Introduction
Osteosarcoma (OS) is the most common primary sarcoma
of the bone and mainly affects adolescents and children (1,2). In
spite of the low incidence rate, OS cause a considerable number of
cancer-related deaths due to its highly aggressive nature and
systemic metastasis occurs at early stages (2,3). With
the development of therapeutic approaches and diagnostic
techniques, the overall survival rate has increased from 20 to
65–75% during the past century (4).
However, cancer metastasis, such as lung metastasis can easily
occur, and only less than 30% of OS patients with metastatic OS can
survive. The high mortality rate is mainly because most OS patients
with cancer metastasis inevitably develop resistance to currently
available chemical drugs (5).
Genetic factors are critical players in the
occurrence and development of OS (6,7).
Non-coding RNAs, such as miRNAs and long (>200 nt) non-coding
RNAs (lncRNAs) encode no proteins but participate in cancer
development by regulating gene expression (8–10).
ncRNA-targeted therapies have shown promising potentials in cancer
diagnosis and prognosis (11), while
the function of most ncRNAs is hardly known, which limits their
clinical applications. LncRNA ELF3-AS1 has been reported to be an
oncogenic lncRNA in bladder cancer (12). Our preliminary data showed that
ELF3-AS1 was inversely correlated with miR-205, which plays
tumor-suppressive or oncogenic roles in different types of cancer
(13,14). It is known miR-205 can directly
target KLF12 in basal-like breast carcinoma (15). The present study was carried out to
investigate the interaction between miR-205, KLF12 and ELF3-AS1 in
OS.
Materials and methods
Research subjects
The First Affiliated Hospital of Wannan Medical
College (Wuhu, China) admitted 79 patients with OS during the
period between December 2015 and December 2018. The current study
selected 40 (25 males and 17 females; range, 19–48 years; mean age,
33.2±5.4 years) of these patients according to strict criteria. The
inclusion criteria were as follows: i) Newly diagnosed patients
with OS; and ii) all major organs showed normal functions. The
exclusion criteria were as follows: i) therapies initiated before
admission; ii) family history of malignancies; or iii) previous
history of malignancies. Patients with OS were staged according to
American Joint Committee on Cancer criteria (16), and there were 8, 13, 11 and 8 cases
at stages I–IV, respectively. All patients were informed of
experimental details and consented to the use of their samples in
this study, and the Ethics Committee of the First Affiliated
Hospital of Wannan Medical College approved the study.
Tissues
Patients with OS were diagnosed by biopsy. During a
biopsy, OS tumor and non-tumor (within 2 cm of the tumor) tissues
were collected from each patient. The weight of tissues were
0.08–0.12 g, and the tissue types were confirmed by
histopathological examinations.
Cells and transient transfection
The human OS cell line U2OS (ATCC) was used in this
study. Eagle's minimum essential medium (American Type Culture
Collection) with 10% FBS (Sigma-Aldrich; Merck KGaA) was used as a
cell culture medium, and cell culture conditions were 37°C and 5%
CO2. KLF12 and ELF3-AS1 expression vectors were
constructed by Sangon Biotech Co., Ltd. using the pcDNA3.1 vector.
Negative control miRNA (5′-UGACGUCAGUCGUAGGUACGUG-3′) and miR-205
mimic (5′-UCCUUCAUUCCACCGGAGUCUG-3′) were purchased from
Sigma-Aldrich (Merck KGaA). KLF12 and ELF3-AS1 expression vector
(10 nM), or 10 nM empty pcDNA3 vector (negative control, NC1), 45
nM miR-205 mimic or negative control miRNA (NC2, targets to a
non-human sequence) were transfected into 106 U2OS cells
using Lipofectamine® 2000 reagent (Sigma-Aldrich; Merck
KGaA). Untransfected cells were control cells (C). The subsequent
experiments were performed using cells collected at 24 h after
transfection.
RT-qPCR
RiboZol™ RNA extraction reagent (VWR International)
was mixed with U2OS cells and tissue powder (ground in liquid
nitrogen before use) to extract total RNA. SensiFAST™ cDNA
Synthesis kit (Bioline) was used to perform RT. Temperature
conditions for RT were 55°C for 30 min and 80°C for 10 min. SYBR
Green Master mix (Bio-Rad Laboratories, Inc.) was used for
preparation of qPCR mixtures. 18S rRNA and GAPDH were used as
endogenous controls to detect the expression of KLF12 and ELF3-AS1,
respectively. To detect miR-205 expression, an miRNA Isolation kit
(Geneaid Biotech Ltd.) was used to extract miRNA. miRNA reverse
transcription and qPCR were performed using All-in-One™ miRNA
qRT-PCR Detection Kit (Genecopoeia, Inc.). U6 was used as an
endogenous control for miR-205. Primer sequences were as follows:
ELF3-AS1 forward 5′-TGAAGTCATCACGAACCG-3′ and reverse,
5′-GAGCCCCAAGTTAATGCG-3′; KLF12 forward 5′-GATGAATTGTTGCAGCCCCA-3′
and reverse, 5′-CATTTGAAATAATGACCAAG-3′; GADPH forward
5′-CCAGGGCTGCTTTTAACTCT-3′ and reverse, 5′-GGACTCCACGACGTACTCA-3′.
The forward primer for miR-205 was 5′-UCCUUCAUUCCACCGGAGU-3′, and
the reverse primer for miR-205 and both U6 primers were from the
kit. Thermocycling conditions for all qPCR reactions were 95°C for
40 sec, followed by 95°C for 10 sec and 60°C for 30 sec for 40
cycles. Each sample was measured in triplicate, and the
2−ΔΔCq method was used to process data (17).
Methylation-specific PCR (MSP)
Genomic DNA was extracted from U2OS cells using a
genomic DNA extraction kit (cat. no. ab156900; Abcam). All DNA
samples were processed using a Methylation-Gold™ kit (Zymo Research
Corp.). Routine Taq DNA polymerase (New England BioLabs, Inc.) was
used to perform the subsequent PCR. Primer sequences were as
follows: Methylation (M) forward, 5′-TTGTTTTGATGATTATGAAGGAATG-3′
and reverse, 5′-ACCCCTAAACTAACTAAACCCAAAC-3′; unmethylation (U)
forward 5′-TCTGCCCTGATGATCATGAAGG-3′ and reverse,
5′-CCTGGGCTGACTGAACCCAAGCC-′. Thermocycling conditions were 95°C
for 40 sec, followed by 95°C for 30 sec, 55°C for 30 sec and 72°C
for 40 sec for 30 cycles. PCR products were subjected to 1.5%
agarose gel electrophoresis (80 v for 15 min) and ethidium bromide
staining (10 min at room temperature).
Measurement of cell proliferation
ability
U2OS cells were harvested at 24 h after
transfections, and 4×104 cells were mixed with 1 ml
Eagle's minimum essential medium with 10% FBS to prepare
single-cell suspensions. A 96-well cell culture plate was used to
cultivate the cells at 37°C and in 5% CO2, and 10 µl
Cell Counting Kit-8 reagent (Dojindo Molecular Technologies, Inc.)
was added at 2 h Prior to cell collection. Cells were collected
every 24 h for a total of 4 days. Subsequently, cell proliferation
rates were calculated based on OD values measured at a wavelength
of 450 nm.
Western blot analysis
RIPA solution (Sangon Biotech Co., Ltd.) was used to
extract total protein from both tissues and transfected cells.
Proteins were denatured in boiling water for 10 min, and SDS-PAGE
was performed using 12% gels with 30 µg protein per lane. Proteins
were transferred to PVDF membranes, followed by blocking in 5%
non-fat milk for 2 h at 25°C. Subsequently, the membranes were
incubated with rabbit polyclonal primary antibodies against GAPDH
(1:1,300; cat. no. ab9485; Abcam) and KLF12 (1:1,200; cat. no.
ab221602; Abcam) overnight at 4°C, followed by incubation with goat
anti-rabbit IgG horseradish peroxidase-conjugated secondary
antibody (1:2,000; cat. no. sc-2004; Santa Cruz Biotechnology,
Inc.) for 2 h at room temperature. ECL (Sigma-Aldrich; Merck KGaA)
was used to develop signals. ImageJ software (version 1.46;
National Institutes of Health) was used to analyze data.
Statistical analysis
Mean values (± SEM) represent data from 3 biological
replicates. GraphPad Prism 6 (GraphPad, Inc.) was used to perform
all statistical analysis. Paired Student's t-test was used to
compare the differences between 2 types of tissues from OS
patients. One-way ANOVA and Tukey's post hoc test were used to
explore the differences between different patient and cell groups.
Correlations were analyzed by linear regression. P<0.05 was
considered to indicate a statistically significant difference.
Results
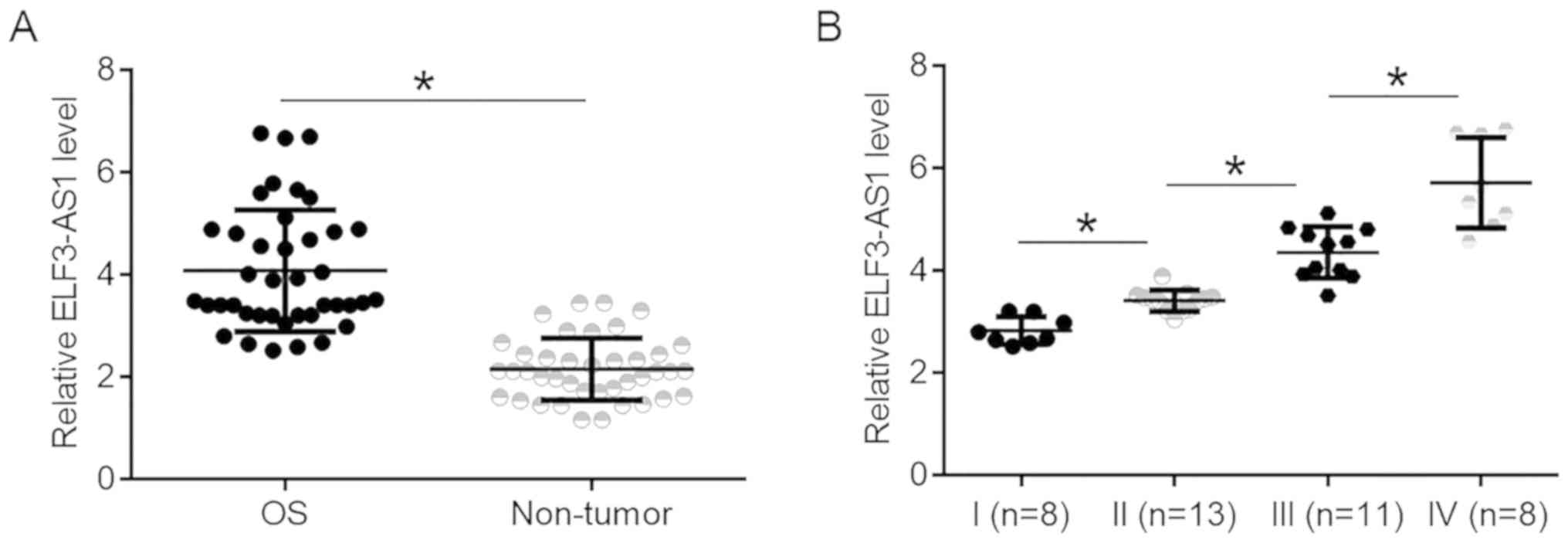
ELF3-AS1 is upregulated in OS and
affected by cancer development
ELF3-AS1 expression was compared between tumor and
non-tumor tissues. It was observed that expression levels of
ELF3-AS1 were significantly higher in OS tissues compared with
non-tumor tissues (Fig. 1A;
P<0.05). ELF3-AS1 expression data in OS tissues were compared
between patients at 4 clinical. It was observed that ELF3-AS1
expression levels in OS tissues increased with increasing clinical
stages (Fig. 1B; P<0.05).
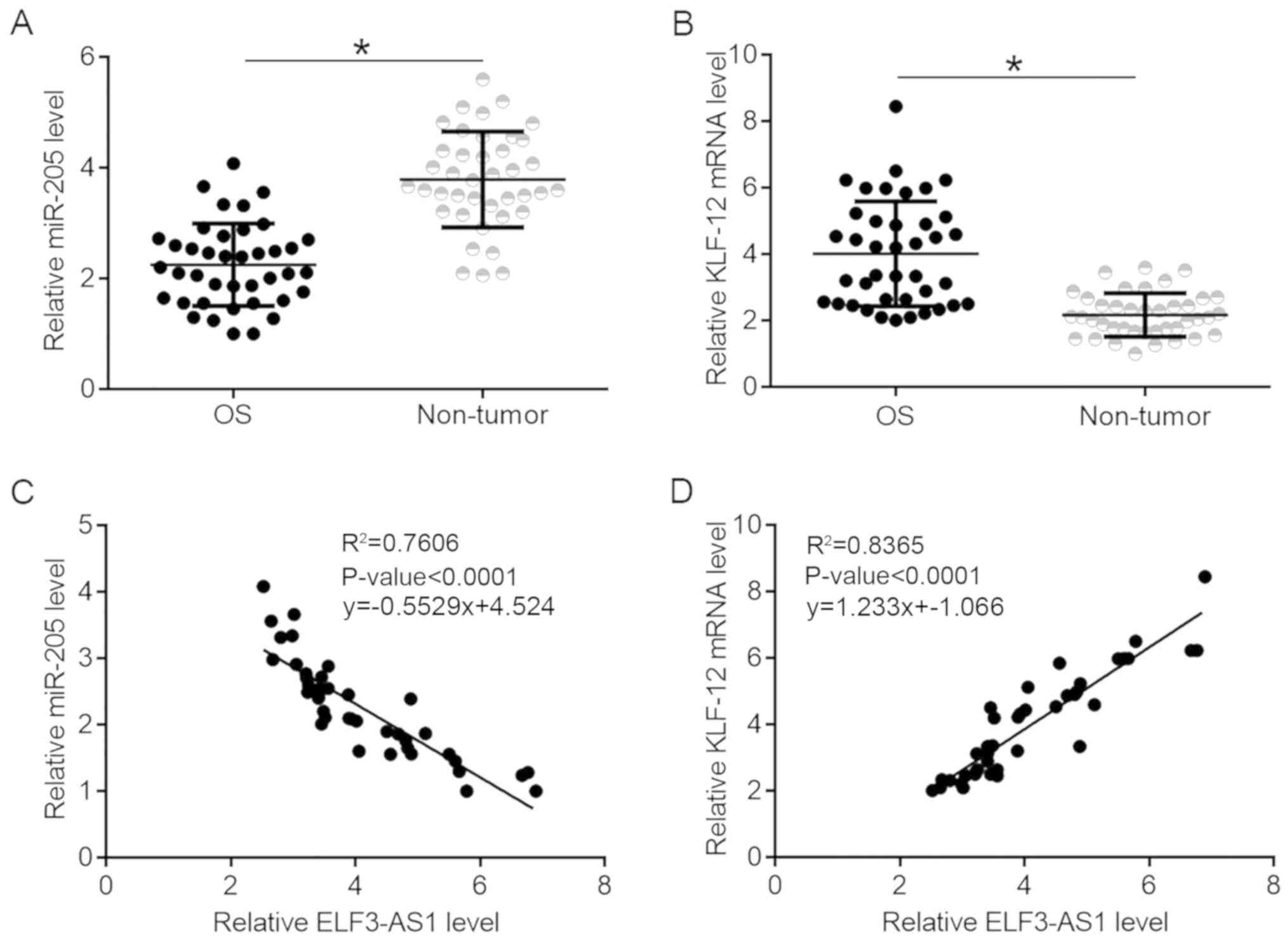
KLF12 and miR-205 are significantly
correlated with ELF3-AS1 in OS
KLF12 and miR-205 expression were also detected in
tumor and non-tumor tissues. It was observed that miR-205 was
significantly downregulated (Fig.
2A; P<0.05), while KLF12 was upregulated (Fig. 2B; P<0.05) in OS tissues compared
with non-tumor tissues. ELF3-AS1 expression was found to be
negatively correlated with miR-205 expression (Fig. 2C), while ELF3-AS1was positively
correlated with KLF12 (Fig. 2D).
ELF3-AS1 upregulates KLF12 through
miR-205
ELF3-AS1 and KLF12 expression vectors, as well as
miR-205 mimic, were transfected into U2OS cells. Compared with C
and NC groups, expression levels of ELF3-AS1, miR-205 and KLF12
were significantly upregulated at 24 h after transfection (Fig. 3A; P<0.05). ELF3-AS1 overexpression
resulted in miR-205 downregulation (P<0.05), while miR-205
overexpression failed to affect ELF3-AS1 (Fig. 3B). ELF3-AS1 overexpression resulted
in upregulated KLF12, while overexpression of miR-205 led to
downregulated KLF12 expression at both mRNA and protein levels
(Fig. 3C; P<0.05). Overexpression
of miR-205 reduced the effects of ELF3-AS1 overexpression in cells
transfected with both the ELF3-AS1 expression vector and miR-205
mimic (Fig. 3C; P<0.05).
Additionally, overexpression of KLF12 exhibited no significant
impact on ELF3-AS1 and miR-205 expression (Fig. 3D).
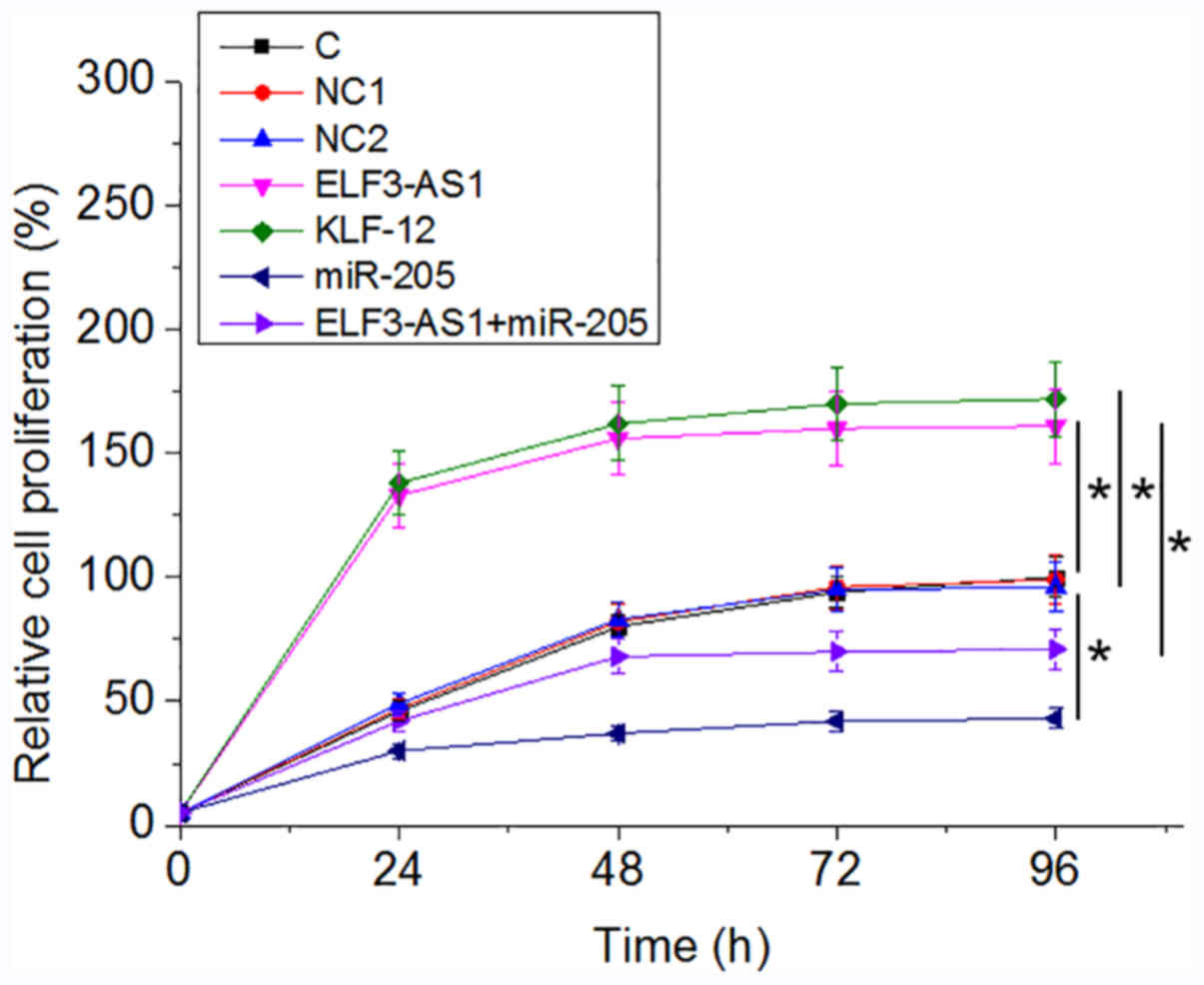
ELF3-AS1 promotes OS cell
proliferation through KLF12 and miR-205
Analysis of cell proliferation revealed that,
compared with the C group, ELF3-AS1 and KLF12 overexpression
resulted in an increased proliferation rate in OS cells at 96 h
after the initiation of cell culture, while overexpression miR-205
played an opposite role and attenuated the effects of ELF3-AS1
overexpression (Fig. 4;
P<0.05).
ELF3-AS1 promotes the methylation of
miR-205 gene
MSP was performed to analyze the effects of ELF3-AS1
overexpression on miR-205 gene methylation. It was observed that
ELF3-AS1 overexpression led to increased methylation of the miR-205
gene (Fig. 5).
Discussion
The expression pattern and function of ELF3-AS1 have
been investigated in the present study. ELF3-AS1 was fond to be
upregulated in OS tissues compared with adjacent non-tumor tissue,
and overexpression of ELF3-AS1 promoted the proliferation of OS
cells by upregulating KLF12 through miR-205. A previous study found
that miR-205 can directly target KLF12 in basal-like breast
carcinoma (15).
miR-205 serves different roles in various types of
cancer (13,14). In esophageal adenocarcinoma, miR-205
is downregulated and inhibits the development of cancer (13). By contrast, in squamous cell
carcinoma of the esophagus and ovarian cancer, miR-205 is
upregulated and promotes cancer progression by targeting tumor
suppressors such as PTEN and SMAD4 (13,14). In
a recent study, miR-205 was found to target KLF12 directly and to
be involved in the regulation of cancer cell apoptosis and invasion
in basal-like breast carcinoma (15). KLF12 is a zinc finger DNA-binding
transcription factor that regulates gene expression (18). In many types of cancer, including
colorectal cancer, KLF12 promotes the growth of the tumor by
regulating cancer-related factors, such as early growth response
protein 1 (18). In the present
study, upregulated KLF12 and downregulated miR-205 were observed in
OS tissues, and the miR-205 overexpression in OC cells resulted in
downregulation of KLF12. Therefore, miR-205 may also directly
target KLF12 in OS.
In the current study, a reduced cell proliferation
rate was observed after miR-205 overexpression and an increased
rate was observed after KLF12 overexpression. Therefore, miR-205
may be a tumor-suppressive miRNA, while KLF12 may be an oncogene in
OS. In bladder cancer, ELF3-AS1 promotes cancer development by
interacting with KLF8 (12). In the
present study, ELF3-AS1 may upregulate KLF12 in OS cells via the
downregulation of miR-205. Therefore, ELF3-AS1 may regulate
different KLFs in various types of cancer, or ELF3-AS1 may regulate
multiple KLFs. However, the mechanism underlying the inhibition of
miR-205 by ELF3-AS1 is unclear. It is known that lncRNA can inhibit
the function of miRNAs by serving as miRNA sponges (19,20).
However, sponges do not downregulate miRNA expression, and in the
current study, downregulated miR-205 was observed following
ELF3-AS1 overexpression. In addition, no target site of miR-205 was
found on ELF3-AS1. However, ELF3-AS1 overexpression led to
increased methylation of the miR-205 gene. Therefore, ELF3-AS1 may
downregulate miR-205 via methylation of its gene. It is worth
noting that only one cell line was used in this study and therefore
the findings should be further validated using other OS cell lines.
In addition, the present study failed to perform KLF12 and miR-205
knockdown experiments. Future studies will try to include these
experiments to further validate the findings. The effects of
co-transfection of miR-205 mimic and KLF12 expression vector on the
proliferation of OS cells should also be investigated to further
analyze the interactions between miR-205 and KLF12 in regulating OS
cell proliferation. The cell phenotypes were also analyzed after
transfections, but no significant changes were observed (data not
shown). Therefore, ELF3-AS1, miR-205 and KLF12 may have no
significant effects on OS cell phenotypes.
In conclusion, ELF3-AS1 was found to be upregulated
in OS, and ELF3-AS1 overexpression may promote the proliferation of
OS cells by upregulating KLF12 potentially through methylation of
the miR-205 gene.
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science
Foundation for Middle-aged and Young Scientists of Wannan Medical
College for 2019 (grant no. WK2019F34) and Funding of ‘Peak’
Training Program and ‘Panfeng’ Innovation Team Project for
Scientifc Research of Yijishan Hospital, Wannan Medical College
(grant. no. PF2019005).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JY and JK designed and performed experiments. MY
collected and analyzed data. JK drafted the manuscript. All authors
approved this manuscript.
Ethics approval and consent to
participate
The Ethics Committee of the First Affiliated
Hospital of Wannan Medical College (Wuhu, China) approved the
study. All patients consented to the use of their samples in this
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Geller DS and Gorlick R: Osteosarcoma: A
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.PubMed/NCBI
|
|
2
|
Messerschmitt PJ, Garcia RM, Abdul-Karim
FW, Greenfield EM and Getty PJ: Osteosarcoma. J Am Acad Orthop
Surg. 17:515–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jaffe N: Osteosarcoma: Review of the past,
impact on the future. The American experience. Cancer Treat Res.
152:239–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Daw NC, Billups CA, Rodriguez-Galindo C,
McCarville MB, Rao BN, Cain AM, Jenkins JJ, Neel MD and Meyer WH:
Metastatic osteosarcoma. Cancer. 106:403–412. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kansara M and Thomas DM: Molecular
pathogenesis of osteosarcoma. DNA Cell Biol. 26:1–18. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hansen MF: Genetic and molecular aspects
of osteosarcoma. J Musculoskelet Neuronal Interact. 2:554–560.
2002.PubMed/NCBI
|
|
8
|
Reddy KB: MicroRNA (miRNA) in cancer.
Cancer Cell Int. 15:382015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McManus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo Y, Chen D, Su X, Chen J and Li Y: The
lncRNA ELF3-AS1 promotes bladder cancer progression by interaction
with Krüppel-like factor 8. Biochem Biophys Res Commun.
508:762–768. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hezova R, Kovarikova A, Srovnal J,
Zemanova M, Harustiak T, Ehrmann J, Hajduch M, Sachlova M, Svoboda
M and Slaby O: MiR-205 functions as a tumor suppressor in
adenocarcinoma and an oncogene in squamous cell carcinoma of
esophagus. Tumuor Biol. 37:8007–8018. 2016. View Article : Google Scholar
|
|
14
|
Li J, Hu K, Gong G, Zhu D, Wang Y, Liu H
and Wu X: Upregulation of MiR-205 transcriptionally suppresses
SMAD4 and PTEN and contributes to human ovarian cancer progression.
Sci Rep. 7:413302017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guan B, Li Q, Shen L, Rao Q, Wang Y, Zhu
Y, Zhou XJ and Li XH: MicroRNA-205 directly targets Krüppel-like
factor 12 and is involved in invasion and apoptosis in basal-like
breast carcinoma. Int J Oncol. 49:720–734. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Steffner RJ and Jang ES: Staging of bone
and soft-tissue sarcomas. J Am Acad Orthop Surg. 26:e269–e278.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim SH, Park YY, Cho SN, Margalit O, Wang
D and DuBois RN: Krüppel-like factor 12 promotes colorectal cancer
growth through early growth response protein 1. PLoS One.
11:e01598992016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|