Introduction
In 2018, a total of 570,000 new cases of cervical
cancer and 311,000 associated mortalities were reported, and this
malignancy is the fourth most common type of cancer in terms of
incidence and mortality rates (1).
Cervical squamous cell carcinoma (CSCC) is the most common
histopathological form of cervical cancer and accounts for ~90% of
all reported cases (2,3). Human papillomavirus (HPV) infection is
the leading cause of CSCC (4). With
the popularization of HPV screening and vaccination program, the
incidence of HPV-positive CSCC has dropped significantly during the
past years (5). However, HPV
vaccines cannot improve the conditions of patients who have already
been infected (5). In addition,
HPV-negative CSCC is more aggressive, and effective prevention and
treatment approaches are currently lacking (6).
Genetic studies have revealed a considerable number
of genetic factors with critical roles in CSCC (7). Cyclin-dependent kinase 6 (CDK6), a
member of the CDK family, mainly regulates cell cycle progression
in G1 phase (8). In CSCC, CDK6 is
overexpressed and accelerates cell cycle progression of cancer
cells to promote cancer progression (9). Therefore, inactivation of CDK6 is a
promising strategy for the treatment of different types of cancer
(10).
Specific tumor-suppressive microRNAs (miRNAs), such
as miR-34a, have been demonstrated to target and cleave CDK6,
thereby inhibiting tumor growth (11). MACC1-AS1 is a long noncoding RNA
(lncRNA), an RNA with a length of >200 nucleotides, that has
been reported to have oncogenic functions only in gastric cancer
(12,13). In the present study, bioinformatics
analysis revealed that MACC1-AS1 may form a base pair with miR-34a.
The study aimed to investigate the interactions among MACC1-AS1,
miR-34a and CDK6 in CSCC.
Materials and methods
CSCC patients
In the present study, a total of 60 CSCC patients
[including 39 males and 21 females; age range, 40–66 years; mean
age, 51.9±6.6 (SD) years] were selected from 111 CSCC cases
diagnosed in Qingdao No. 6 People's Hospital (Qingdao) between
March 2015 and April 2018. The present study was approved by the
review board of the Ethics Committee of Qingdao No. 6 People's
Hospital. Patients were included into the present study if they
were newly diagnosed CSCC cases, had not received any prior cancer
therapies, and no other therapies were initiated with 100 days
prior to the admission day. The exclusion criteria included
diagnosis of multiple clinical disorders, recurrent cases and
history of other malignancies. Based on the clinical findings and
American Joint Committee on Cancer (AJCC) staging system (14), a total of 12, 12, 16 and 20 of the
included cases were classified as clinical stage I, II, III, and
IV, respectively. Among the 60 CSCC patients, 46 cases were
HPV-positive (including infection with HPV types 11, 16, and 18).
All patients were informed of the contents of the present study and
the potential publication of this paper, and all participants
signed an informed consent form.
CSCC tissues and cells
Prior to the initiation of any therapies, a cervical
biopsy was performed under the guidance of magnetic resonance
imaging. During the biopsy, CSCC and adjacent non-tumor (within 2
cm around the tumors) tissues were obtained from the patients. The
weight of each sample ranged between 0.014 and 0.019 g. All tissue
samples were subjected to histopathological tests to confirm that
they were tumor or non-tumor samples. All tissue samples were
stored at −80°C before RNA extractions.
In addition, the SiHa human CSCC cell line (ATCC)
was used in the present study. SiHa cells were cultured in a
mixture of 10% fetal bovine serum and 90% Eagle's minimum essential
medium under the conditions of 5% CO2, 95% humidity and
37°C.
Cell transfection
miR-34a mimic (5′-UGGCAGUGUCUUAGCUGGUUGU-3′) and
negative control (NC) mimic (5′-CGCGAUUGUAAACUUGCCGCG-3′) were
obtained from GenePharma Co., Ltd. MACC1-AS1 and CDK6 expression
vectors were established using the pcDNA3.1 vector (GenePharma Co.,
Ltd.). In order to perform transient transfections, SiHa cells were
harvested when 80% confluence was reached and counted. Next,
2×106 cells in 2 ml medium (10% fetal bovine serum and
90% Eagle's minimum essential medium) were transferred to each well
of a 6-well plate and were incubated with the transfection mixture,
containing Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) and 40 nM miR-34a mimic or 10 nM overexpression
vector, for 5 h at 37°C. NC mimic or empty vector served as the NC
groups. Following transfection, the cells were washed with fresh
cell culture medium. Untransfected cells served as the normal
control cells in all transfection experiments. All subsequent
experiments were performed using cells collected at 24 h
post-transfection.
RNA extraction
Total RNAs were extracted from 2×105
cells (harvested at 24 h post-transfection) or 0.015 g tissue
samples (ground in liquid nitrogen) using RiboZol (Sigma-Aldrich;
Merck KGaA). To retain the miRNAs in RNA samples, 85% of ethanol
was used to precipitate and wash the RNAs. All RNA samples were
subjected to digestion with DNase I for 2 h at 37°C to remove
genomic DNA.
Reverse transcription-quantitative
polymerase chain reaction (qPCR)
For mRNA detection, the digested RNA samples were
reverse transcribed into cDNAs using Tetro Reverse Transcriptase
(Bioline), and TB Green Advantage qPCR Premix (Clontech; Takara Bio
USA, Inc.) was used in the qPCR mixture. The mRNA expression levels
of MACC1-AS1 and CDK6 were measured by qPCR, with GAPDH serving as
the endogenous control. For miRNA detection, the
All-in-One™ miRNA qRT-PCR Detection kit (GeneCopoeia,
Inc.) was used to perform addition of poly (A), reverse
transcription and qPCR assays. The expression levels of miR-34a
were measured, with U6 serving as the endogenous control. The
thermal cycling conditions for all reactions were as follows: 95°C
for 30 min, followed by 40 cycles of 95°C for 10 sec and 55°C for
40 sec. The following primer sequences were used in qPCR assays:
MACC1-AS1 forward 5′-GCCAGTCAGAAAATGAGGAAC-3′ and reverse,
5′-CCAGTTGGGTGAACAGGAC-3′; CDK6 forward 5′-TGGAGACCTTCGAGCACC-3′
and reverse, 5′-CACTCCAGGCTCTGGAACTT-3′; GAPDH forward
5′-CATCACTGCCACCCAG-3′ and reverse 5′-ATGCCAGTGAGCTTCCC-3′; miR-34a
forward 5′-CCGGCATGGCAGTGTCTTAGCT-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGTA-3′; and U6 forward 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTGCGT-3′. The 2−ΔΔCq
method (15) was used for gene
expression normalization, and all qPCR experiments were performed
in three replicates.
LncRNA-miRNA interaction
prediction
The interacton between MACC1-AS1 and miR-34a was
analyzed using IntaRNA 2.0 (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp)
(16). In the analysis, the sequence
of miR-34a was used as short sequence and sequence of MACC1-AS1 was
used as long sequence. All other parameters were set as the
default.
Western blot analysis
Total proteins were extracted from 2×105
cells (harvested at 24 h post-transfection) using RIPA solution and
quantified using a BCA kit (both from GenePharma Co., Ltd.). For
protein denaturation, all protein samples were incubated in boiling
water for 8 min. The denatured protein samples were then subjected
to 10% SDS-PAGE, followed by gel transfer to PVDF membranes and
blocking in 5% non-fat milk in PBS for 90 min at room temperature.
To detect the protein expression of CDK6, the membranes were
incubated with rabbit anti-CDK6 (1:1,300; ab226349; Abcam) and
anti-GAPDH (serving as an endogenous control; 1:1,300; ab37168;
Abcam) primary antibodies at 4°C for 18 h. Next, the membranes were
further incubated with horseradish peroxidase-conjugated IgG
sercondary antibody (1:1,300; ab6721; Abcam), and this incubation
was performed for 2 h at 24°C. An enhanced chemiluminescence
reagent (Sigma-Aldrich; Merck KGaA) was used to incubate the
membranes for 10 min to develop the signals, and all data were
quantified and normalized using ImageJ software, version 1.46
(National Institutes of Health, Bethesda, MD, USA).
Cell Counting Kit-8 (CCK-8) cell
proliferation analysis
Single-cell suspensions were prepared by mixing
4×103 cells with 1 ml aforementioned culture medium (10%
fetal bovine serum and 90% Eagle's minimum essential medium). Cells
were incubated in a 96-well culture plate with 0.1 ml per well
under the aforementioned cell culture conditions. Cells were
collected every 24 h for a total of 4 days. At 4 h before the
collection of cells, 10 µl CCK-8 solution (Sigma-Aldrich; Merck
KGaA) was added to each well. Finally, the optical density values
at 450 nM were measured.
Cell cycle analysis
SiHa cells were collected at 24 h post-transfection
and were subjected to trypsinization. Cells were resuspended in
pre-cold PBS, followed by centrifugation at 1,200 × g for 10 min at
4°C. The supernatant was removed, and the cell pellets were
resuspended in 75% ethanol. Following incubation in 75% ethanol for
4 h at 4°C, the cells were centrifuged at 1,200 × g for 10 min at
4°C. Next, the supernatant was removed, and the cell pellets were
resuspended in pre-cold PBS. BD Pharmingen™ PI/RNase staining was
then performed for 3 min, and flow cytometer was conducted to
separate the cells. The gating strategy was as follows: i) Define
Gate 1: X-FSC; Y-SSC; Gate 2: X width; Y-FL2A; ii) cell debris and
dead cells were excluded with ModFit LT (Verity Software House);
and iii) a fluorescence analysis dot plot was then set up. ‘Region
1-FSC vs. SSC’, ‘Zone 2 feasible’ and ‘Zone 3-Shared Mark’ are on
each corresponding fluorescent dot map. In each experiment,
105 events were counted. Subsequently, Infinicyt™ flow
cytometry data analysis software (ALPCO, Macedon, NY, USA) was used
to analyze the data. A figure presenting the cell cycle data was
plotted using Origin software, version 9.5 (OriginLab
Corporation).
Statistical analysis
Experiments were performed in three replicates. Mean
values were calculated and used in all data analyses. Assessment of
differences between tissue types (non-tumor vs. CSCC) and among
multiple cell transfection groups was performed by performing
paired t-test or one-way analysis of variance (ANOVA) in
combination with Tukey's test, respectively. Comparison between
HPV-negative and HPV-positive CSCC patients was performed by
unpaired t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Upregulation of MACC1-AS1 is affected
by clinical stage, but not by HPV infection status
The levels of MACC1-AS1 expression in the non-tumor
and CSCC tissue samples were measured by RT-qPCR and compared by
conducting paired t-test. It was observed that, compared with the
non-tumor tissues, MACC1-AS1 expression was significantly higher in
CSCC tissues (Fig. 1A; P<0.05).
Among the 60 CSCC patients included in the present study, a total
of 12, 12, 16 and 20 cases were classified as clinical stage I–IV,
respectively. The expression levels of MACC1-AS1 were compared
among the four stages by performing one-way ANOVA in combination
with Tukey's test. With the increase of clinical stage,
significantly increased expression levels of MACC1-AS1 were
observed in CSCC tissues (Fig. 1B;
P<0.05). Among the 60 CSCC patients, 46 cases were HPV-positive
(including infection with HPV types 11, 16 and 18). Comparison of
expression levels of MACC1-AS1 in CSCC tissues between HPV-negative
and HPV-positive patients was performed by unpaired t-test. The
results revealed no significant difference between these two
patient groups (Fig. 1C).
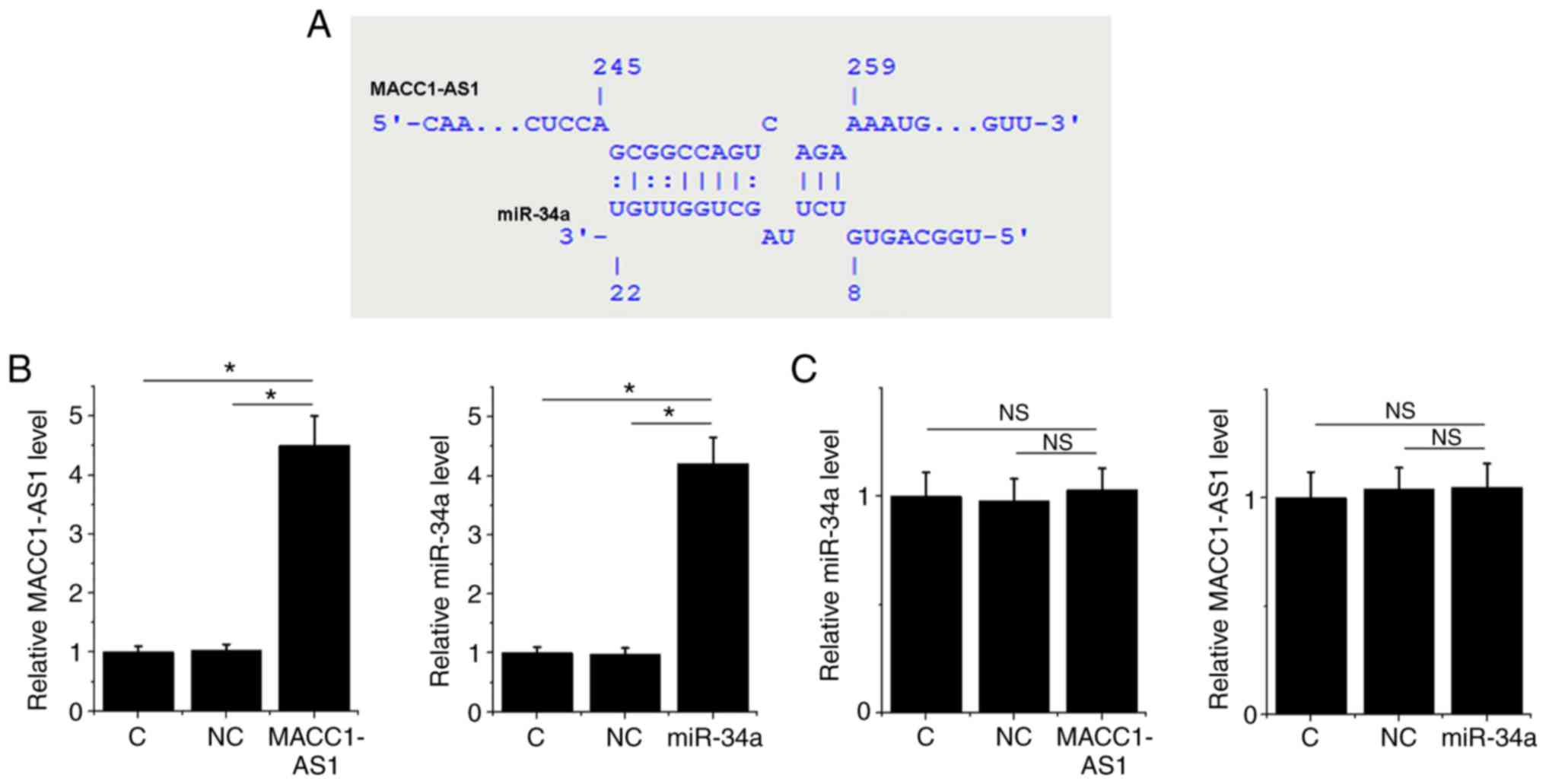
MACC1-AS1 may be a sponge of
miR-34a
Bioinformatics analysis using the IntaRNA tool
demonstrated that MACC1-AS1 forms base pairing with miR-34a
(Fig. 2A). To further investigate
the interaction between MACC1-AS1 and miR-34a, a MACC1-AS1
expression vector or miR-34a mimic was transfected into SiHa cells.
At 24 h post-transfection, the RT-qPCR results revealed that,
compared to the normal control and NC groups, the expression levels
of MACC1-AS1 and miR-34a were significantly increased in their
corresponding transfected SiHa cells, indicating successful
overexpression (Fig. 2B; P<0.05).
However, MACC1-AS1 overexpression did not affect the level of
miR-34a, and similarly miR-34a overexpression did not significantly
change the level of MACC1-AS1 (Fig.
2C; P>0.05).
MACC1-AS1 may sponge miR-34a to
upregulate CDK6
It is known that CDK6 is a direct target of miR-34a
(11). Western blot and RT-qPCR
assays were performed to analyze the effects of the overexpression
of miR-34a and CDK6 on CDK6 expression. As compared with the normal
control and NC groups, MACC1-AS1 overexpression led to upregulated
CDK6 at the mRNA (Fig. 3A) and
protein (Fig. 3B) levels. By
contrast, miR-34a overexpression had the opposite effect on CDK6
levels, while it reduced the effects of MACC1-AS1 overexpression in
co-transfected cells (Fig. 3A and
B).
MACC1-AS1 promoted SiHa cell
proliferation and cell cycle progression via CDK6 and miR-34a
Cell proliferation and cell cycle assays were
performed to analyze the effect of transfection on the
proliferation and cell cycle progression of SiHa cells.
Overexpression of CDK6 in vector-transfected cells was confirmed by
western blot analysis (supplementary figure Fig. S1). Compared with the normal control
and NC groups, overexpression of MACC1-AS1 or CDK6 significantly
promoted cell proliferation (Fig.
4A; P<0.05), while it also led to an increased cell
percentage at G2 phase and decreased cell percentage at G1 phase
(Fig. 4B and C; P<0.05). By
contrast, miR-34a overexpression had the opposite effects on cell
cycle progression and cell proliferation. In addition, miR-34a
reduced the effects of MACC1-AS1 overexpression in cells
co-transfected with MACC1-AS1 vector and miR-34a mimic (Fig. 4A-C). Therefore, MACC1-AS1 may
upregulate CDK6 via miR-134a to promote SiHa cell proliferation and
cell cycle progression.
Discussion
To date, the oncogenic function of MACC1-AS1 has
only been analyzed in gastric cancer. It has been reported that
MACC1-AS1 was upregulated in gastric cancer, and that
overexpression of MACC1-AS1 was at least partially responsible for
cancer cell metabolic plasticity and stemness (12,13).
However, to the best of our knowledge, the role of this lncRNA in
other types of cancer remains unknown. The present study is, thus,
the first to report the upregulation of MACC1-AS1 in CSCC tissues,
and that MACC1-AS1 overexpression led to accelerated cell cycle
progression and cell proliferation. In addition, the current study
further revealed that MACC1-AS1 may serve as a sponge of miR-34a to
upregulate CDK6, thereby promoting cell cycle progression and cell
proliferation. Therefore, MACC1-AS1 may serve an oncogenic role in
CSCC by promoting cell division.
The cancer stage is significantly associated with
the survival of cancer patients (16). In the present study, increased
expression levels of MACC1-AS1 were observed with the increase in
CSCC clinical stage. This observation suggested that MACC1-AS1
expression may have a prognostic value in CSCC, and our future
studies will perform follow-up analysis to evaluate this potential
prognostic value of MACC1-AS1. Although HPV infection is the main
cause of CSCC (17), the present
study data suggested that MACC1-AS1 expression was not affected by
HPV infection. Therefore, MACC1-AS1 may be involved in CSCC through
HPV-independent pathways.
Previously, it has been reported that miR-34a can
target CDK6 to suppress glioblastoma (11). In the present study, downregulation
of CDK6 was observed in cells transfected with miR-34a mimic.
Therefore, miR-34a may also target CDK6 in CSCC. Bioinformatics
analysis further demonstrated that MACC1-AS1 forms base pairing
with miR-34a. However, overexpression experiments revealed that
MACC1-AS1 and miR-34a did not significantly affect the expression
of each other, suggesting that miR-34a may not target MACC1-AS1.
Previous studies on the interaction between lncRNAs and miRNAs
indicated that lncRNAs may sponge miRNAs to upregulate their
targets (18,19). The data of the current study further
revealed that MACC1-AS1 may sponge miR-34a to upregulate CDK6. It
is worth noting that the bioinformatics analysis results indicated
that MACC1-AS1 may sponge multiple miRNAs (data not shown), while
most of these miRNAs are unlikely to be involved in CSCC.
According to the results, the present study revealed
a novel MACC1-AS1/miR-34a/CDK6 axis in CSCC. The regulation of
MACC1-AS1 expression may provide novel insights in CSCC treatment.
It is worth noting that miR-34a in cancer biology may target
multiple oncogenes, such as CD44 and MYCN (20,21).
Therefore, future studies will focus on the interactions between
MACC1-AS1 and other targets of miR-34a. In addition, the
bioinformatics analysis performed in the present study revealed
that MACC1-AS1 interacted with multiple miRNAs, and these
interactions should also be further investigated in future
studies.
In conclusion, the current study revealed that
MACC1-AS1 was upregulated in CSCC and that it may serve as a sponge
of miR-34a to upregulate CDK6, thereby promoting the progression of
CSCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JQJ and XMC performed the clinical studies,
experimental work, data analysis and manuscript writing. JC
performed the experimental work and literature research. XG was
responsible for the study concept, research design and manuscript
editing. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the Ethics
Committee of Qingdao No. 6 People's Hospital (Qingdao, China).
Written informed consent was signed by all participants.
Patient consent for publication
All patients provided written informed consent and
the study was approved by the Ethics committee of the Qingdao No. 6
People's Hospital (Qingdao, China).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Koh WJ, Greer BE, Abu-Rustum NR, Apte SM,
Campos SM, Cho KR, Chu C, Cohn D, Crispens MA, Dorigo O, et al:
Cervical cancer, version 2.2015. J Natl Compr Canc Netw.
13:395–404. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Small W Jr, Bacon MA, Bajaj A, Chuang LT,
Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR,
Viswanathan AN and Gaffney DK: Cervical cancer: A global health
crisis. Cancer. 123:2404–2412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wentzensen N, Schiffman M, Palmer T and
Arbyn M: Triage of HPV positive women in cervical cancer screening.
J Clin Virol. 76 (Suppl 1):S49–S55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lowy DR: HPV vaccination to prevent
cervical cancer and other HPV-associated disease: From basic
science to effective interventions. J Clin Invest. 126:5–11. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walboomers JM and Meijer CJ: Do
HPV-negative cervical carcinomas exist? J Pathol. 181:253–254.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cancer Genome Atlas Research Network;
Albert Einstein College of Medicine; Analytical Biological
Services; Barretos Cancer Hospital; Baylor College of Medicine;
Beckman Research Institute of City of Hope; Buck Institute for
Research on Aging; Canada's Michael Smith Genome Sciences Centre;
Harvard Medical School; Helen F. Graham Cancer Center &Research
Institute at Christiana Care Health Services, ; et al: Integrated
genomic and molecular characterization of cervical cancer. Nature.
543:378–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Swaffer MP, Jones AW, Flynn HR, Snijders
AP and Nurse P: CDK substrate phosphorylation and ordering the cell
cycle. Cell. 167:1750–1761.e16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen SN, Wang H and Gong BL: Expressions
of miRNA29 target genes CCND2 and CDK6 in cervical cancer. Disc
Clin Cases. 5:14–18. 2018. View Article : Google Scholar
|
|
10
|
Sherr CJ, Beach D and Shapiro GI:
Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov.
6:353–367. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Guessous F, Zhang Y, Dipierro C,
Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen
TD, et al: MicroRNA-34a inhibits glioblastoma growth by targeting
multiple oncogenes. Cancer Res. 69:7569–7576. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang
S, Dong S, Wen Z, Rao J, Liao W and Shi M: The lncRNA MACC1-AS1
promotes gastric cancer cell metabolic plasticity via AMPK/Lin28
mediated mRNA stability of MACC1. Mol Cancer. 17:692018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He W, Liang B, Wang C, Li S, Zhao Y, Huang
Q, Liu Z, Yao Z, Wu Q, Liao W, et al: MSC-regulated lncRNA
MACC1-AS1 promotes stemness and chemoresistance through fatty acid
oxidation in gastric cancer. Oncogene. 38:4637–4654. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Busch A, Richter AS and Backofen R:
IntaRNA: Efficient prediction of bacterial sRNA targets
incorporating target site accessibility and seed regions.
Bioinformatics. 24:2849–2856. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Strohl AE, Mendoza G, Ghant MS, Cameron
KA, Simon MA, Schink JC and Marsh EE: Barriers to prevention:
Knowledge of HPV, cervical cancer, and HPV vaccinations among
African American women. Am J Obstet Gynecol. 212:65 e1–e5. 2015.
View Article : Google Scholar
|
|
18
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing MiRNA- LncRNA Interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jalali S, Bhartiya D, Lalwani MK,
Sivasubbu S and Scaria V: Systematic transcriptome wide analysis of
lncRNA-miRNA interactions. PLoS One. 8:e538232013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei JS, Song YK, Durinck S, Chen QR, Cheuk
AT, Tsang P, Zhang Q, Thiele CJ, Slack A, Shohet J and Khan J: The
MYCN oncogene is a direct target of miR-34a. Oncogene.
27:5204–5213. 2008. View Article : Google Scholar : PubMed/NCBI
|