9). MAPK activation is a common event in
tumor progression and metastasis. Inhibition of ERK1/2 and p38 MAP
kinase pathways in BC could inhibit proliferation and growth
(28). The key target in this signal
transduction pathway is EGFR, a receptor tyrosine kinase (29). Binding of EGF to EGFR in BC activates
EGFR, which is already overexpressed; furthermore, the Ras-MAPK
pathway is activated through the MAPK/ERK pathway. This continuous
‘ON’ status of MAPK signaling results in overexpression of MEK2 and
MKK3, 4, and 6, which lie upstream of MAP kinase (i.e., ERK2 and
p38α) and activate ERK2 and p38α, leading to reduced interaction
between ARHGAP9 and ERK2 or p38α in BC (this is probably
attributable to competitive displacement by overexpressed docking
proteins) (Fig. 4). The microarray
data revealed a competitive correlation between expression of
ARHGAP9 mRNA and that of MKK6, and a positive
correlation between ARHGAP9 and ERK2 (Table VI). These findings suggest that
ARHGAP9 acts as a tumor suppressor gene in BC. EGFR acts as
a receptor molecule in the MAPK signaling pathway, and is a
prognostic marker for many cancer types, including BC (30). Our previous study showed that
EGFR is a progression-related gene in MIBC; increased
expression of EGFR is associated with a poor prognosis
(31). Here, we found that lower
expression of ARHGAP9 was related to poor PFS and CSS
(Fig. 3A and B), which is consistent
with previous results. However, no definitive evidence has been
demonstrated on the recurrence rate of MIBC after radical
cystectomy, and the definition of local and distant recurrence is
not standardized (32). In our
preliminary study, twenty-six MIBC patients received radical
cystectomy and only three of them were manifested recurrence, such
result should be examined in further study with more samples for
the statistically significant validation of the survival
analysis.
Furthermore, the ARHGAP9 mRNA expression
could predict the recurrence of NMIBC, that is, lower expression of
ARHGAP9 was related to poor RFS (Fig. 2A). In particular, T1HG BC patients
with higher expression of ARHGAP9 experienced less
recurrence and progression (Fig. 2B and
C). A more careful monitoring and optimal treatment
recommendation should be implemented for T1HG BCs because of their
highly recurrent nature and risk of progression to MIBC (33), which highlights the strategy for
predicting prognosis. This study indicates that ARHGAP9 gene
has a good performance in predicting prognosis of T1HG BC
patients.
In addition, TCGA data from the Human Pathology
Atlas (https://www.proteinatlas.org/ENSG00000123329-ARHGAP9/pathology/tissue/urothelial+cancer)
show that BC patients with higher expression of ARHGAP9 mRNA
tend to survive longer, though it is not statistically significant
(P=0.069). On the basis of the results of this study, we can
conclude that ARHGAP9 regulates growth and proliferation of
BC by regulating the MAPK signaling pathway. Future studies should
use real-time PCR assays to validate the results of microarray
tests to confirm reliability of the data. For a better
understanding of ARHGAP9, its protein levels in BC should be
evaluated and the experimental samples should be increased to
reduce the statistical limitations in the future. Moreover, the
function of miR-3620, which interacted with ARHGAP9
mRNA, could be clarified by validating the function of
ARHGAP9 in the future.
In conclusion, our findings provide a novel tumor
suppressor gene in BC, which could be served as an independent
prognostic marker for stratification of NMIBC and MIBC patients
into favorable and poor prognosis. Moreover, a new paradigm in BC
tumorigenesis and pathogenesis is estimated, since this novel gene
seems to involve in the crucial tumorigenesis signaling
pathways.
Supplementary Material
Supporting Data
Acknowledgements
The biospecimens used in the present study were
provided by the Chungbuk National University Hospital, a member of
the National Biobank of Korea, which is supported by the Ministry
of Health, Welfare, and Family Affairs. All samples derived from
the National Biobank of Korea were obtained with informed consent
under institutional review board-approved protocols. The authors
would like to thank Ms. Eun-Ju Shim from the National Biobank of
Korea at Chungbuk National University Hospital for preparing
samples and her excellent technical assistance.
Funding
The present study was supported by the International
Science and Business Belt Program of the Ministry of Science, ICT
and Future Planning (grant no. 2015-DD-RD-0070); the National
Research Foundation of Korea funded by the Korean government (grant
no. 2018R1A2B2005473); and the Basic Science Research Program of
the National Research Foundation of Korea, funded by the Ministry
of Education (grant no. 2017R1D1A1B03033629).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XMP, PJ, SJY and WJK designed the study and all
experiments. XMP performed the experiments. YHK, YJB, YX, SPS and
SKM collected patient samples. XMP, CY, HWK and WTK assisted with
data collection. XMP, JYL, IYK, YHC, EJC and SJY analyzed the data.
WJK provided funding. XMP, SJY and WJK wrote the manuscript.
Ethics approval and consent to
participate
The collection and analysis of all samples were
approved by the Institutional Review Board at Chungbuk National
University (approval no. GR2010-12-010). The study methodologies
conformed with the standards set by the Declaration of Helsinki.
All samples derived from the National Biobank of Korea were
obtained with informed consent under institutional review
board-approved protocols.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ARHGAP9
|
Rho GTPase-activating protein 9
|
|
BC
|
Bladder cancer
|
|
CI
|
confidence interval
|
|
CIS
|
carcinoma in situ
|
|
CSS
|
cancer-specific survival
|
|
EGFR
|
epidermal growth factor receptor
|
|
Gli1
|
glioma-associated oncogene homolog
1
|
|
HR
|
hazard ratio
|
|
MAPK1
|
mitogen-activated protein kinase 1
(also known as ERK2)
|
|
MAPK14
|
mitogen-activated protein kinase 14
(also known as p38α)
|
|
MIBC
|
muscle invasive BC
|
|
MKK3
|
mitogen-activated protein kinase
kinase 3
|
|
MKK6
|
mitogen-activated protein kinase
kinase 6
|
|
NGS
|
next generation sequencing
|
|
NMIBC
|
non-muscle invasive BC
|
|
PFS
|
progression-free survival
|
|
Real-time PCR
|
real-time polymerase chain
reaction
|
|
RFS
|
recurrence-free survival
|
|
T1HG
|
T1 high grade
|
References
|
1
|
Lotan Y, Black PC, Caba L, Chang SS,
Cookson MS, Daneshmand S, Kamat AM, McKiernan JM, Pruthi RS, Ritch
CR, et al: Optimal trial design for studying urinary markers in
bladder cancer: A collaborative review. Eur Urol Oncol. 1:223–230.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soukup V, Čapoun O, Cohen D, Hernandez V,
Babjuk M, Burger M, Compérat E, Gontero P, Lam T, MacLennan S, et
al: Prognostic performance and reproducibility of the 1973 and
2004/2016 World Health Organization grading classification systems
in non-muscle-invasive bladder cancer: A European Association of
Urology non-muscle invasive bladder cancer guidelines panel
systematic review. Eur Urol Suppl. 72:801–813. 2017. View Article : Google Scholar
|
|
3
|
Westhoff E, Witjes JA, Fleshner NE, Lerner
SP, Shariat SF, Steineck G, Kampman E, Kiemeney LA and Vrieling A:
Body mass index, diet-related factors, and bladder cancer
prognosis: A systematic review and meta-analysis. Bladder Cancer.
4:91–112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sethi S, Kong D, Land S, Dyson G, Sakr WA
and Sarkar FH: Comprehensive molecular oncogenomic profiling and
miRNA analysis of prostate cancer. Am J Transl Res. 5:200–211.
2013.PubMed/NCBI
|
|
5
|
Lee JY, Yun SJ, Jeong P, Piao XM, Kim YH,
Kim J, Subramaniyam S, Byun YJ, Kang HW, Seo SP, et al:
Identification of differentially expressed miRNAs and
miRNA-targeted genes in bladder cancer. Oncotarget. 9:27656–27666.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Furukawa Y, Kawasoe T, Daigo Y, Nishiwaki
T, Ishiguro H, Takahashi M, Kitayama J and Nakamura Y: Isolation of
a novel human gene, ARHGAP9, encoding a rho-GTPase activating
protein. Biochem Bioph Res Commun. 284:643–649. 2001. View Article : Google Scholar
|
|
7
|
Hall A: Rho GTPases and the control of
cell behaviour. Biochem Soc Trans. 33:891–895. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ang BK, Lim CY, Koh SS, Sivakumar N, Taib
S, Lim KB, Ahmed S, Rajagopal G and Ong SH: ArhGAP9, a novel MAP
kinase docking protein, inhibits Erk and p38 activation through WW
domain binding. J Mol Signal. 2:12007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sahai E and Marshall CJ: RHO-GTPases and
cancer. Nat Rev Cancer. 2:133–142. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jaffe AB and Hall A: Rho GTPases in
transformation and metastasis. Adv Cancer Res. 84:57–80. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang T and Ha M: Silencing ARHGAP9
correlates with the risk of breast cancer and inhibits the
proliferation, migration, and invasion of breast cancer. J Cell
Biochem. 119:7747–7756. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Katoh Y and Katoh M: Integrative genomic
analyses on GLI1: Positive regulation of GLI1 by Hedgehog-GLI,
TGFβ-Smads, and RTK-PI3K-AKT signals, and negative regulation of
GLI1 by Notch-CSL-HES/HEY, and GPCR-Gs-PKA signals. Int J Oncol.
35:187–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sharma S, Ksheersagar P and Sharma P:
Diagnosis and treatment of bladder cancer. Am Fam Physician.
80:717–723. 2009.PubMed/NCBI
|
|
15
|
Hall MC, Chang SS, Dalbagni G, Pruthi RS,
Seigne JD, Skinner EC, Wolf JS Jr and Schellhammer PF: Guideline
for the management of nonmuscle invasive bladder cancer (stages Ta,
T1, and Tis): 2007 update. J Urology. 178:2314–2330. 2007.
View Article : Google Scholar
|
|
16
|
Babjuk M, Böhle A, Burger M, Capoun O,
Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M,
et al: EAU guidelines on non-muscle-invasive urothelial carcinoma
of the bladder: Update 2016. Eur Urol. 71:447–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Witjes JA, Compérat E, Cowan NC, De Santis
M, Gakis G, Lebret T, Ribal MJ, Van der Heijden AG and Sherif A;
European Association of Urology, : EAU guidelines on
muscle-invasive and metastatic bladder cancer: Summary of the 2013
guidelines. Eur Urol. 65:778–792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim WT, Kim J, Yan C, Jeong P, Choi SY,
Lee OJ, Chae YB, Yun SJ, Lee SC and Kim WJ: S100A9 and EGFR gene
signatures predict disease progression in muscle invasive bladder
cancer patients after chemotherapy. Ann Oncol. 25:974–979. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bolstad BM, Irizarry RA, Åstrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS,
Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al: Predictive value
of progression-related gene classifier in primary non-muscle
invasive bladder cancer. Mol Cancer. 9:32010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun J, Nishiyama T, Shimizu K and Kadota
K: TCC: An R package for comparing tag count data with robust
normalization strategies. BMC Bioinformatics. 14:2192013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pignot G, Vieillefond A, Vacher S, Zerbib
M, Debre B, Lidereau R, Amsellem-Ouazana D and Bieche I: Hedgehog
pathway activation in human transitional cell carcinoma of the
bladder. Br J Cancer. 106:1177–1186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee J, Platt KA, Censullo P and Ruiz i
Altaba A: Gli1 is a target of Sonic hedgehog that induces ventral
neural tube development. Development. 124:2537–2552.
1997.PubMed/NCBI
|
|
27
|
Kimura H, Stephen D, Joyner A and Curran
T: Gli1 is important for medulloblastoma formation in Ptc1+/- mice.
Oncogene. 24:4026–4036. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kumar B, Sinclair J, Khandrika L, Koul S,
Wilson S and Koul HK: Differential effects of MAPKs signaling on
the growth of invasive bladder cancer cells. Int J Oncol.
34:1557–1564. 2009.PubMed/NCBI
|
|
29
|
Spiess PE and Czerniak B: Dual-track
pathway of bladder carcinogenesis: Practical implications. Arch
Pathol Lab Med. 130:844–852. 2006.PubMed/NCBI
|
|
30
|
Nicholson RI, Gee JM and Harper ME: EGFR
and cancer prognosis. Eur J Cancer. 37 (Suppl 4):S9–S15. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim WJ, Kim SK, Jeong P, Yun SJ, Cho IC,
Kim IY, Moon SK, Um HD and Choi YH: A four-gene signature predicts
disease progression in muscle invasive bladder cancer. Mol Med.
17:478–485. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mari A, Campi R, Tellini R, Gandaglia G,
Albisinni S, Abufaraj M, Hatzichristodoulou G, Montorsi F, van
Velthoven R, Carini M, et al: Patterns and predictors of recurrence
after open radical cystectomy for bladder cancer: A comprehensive
review of the literature. World J Urol. 36:157–170. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yun SJ, Kim SK and Kim WJ: How do we
manage high-grade T1 bladder cancer? Conservative or aggressive
therapy? Investig Clin Urol. 57 (Suppl 1):S44–S51. 2016. View Article : Google Scholar : PubMed/NCBI
|