Introduction
Primary liver cancer is one of the six most common
cancers, and the number of deaths ranks 2nd in the total
cancer-related deaths (1).
Hepatocellular carcinoma, the most important primary liver cancer,
accounts for approximately 90% in primary liver tumors, making it a
major international public health issue (2). The occurrence and development of liver
cancer is a progressive cumulative complex process covering
multiple factors, stages, mechanisms, links and genetic changes,
which involves a variety of abnormal cellular or molecular changes,
such as oxidative stress, endoplasmic reticulum stress and cell
cycle disorder (3,4). Therefore, clarifying the molecular
mechanism for the occurrence and development of liver cancer has
great significance in its early diagnosis and treatment.
The polycomb group (PcG) protein is an important
epigenetic regulatory factor, which can serve as a transcription
inhibitor silencing specific genomes via chromatin modification
(5). The PcG protein belongs to the
polycomb repressive complex (PRC) family. PRC2 includes the Zeste2
enhancer (EZH2), suppressor of Zeste12 (SUZ12) and embryonic
ectoderm development (EED) (6). EZH2
is a catalytically active component of PRC2, which can catalyze the
histone H3 lysine 27 (H3K27) for trimethylation after forming the
complex with EED (7). Recently,
increasingly more studies have revealed that EZH2 has a
cancer-promoting effect, including the induction of the abnormal
cell differentiation and promotion of cancer cell proliferation
(8). EZH2 is overexpressed in
various tumors, showing a close correlation with the poor prognosis
of patients (9). It is reported in
studies that the low expression of miR-101 in glioma cells can lead
to the upregulation of EZH2, thereby enhancing the proliferation,
invasion and migration of glioma cells (10).
miRNAs are a group of single-stranded non-coding
RNAs existing in eukaryotes, with 20–24 nt in length and various
regulatory functions (11). miRNAs
can regulate the expression of a variety of genes through targeted
binding to specific genes, thus playing an important role in the
physiological activities of cells, such as proliferation,
differentiation and apoptosis (12).
To the best of our knowledge, the expression of miR-101 in liver
cancer and its mechanism have not been reported yet. In the present
study, the expression level of miR-101 in carcinoma and
para-carcinoma tissues of liver cancer patients was detected, and
the liver cancer cell lines with miR-101 overexpression were
constructed using miR-101 mimics, so as to observe the influence of
miR-101 overexpression on the proliferation of liver cancer cells
and further explore the potential mechanism of miR-101 in affecting
the proliferation of liver cancer cells.
Materials and methods
Tissue specimens
A total of 67 pairs of liver cancer tissues and the
corresponding para-carcinoma tissues surgically resected in Chinese
PLA General Hospital (Beijing, China) from December 2016 to June
2018 were collected. After the blood stains were washed away with
normal saline, all specimens were cut into pieces, placed into an
Eppendorf (EP) tube and stored in a refrigerator at −80°C. All the
above procedures were approved by the Medical Ethics Committee of
Chinese PLA General Hospital and informed consents were signed by
the patients or the guardians.
Cell culture
The liver cancer HepG2 cell line was purchased from
the Biological Research Institute of the Chinese Academy of
Sciences (cat. no. TCHu106). Phosphate buffered saline (PBS),
trypsin, fetal bovine serum (FBS) and RPMI-1640 medium were
purchased from Gibco; Thermo Fisher Scientific, Inc. Small
interfering RNAs (siRNAs) were from Google Biology. HepG2 cells
were cultured in an incubator with 5% CO2 at 37°C, and
then digested with 0.25% trypsin-EDTA and passaged when they fully
covered the culture dish.
Detection of expression of related
genes via reverse transcription-polymerase chain reaction
(RT-PCR)
i) The total RNA was extracted from liver cancer and
para-carcinoma tissues using TRIzol (RT kit cat. no. 10928042), the
concentration and purity of the RNA extracted were detected using
an ultraviolet spectrophotometer (Mettler Toledo) and the RNA with
absorbance (A)260/A280 of 1.8–2.0 was used. ii) The messenger RNA
(mRNA) was synthesized into the complementary deoxyribonucleic acid
(cDNA) through RT kit (cat no. 4366596; Thermo Fisher), and stored
in the refrigerator at −80°C. iii) RT-PCR system: 2.5 µl 10X
buffer, 2 µl cDNAs, 0.25 µl forward primers (20 µmol/l), 0.25 µl
reverse primers (20 µmol/l), 0.5 µl dNTPs (10 mmol/l), 0.5 µl Taq
polymerase enzymes (2×106 U/l) and 19 µl
ddH2O. The amplification system of RT-PCR was the same
as above. It was synthesized at 50°C and amplified (40 cycles). The
final Cq value was measured by LightCycler 480 system. The internal
reference was GAPDH. Primer sequence are shown in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Target gene | Primer sequence |
|---|
| GAPDH |
|
|
Forward |
5′-GACATGCCGCCTGGAGAAAC-3′ |
|
Reverse |
5′-AGCCCAGGATGCCCTTTAGT-3′ |
| miR-101 |
|
|
Forward |
5′-AAAGCTGATCGTAGGCTGTTCCTT-3′ |
|
Reverse |
5′-AGTCGATGCCAAAGAAGT-3′ |
Construction of cell lines with
miR-101 overexpression
When HepG2 cells were in the logarithmic growth
phase, they were immediately digested and inoculated into a 6-well
plate. After 12 h (60–80% cells were fused), the complete medium
was discarded, and cells were washed with the serum-free medium 2–3
times and starved in the incubator for synchronous growth. miR-101
mimics were dissolved in RNase deionized water to be prepared into
transfection solution at a final concentration of 20 µmol/l. The
cells were divided into the blank control group (NC group) and
miR-101 overexpression group (miR-101 mimics group). The
transfection solution prepared already was added into each well and
fully mixed, followed by cell culture for another 6 h. Then the
solution was replaced with complete medium.
Western blotting
The medium was discarded and washed by PBS three
times. Each dish was filled with 1,000 µl RIPA lysis buffer (Thermo
Fisher) and shaken for 20 min. Then the cells on the bottom of the
dish were fully scraped off by a brush and put into the EP tube.
The collected cells were lysed for about 15 sec with an ultrasound
lyser and centrifuged for 0.5 h (at 10,000 × g) after 15 min at
4°C. The supernatant was separated into EP tubes, and the protein
concentration was measured by BCA and ultraviolet
spectrophotometry. The protein concentration of all samples was
fixed to the same concentration. After packing, they were put in
−80°C refrigerator. Then, 15% SDS-PAGE electrophoresis was
performed after the total protein was extracted from liver cancer
cells prior to the addition of 10 μg protein per lane. The protein
was transferred to PVDF membrane. The membrane was blocked with 5%
milk at 23°C for 1 h. Enhanced chemiluminescent (ECL) kit
(Beyotime) was used for visualisation. Western blot analysis was
carried out. An Odyssey scanner (Odyssey) was used to scan and
quantify protein bands, and GAPDH was used to correct the protein
level. Image J (NIH) was used for densitometry of the bands.
Cell Counting Kit-8 (CCK-8) cell
proliferation assay
The cells in the logarithmic growth phase in each
group were inoculated into a 96-well plate and cultured in the
incubator with 5% CO2 at 37°C for 0, 24, 48, 72 and 96
h. The blank control group was used as the negative control group
(NC group). Then the medium was discarded, and the developing
solution was prepared in a dark place using RPMI-1640 medium and
CCK-8 (10:1). Then, 110 µl developing solution was added into each
well of the 96-well plate, followed by incubation at 37°C for 2 h,
and the absorbance in each group was detected at 540 nm using the
ultraviolet spectrophotometer. The experiment was repeated three
times.
Ki67 staining
At 48 h after transfection with miR-101 mimics,
liver cancer cell lines were stained using the Ki67 staining kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to
manufacturers instructions. After staining, the cells were
photographed under a fluorescence microscope (Olympus) and three
fields of view were randomly selected in each glass slide. Finally,
the Ki67-positive cells were counted and quantified.
Detection of cell cycle via flow
cytometry
The cells in the logarithmic growth phase were
taken, digested with 0.25% trypsin-EDTA, prepared into suspension
and inoculated into the 6-well medium. The cells were loaded and
the proportions of cells in different phases were detected
according to the instructions of the Annexin V-FITC PI cell cycle
assay kit (Beyotime Institute of Biotechnology).
Luciferase reporter gene assay
First, the possible binding sites of transcription
factors in the promoter region were predicted using bioinformatics
method (TargetScan). The primers were designed, and the EZH2 gene
segment was cloned from genomic DNAs via PCR and inserted into the
luciferase reporter gene plasmid. The positive clones were
screened. The miR-101 plasmid was amplified and purified for later
use. At the same time, the corresponding empty plasmid control was
set up. The reporter gene plasmid and transcription
factor-expressing plasmid were co-transfected into cells. The
specific fluorescein substrate enzyme was added, and the
fluorescence intensity was detected to determine whether there was
a targeted effect.
Statistical analysis
SPSS22.0 software (IBM Corp.) was used for the
analysis of all data. Measurement data were expressed as mean ±
standard deviation, and t-test was used for the comparison of data
between the two groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-101 in liver cancer
and para-carcinoma tissues
The expression of miR-101 in carcinoma and
para-carcinoma tissues was detected via RT-PCR, and the results
revealed that the expression level of miR-101 in liver cancer
tissues was significantly lower than that in para-carcinoma tissues
(P<0.05; Fig. 1).
Influence of miR-101 overexpression on
proliferation of liver cancer cells
At 0, 24, 48, 72 and 96 h after miR-101 mimics were
transfected into liver cancer cells, the cell proliferation in each
group was detected using the CCK-8 kit, and the optical density
(OD) at 540 nm was used to reflect the proliferation ability. The
results showed that the proliferation of liver cancer cells was
significantly reduced at 0, 24, 48, 72 and 96 h after transfection
of miR-101 mimics (P<0.05; Fig.
2).
miR-101 overexpression inhibits colony
formation of liver cancer cells
At 10 days after miR-101 overexpression, the colony
formation ability in each group was detected. It was found that the
number of colonies formed was 150.45±3.88 in NC group and
32.12±2.08 in miR-101 mimics group (P<0.05; Fig. 3), suggesting that the miR-101
overexpression can significantly inhibit the colony formation
ability of liver cancer cells.
Ki67 staining results of miR-101
overexpression on liver cancer cells
Furthermore, the cell proliferation ability in each
group was evaluated using Ki67 staining. As shown in Fig. 4, the transfection of miR-101 mimics
was able to reduce the number of Ki67-positive cells by
approximately 5.32 times (P<0.05).
Influence of miR-101 mimics on liver
cancer cell cycle
As shown in Fig. 5,
the liver cancer cell cycle was obviously changed after miR-101
mimics were added. In miR-101 mimics group, the proportion of liver
cancer cells in G0/G1 phase was obviously increased, while the
proportion of cells in G2/M and S phases was obviously decreased
(P<0.05), indicating that miR-101 mimics can significantly
inhibit the cycle of liver cancer cells.
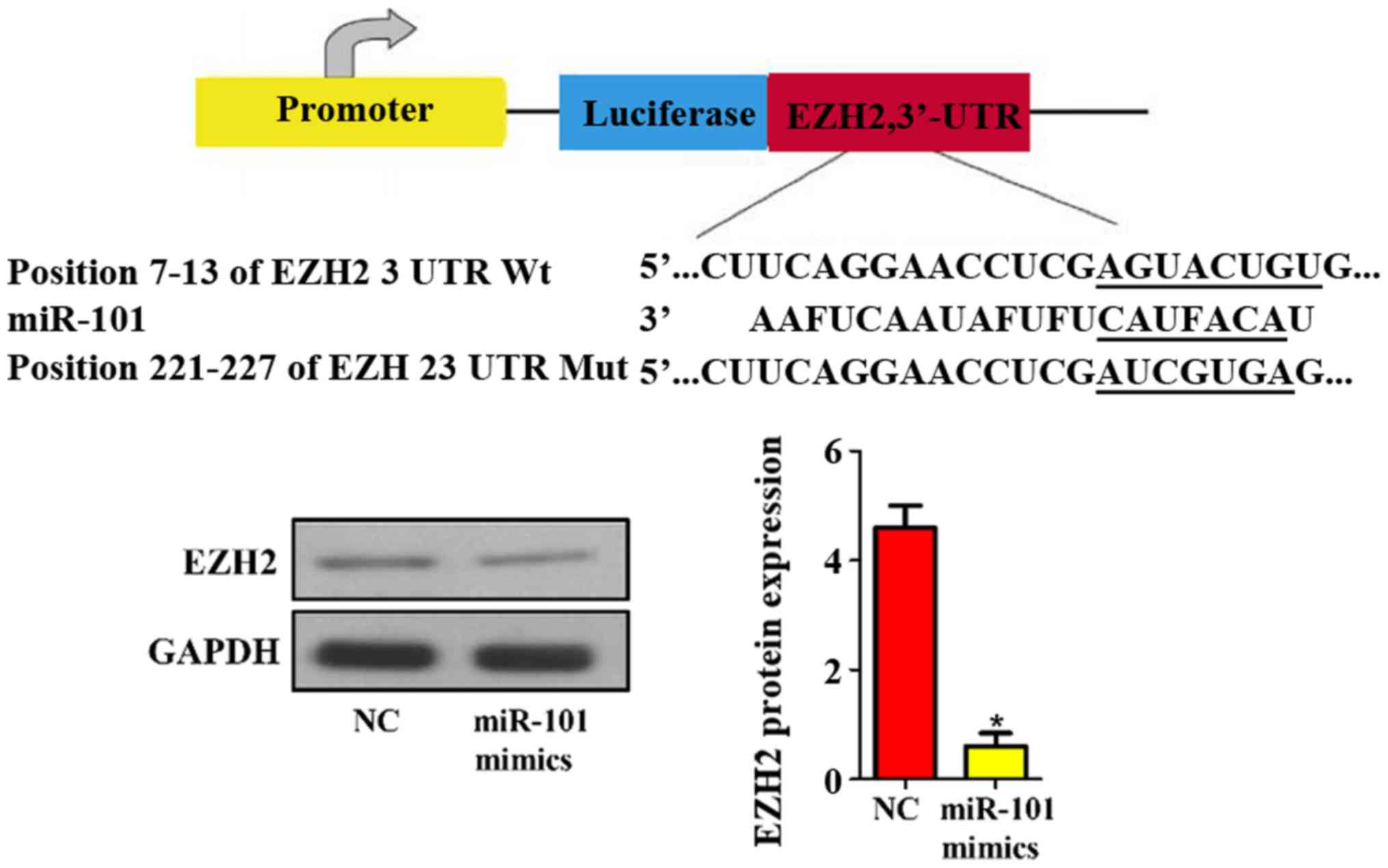
Prediction and verification of miR-101
target genes
In addition, the target genes of mouse miR-101 were
predicted using bioinformatics technique. The results revealed that
EZH2 was one of the target genes of miR-101 (Fig. 6). Then the protein expression level
of EZH2 in NC and miR-101 mimic groups was detected via western
blotting, and it was found that miR-101 mimics remarkably
suppressed the expression level of EZH2 in liver cancer cells
compared with that in NC group (P<0.05; Fig. 6).
Influence of miR-101 overexpression on
the mitogen-activated protein kinase (MAPK)/extracellular
signal-regulated kinase (ERK) signaling pathway
Considering the important role of the MAPK/ERK
signaling pathway in the occurrence and development of liver
cancer, whether the activation of the MAPK/ERK signaling pathway in
liver cancer can be regulated by miR-101 was detected. The ERK1/2
phosphorylated protein and total protein in each group were
quantified using western blotting. As shown in Fig. 7, the phosphorylation of ERK1/2 was
also significantly inhibited after miR-101 overexpression in liver
cancer cells (P<0.05), further revealing that the inhibitory
effect of miR-101 on the proliferation of liver cancer cells is
realized by its inhibition on the MAPK/ERK signaling pathway
through targeted binding to EZH2.
Discussion
In recent years, the morbidity and mortality rates
of liver cancer have increased year by year around the world, and
the number of deaths is up to 662,000 every year, about half of
which are from China (13). The main
causes of liver cancer include hepatitis B virus infection, smoking
and drinking. Therefore, many research efforts have been made to
search for the pathogenic genes and diagnostic markers of liver
cancer, such as the tumor size, alpha fetoprotein level and various
differentially-expressed genes in primary liver cancer tissues
(14). Despite the significant
improvement in the diagnosis and treatment strategies, the overall
prognosis of liver cancer is still poor (15). Therefore, it is of great significance
to search for the key genes, proteins or RNAs causing liver cancer
for the precise treatment of liver cancer.
With the rapid development of transcriptomics,
increasingly more disease-related differentially-expressed genes
have been revealed. Similarly, many studies have confirmed the role
of miRNAs in the occurrence and development of liver cancer
(16). For example, miR-139 can
inhibit the invasion and metastasis of liver cancer cells through
downregulating the expression of Rho-kinase 2 (17). On the contrary, miR-21 can promote
the proliferation of liver cancer, whose mechanism may be related
to the direct targeted inhibition of miR-21 on MAP kinase-kinase 3
(MAP2K3) (18). Moreover, miR-346
also serves as a cancer-promoting gene, which can facilitate the
proliferation, invasion and metastasis of liver cancer cells
through targeted inhibition on F-Box and leucine-rich repeat
protein (FBXL2) (19). In the
present study, it was found for the first time, to the best of our
knowledge, that the miR-101 expression level in liver cancer
tissues was significantly lower than that in para-carcinoma
tissues, indicating that miR-101 may play a role as a cancer
suppressor gene. In addition, the influence of miR-101
overexpression on the proliferation of liver cancer cells was
detected using various molecular biological methods. It was proved
in Ki67 staining, flow cytometry and colony formation assay that
the miR-101 overexpression inhibited the cycle, DNA replication and
division of liver cancer cells. EZH2 is a human gene discovered in
recent years, which has a close correlation with the cell life
activity. EZH2 can promote the proliferation and spread of tumor
cells through inhibiting the characteristic target genes in
chromatin, and the mechanism of its transcriptional inhibition may
be related to its regulatory effect on histone methyltransferase
(20). Cardenas et al found
that inhibiting EZH2 can also promote the endothelial-mesenchymal
transition of ovarian cancer, thereby inhibiting the invasion of
ovarian cancer cells (21). It has
been reported in previous studies that miR-101/EZH2 is expressed
abnormally in a variety of tumors, including prostate cancer,
bladder cancer, gastric cancer and glioma. Moreover, the abnormal
expression of miR-101/EZH2 is closely related to migration,
invasion and metastasis of these tumors (22–24). In
the present study, it was primarily revealed, using bioinformatics
and molecular biological methods, that EZH2 was one of the
potential direct targets of miR-101. In fact, EZH2 can serve as a
target gene for various miRNAs. For example, miR-98 can
downregulate the Wnt/β-catenin signaling pathway through targeted
inhibition on EZH2, ultimately suppressing the proliferation of
liver cancer cells (25).
The MAPK/ERK signaling pathway plays an
indispensable role in the biological behavior of liver cancer
cells. It is reported that EZH2 can affect the activation of the
MAPK/ERK signaling pathway, and the MAPK/ERK can also in turn
affect the EZH2 expression, indicating that there may be a
potential negative feedback regulatory correlation between EZH2 and
MAPK/ERK. This study revealed that miR-101 also inhibited the
phosphorylation activation of ERK1/2, but whether the activation of
ERK1/2 depends on the expression of EZH2 remains to be further
investigated. However, there are still some limitations in this
study: i) only one kind of cell line was used; and ii) the
subcutaneous tumor formation assay was not performed.
In conclusion, this study indicates for the first
time to the best of our knowledge, that miR-101 can inhibit the
phosphorylation level of ERK1/2 through targeted inhibition on
EZH2, ultimately suppressing the proliferation of liver cancer
cells.
Acknowledgements
Not applicable.
Funding
The study was funded by Wu Jieping Medical
Foundation (grant no. 320.6750.11010).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XM wrote the manuscript. XM and YS performed PCR and
western blotting. XX and CL assisted with CCK-8 assay. XG and KP
were responsible for cell culture and construction of cell lines
with miR-101 overexpression. YL contributed to Ki67 staining. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Medical Ethics
Committee of Chinese PLA General Hospital (Beijing, China) and
informed consents were signed by the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Golob-Schwarzl N, Krassnig S, Toeglhofer
AM, Park YN, Gogg-Kamerer M, Vierlinger K, Schröder F, Rhee H,
Schicho R, Fickert P, et al: New liver cancer biomarkers:
PI3K/AKT/mTOR pathway members and eukaryotic translation initiation
factors. Eur J Cancer. 83:56–70. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang H, Song Y, Yang H, Liu Z, Gao L,
Liang X and Ma C: Tumor cell-intrinsic Tim-3 promotes liver cancer
via NF-κB/IL-6/STAT3 axis. Oncogene. 37:2456–2468. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maucort-Boulch D, de Martel C, Franceschi
S and Plummer M: Fraction and incidence of liver cancer
attributable to hepatitis B and C viruses worldwide. Int J Cancer.
142:2471–2477. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qiu WQ, Shi JF, Guo LW, Mao AY, Huang HY,
Hu GY, Dong P, Bai FZ, Yan XL, Liao XZ, et al: Medical expenditure
for liver cancer in urban China: A 10-year multicenter
retrospective survey (2002–2011). J Cancer Res Ther. 14:163–170.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao SB, Li KL, Qiu H, Zhu LY, Pan CB, Zhao
Y, Wei SH, Shi S, Jin GH and Xue LX: Enhancing chemotherapy
sensitivity by targeting PcG via the ATM/p53 pathway. Am J Cancer
Res. 7:1874–1883. 2017.PubMed/NCBI
|
|
6
|
Laugesen A, Højfeldt JW and Helin K: Role
of the polycomb repressive complex 2 (PRC2) in transcriptional
regulation and cancer. Cold Spring Harb Perspect Med. 6:62016.
View Article : Google Scholar
|
|
7
|
Hu G, Gupta SK, Troska TP, Nair A and
Gupta M: Long non-coding RNA profile in mantle cell lymphoma
identifies a functional lncRNA ROR1-AS1 associated with EZH2/PRC2
complex. Oncotarget. 8:80223–80234. 2017.PubMed/NCBI
|
|
8
|
Yoshida K, Toden S, Ravindranathan P, Han
H and Goel A: Curcumin sensitizes pancreatic cancer cells to
gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1
expression. Carcinogenesis. 38:1036–1046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wan L, Xu K, Wei Y, Zhang J, Han T, Fry C,
Zhang Z, Wang YV, Huang L, Yuan M, et al: Phosphorylation of EZH2
by AMPK suppresses PRC2 methyltransferase activity and oncogenic
function. Mol Cell. 69:279–291.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smits M, Nilsson J, Mir SE, van der Stoop
PM, Hulleman E, Niers JM, de Witt Hamer PC, Marquez VE, Cloos J,
Krichevsky AM, et al: miR-101 is down-regulated in glioblastoma
resulting in EZH2-induced proliferation, migration, and
angiogenesis. Oncotarget. 1:710–720. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Naga Prasad SV, Gupta MK, Duan ZH,
Surampudi VS, Liu CG, Kotwal A, Moravec CS, Starling RC, Perez DM,
Sen S, et al: A unique microRNA profile in end-stage heart failure
indicates alterations in specific cardiovascular signaling
networks. PLoS One. 12:e01704562017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valery PC, Laversanne M, Clark PJ, Petrick
JL, McGlynn KA and Bray F: Projections of primary liver cancer to
2030 in 30 countries worldwide. Hepatology. 67:600–611. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang CH, Lu CM, Huang YP, Li XF, Fan YF,
Yang J, Xiang N and Pan JH: Study on the application value of
digital medical technology in the operation on primary liver
cancer. Zhonghua Wai Ke Za Zhi. 47:523–526. 2009.(In Chinese).
PubMed/NCBI
|
|
15
|
Wong MC, Jiang JY, Goggins WB, Liang M,
Fang Y, Fung FD, Leung C, Wang HH, Wong GL, Wong VW, et al:
International incidence and mortality trends of liver cancer: a
global profile. Sci Rep. 7:458462017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wong CC, Wong CM, Tung EK, Au SL, Lee JM,
Poon RT, Man K and Ng IO: The microRNA miR-139 suppresses
metastasis and progression of hepatocellular carcinoma by
down-regulating Rho-kinase 2. Gastroenterology. 140:322–331. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z
and Liu X: MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell
proliferation through repression of mitogen-activated protein
kinase-kinase 3. BMC Cancer. 13:4692013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu Q, Yang X, Duan W, Li C, Luo Y and Lu
S: miRNA-346 promotes proliferation, migration and invasion in
liver cancer. Oncol Lett. 14:3255–3260. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim KH and Roberts CW: Targeting EZH2 in
cancer. Nat Med. 22:128–134. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cardenas H, Zhao J, Vieth E, Nephew KP and
Matei D: EZH2 inhibition promotes epithelial-to-mesenchymal
transition in ovarian cancer cells. Oncotarget. 7:84453–84467.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang D, Wang X, Zhuang C, Shi W, Liu M,
Tu Q, Zhang D and Hu L: Reciprocal negative feedback loop between
EZH2 and miR-101-1 contributes to miR-101 deregulation in
hepatocellular carcinoma. Oncol Rep. 35:1083–1090. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kottakis F, Polytarchou C, Foltopoulou P,
Sanidas I, Kampranis SC and Tsichlis PN: FGF-2 regulates cell
proliferation, migration, and angiogenesis through an
NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell. 43:285–298. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo C, Merz PR, Chen Y, Dickes E, Pscherer
A, Schadendorf D and Eichmüller SB: MiR-101 inhibits melanoma cell
invasion and proliferation by targeting MITF and EZH2. Cancer Lett.
341:240–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang JJ, Chen JT, Hua L, Yao KH and Wang
CY: miR-98 inhibits hepatocellular carcinoma cell proliferation via
targeting EZH2 and suppressing Wnt/β-catenin signaling pathway.
Biomed Pharmacother. 85:472–478. 2017. View Article : Google Scholar : PubMed/NCBI
|