Introduction
Despite the overall reduction in the number of
cancer-related deaths following improved therapeutic options, the
number of pancreatic ductal adenocarcinoma (PDAC)-related deaths
continues to increase; it is associated with a median overall
survival of <1 year and <10% survival at 5 years (1). Thus, an improved understanding of the
mechanisms mediating PDAC tumourigenesis is required.
Plakoglobin, also known as γ-catenin, is a member of
the catenin family and is an important component of desmosomes and
adherens junctions in mammalian cells, where it facilitates
cell-cell adhesion by linking desmosomal cadherins to the
cytoskeleton (2,3). Plakoglobin can also be found localised
in the nucleus, where it exerts nuclear functions, such as
inhibiting the Wnt/β-catenin signalling pathway through regulating
the transcription of specific components within the pathway
(4,5). Nonetheless, the role that plakoglobin
serves in tumorigenesis is disputed; whilst the majority of
previous studies reported that plakoglobin suppressed tumour
growth, others observed that plakoglobin exhibited pro-oncogenic
features (5–9). For example, the overexpression of
plakoglobin in SV40-transformed 3T3 cells suppressed tumour
formation upon transplantation into syngeneic mice, and upon
co-transfection of plakoglobin with N-cadherin, this tumour
suppressive effect was considerably more effective than plakoglobin
overexpression alone. Similarly, the overexpression of plakoglobin
in a human renal carcinoma cell line that lacked the expression of
cadherins or β-catenin also suppressed tumour formation in nude
mice (6). In addition, plakoglobin
reportedly affected several downstream signalling molecules
involved in tumour suppressive pathways; plakoglobin reportedly
regulated the expression and stability of the metastasis suppressor
protein, Nm23, as well as the p53-regulated tumour suppressor,
14-3-3σ (4,7,10). On
the other hand, a recent study demonstrated that high expression
levels of plakoglobin promoted metastasis in invasive
micropapillary carcinoma of the breast by promoting tumour cluster
formation through activating PI3K/AKT/Bcl-2 signalling (9).
As a closely related homologue of β-catenin, a
crucial transcriptional regulator of the canonical Wnt signalling
pathway, plakoglobin shares structural and functional similarities
with β-catenin (11). It has
previously been demonstrated that plakoglobin competes with
β-catenin, which leads to aberrations in Wnt signalling; both
β-catenin and plakoglobin were observed to bind to the
transcriptional co-activator, transcription factor 7-like 2 (also
known as Tcf4), although the latter bound with stronger affinity,
and their co-expression led to the overall net inhibition of Wnt
signalling (12,13). Plakoglobin was also discovered to
activate T-cell factor/lymphocyte enhancer factor (TCF/LEF)
transcriptional activity in the absence of β-catenin (11), and the decrease in plakoglobin
expression levels correlated with poorer prognosis in malignancies
such as non-small lung cancer (14).
PDAC originates from both acinar and ductal cells
and the well-described mechanism of tumorigenesis involves the
early genetic alteration in the oncogene, K-RAS, whereby the
expression of K-RASG12D is thought to induce the
disease (15–17). However, studies in mice models have
demonstrated that the expression of KRASG12D in
mature pancreatic acinar cells was insufficient to induce the PDAC
precursor, pancreatic intraepithelial neoplasia (18), and that the accumulation of mutations
in P16, TP53 and SMAD4 (15–17) are
also required for the cancer to progress. Thus, nodal regulators of
cellular responses have been proposed as potential strategies for
inhibiting PDAC formation (18). It
was noted in a previous study that compared with pancreatic acinar
cells, ductal cell developed more rapidly into PDAC in the presence
of K-RAS and TP53 mutations (19). Thus, the discovery of other
mechanisms dependent or independent of K-RAS is required to
understand the induction, development and progression of
early-stage PDAC. Genetic analysis of 24 patients with advanced
PDAC revealed that twelve core signalling pathways were
dysregulated in 67% of PDAC tumours, including Hedgehog, Wnt/Notch,
K-RAS, small GTPase, transforming growth factor (TGF)-β and
integrin signalling. Notably, despite the heterogeneity of these
altered genes amongst the patients, all PDAC tumours demonstrated
alterations in the Wnt/Notch and Hedgehog signalling pathways
(20). Further investigations have
observed the induction of several mitogenic signalling pathways by
various growth factors in PDAC (21–24).
Altogether, these studies suggested that alterations in single
molecules in PDAC present little opportunity for drug development,
but targeting downstream effectors at nodal points, which control
biological processes such as metabolism, cell migration and
apoptosis, may be more feasible interventions for drug development.
In the present study, the expression of dysregulated genes in early
PDAC tumours, as well as differentially regulated signalling
pathways in the PDAC tumour microenvironment, were investigated
compared with normal pancreatic tissue.
Materials and methods
Patient studies
The present study was approved by The Human Research
Ethics Committee of The University of Witwatersrand (approval no.
M150778; Johannesburg, South Africa). Informed, voluntary consent
was obtained from all patients. Samples were obtained from
consented patients (age range 52–67 years) at Chris Hani
Baragwanath Hospital, Johannesburg South Africa from January 2014
to June 2016. PDAC tumour samples and paired non-malignant
pancreatic tissue samples (≥2 cm away from the tumour) were
obtained from nine patients who underwent a pancreaticoduodenectomy
for clinically resectable, early stage PDAC (Table I). It is important to note that
patients recruited for this study were representative of different
clinical stages, thus observations made within the study were
attributed to PDAC in general. Biopsies were stored in 1 ml
RNAlater RNA stabilization reagent (cat. no. 76106; Qiagen, Inc.)
and subsequently homogenized in 600 µl AllPrep DNA/RNA Mini kit
lysis buffer (cat. no. 80204; Qiagen, Inc.) using a TissueRuptor
(cat. no. 9001272; Qiagen, Inc.).
 | Table I.Characteristics of patients enrolled
in the present study. |
Table I.
Characteristics of patients enrolled
in the present study.
| No. | Sex | Age | Alcohol use | Diabetes |
Differentiation | TNM | Stage | Alive 12/12 |
|---|
| 1 | Male | 67.1 | No | Yes | Mod diff | T3N1M0 | IIB | Yes |
| 2 | Male | 55.8 | Yes | No | Well diff | T1N0M0 | IA | Yes |
| 3 | Female | 62.7 | No | Yes | Well diff | T2N1M0 | IIB | Yes |
| 4 | Male | 63.5 | Yes | No | Mod diff | T3N1M0 | IIB | Yes |
| 5 | Male | 54.3 | Yes | No | Poor diff | T2N1M0 | IIB | Yes |
| 6 | Male | 52.2 | Yes | Yes | Well diff | T3N1M0 | IIB | Yes |
| 7 | Femalea,b | 57.3 | No | No | Mod diff | T3N1M0 | IIB | Yes |
| 8 | Malea,b | 43.4 | No | No | Mod diff | T3N1M0 | IIB | Yes |
| 9 | Femaleb | 52.6 | No | No | Mod diff | T2N1M0 | IIA | Yes |
RNA extraction
Total RNA was extracted from PDAC tumour tissue and
non-malignant pancreatic tissue using an AllPrep DNA/RNA Mini kit
(cat. no. 80201; Qiagen, Inc.) and DNase digestion was performed
using a DNase kit (cat. no. AMPD1; Sigma-Aldrich; Merck KGaA),
according to the manufacturers' protocols. Total RNA was quantified
using a NanoDrop ND-1000 UV spectrophotometer and a ratio of
absorbance at 260 and 280 nm of >1.8 was observed in all
samples.
Gene expression profiling using RNA
sequencing
Total RNA extracted from tissue samples obtained
from patients 7 and 8 (Table I) were
treated with DNase to remove genomic DNA. The NEB Next rRNA
Depletion kit (human/mouse/rat) (New England Biolabs Inc.; cat no.
E6310S) was subsequently used to remove the ribosomal RNA from the
samples. The mRNA was converted into cDNA using the Ultra RNA
Library Prep kit (New England Biolabs Inc.; cat no. 7530L) and
subsequently sequenced using the Illumina MiSeq system. The CLC
Genomics Workbench version 11 software (www.qiagenbioinformatics.com/products/clc-genomics-workbench)
was used for data normalization and differential gene analysis,
with HG19 serving as the reference genome.
Biomarker filtering analysis
The biomarker filtering analysis tool available in
the Ingenuity pathway analysis (IPA) software v01-08 (https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/
was used to identify novel potential biomarkers. Microsoft Excel
files containing the differentially expressed genes (DEGs) obtained
from RNA sequencing of tissue samples were uploaded onto the IPA
platform and the biomarker filtering tool facilitated the
identification of relevant and novel biomarkers based on the
applied filters. The following filters were used: Species (Human);
Tissues and cell lines (Pancreatic cancer cell lines); Node types
(All nodes except complexes and groups): Diseases (Cancer);
Biofluids (Blood); Molecules (All types); Biomarker (Not a known
biomarker). Manual filtering was also conducted, including
searching the NCBI database (https://www.ncbi.nlm.nih.gov/pubmed/) and pancreatic
cancer database (http://pancreaticcancerdatabase.org/index.php) to
further refine the biomarker analysis.
Validation by reverse
transcription-quantitative PCR (RT-qPCR)
The mRNA expression levels of potential biomarkers
were validated using RT2-qPCR primer assays (Qiagen,
Inc.), according to the manufacturer's protocol. The cycling
condition was set as follows: 95°C for 10 min (1 cycle), 95°C for
15 secs (45 cycles) and 60°C for 1 min (45 cycles). At the end of
the cycles the Cp values were obtained to be used for analysis.
Kits were purchased targeting JUP (cat on pancreatic
tissue. no. PPH07138F), COPG (cat. no. PPH07138F),
TCIRG1 (cat. no. PPH09294A) and the reference gene
MRPL19 (cat. no. PPH09294A), a housekeeping gene whose
expression remains unchanged in PDAC carcinogenesis (25). A Roche LightCycler-480 was used with
the cut off Cp set at 3s, according to the manufacturer's protocol.
Experiments adhered to the Minimum Information for Publication of
Quantitative Real-Time PCR Experiments guidelines, including
performing three independent experiments (26). Expression levels were quantified by
uploading the data into the Relative Expression Software Tool
(REST) software (27).
Protein extraction and
quantification
Total protein from 20 mg PDAC and non-cancerous
pancreatic tissue was extracted using a lysis buffer [(0.5% igepal,
0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 150 mM
sodium chloride and 50 mM Tris-HCl (pH 7.5) containing a protease
inhibitor cocktail of aprotinin (0.5 µg/ml) and PMSF (1 mM)].
Tissues were homogenized using TissueRuptor and centrifuged at
2,000 × g for 10 min at 4°C. The supernatant was transferred into a
new tube and the total protein was quantified using the Quick
Start™ Bradford protein assay kit (cat. no. 5000006; Bio-Rad
Laboratories, Inc.), according to the manufacturer's protocol.
Focused proteomic profiling on
pancreatic tissue using Human Oncology Proteome Profiler Array
Protein expression profiling was conducted to
determine the relative expression levels of 84 cancer-related
proteins using the Proteome Profiler Human XL Oncology Array kit
(cat. no. ARY026; R&D Systems Europe, Ltd.), according to the
manufacturer's protocol. Quick Spots image analysis software
vPCM.22.0.0.i (R&D Systems Europe, Ltd.) was used to measure
the intensity of each spot and identify differentially expressed
proteins (DEPs). The intensity of each protein was compared between
the tumour and corresponding normal pancreatic tissues, and
differentially expressed proteins (DEPs) with P<0.05 were
selected. Briefly, the membranes were incubated at room temperature
in 2 ml block buffer and placed on a rocking platform for 1 h.
Subsequently, 500 µl Array Buffer 4 was added and the membranes
were incubated overnight at 4°C. Following incubation, the
membranes were washed three times with 1 ml wash buffer. Then, 30
ml detection antibody cocktail was added to 1.5 ml 1X Array buffer
4/6 and incubated at room temperature with the membrane on a
rocking platform shaker for 1 h. Subsequently, 2 ml diluted
horseradish peroxidase conjugated-streptavidin was added to the
membrane and incubated at room temperature for 30 min. Finally, the
membrane was washed as previously described and 1 ml Chemi reagent
mix was added to the membrane. Membranes were visualised using a
Bio-Rad ChemiDoc imaging system (Bio-Rad Laboratories, Inc.).
Statistical and bioinformatics
analysis
Statistical analysis on three biological replicates
was performed using Microsoft Office Excel (2016) and data are
presented as the mean ± SEM. Differences between groups were
analysed using Student's t-test. P<0.05 was considered to
indicate a statistically significant difference. The Reactome
database (28) and PANTHER version 6
software (29) were used to identify
dysregulated pathways (P<0.05). For statistical analysis of the
real-time PCR data, the Pair Wise Fixed Reallocation Randomisation
Test was performed by the REST software.
Results
JUP is upregulated in early resectable
PDAC tumours
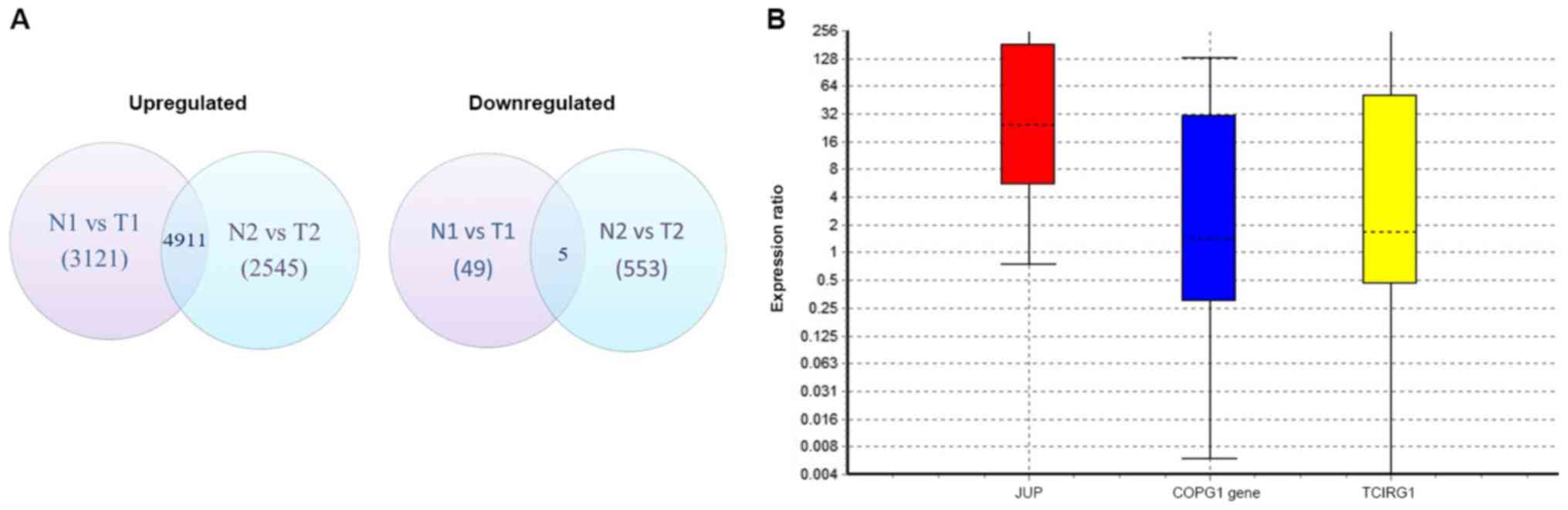
Differential gene expression analysis of the
sequenced transcriptomes from patients 1 and 2 revealed that 4,911
genes were commonly upregulated and 5 were downregulated (Fig. 1A). Using the IPA biomarker module and
aforementioned filter parameters, JUP encoding plakoglobin,
COPG encoding coatomer protein complex, subunit gamma and
TCIRG1 encoding T-cell immune regulator 1 were identified as
potential biomarkers. Validation of these genes using RT-qPCR
demonstrated that JUP expression levels were increased in
all PDAC samples compared with the corresponding normal pancreatic
tissue [p(H1)=0.012] (Fig. 1B).
However, the expression levels of COPG in PDAC tissues were
not significantly different compared with normal tissues, with a
probability of the alternate hypothesis [p(H1)=0.703].
TCIRG1 expression levels were also unchanged between normal
and PDAC tissue samples [p(H1)=0.597]. Although the expression
levels of JUP were significantly increased in tumour
tissues, it was observed to have a wide 95% confidence interval of
1.25–1,856.39.
Dysregulated pathways in PDAC
identified by gene expression analysis
Bioinformatics analysis predicated that the insulin
growth factor-1 (IGF-1) pathway and mitogen-activated protein
kinase (MAPK) kinase signalling cascade were the most upregulated
pathway in PDAC tumours (Tables II
and SI). The other most upregulated
and active signalling pathways were the integrin
(P=9.65×10−15), RAS (P=1.10×10−3),
platelet-derived growth factor (P=1.06×10−6), epidermal
growth factor receptor (EGF; P=7.72×10−6), angiogenesis
(P=5.58×10−7), cholecystokinin receptor
(P=1.16×10−6), Vascular endothelial growth factor
(P=4.11×10−2), B-cell activation
(P=4.11×10−2) and Notch (P=2.6×10−6)
signalling pathways (Tables II and
SII–SX). Pathways found to be downregulated in
PDAC included the proton/oligopeptide co-transportation system,
Golgi-ER trafficking and MHC II antigen presentation pathways
(Table III). Additional analysis
using Reactome confirmed the dysregulation of these pathways.
 | Table II.Top upregulated pathways identified
in tumours using the Reactome and PANTHER databases following RNA
sequencing. |
Table II.
Top upregulated pathways identified
in tumours using the Reactome and PANTHER databases following RNA
sequencing.
| Pathways | P-value | Number of genes
involved |
|---|
| Insulin/IGF
pathway-mitogen activated protein kinase/MAP kinase cascade |
2.19×10−2 | 19 |
| Integrin signaling
pathway |
9.65×10−15 | 112 |
| Ras pathway |
1.10×10−3 | 39 |
| PDGF signaling
pathway |
1.06×10−6 | 74 |
| EGF receptor
signaling pathway |
7.72×10−6 | 70 |
| Angiogenesis |
5.58×10−7 | 88 |
| CCKR signaling
map |
1.16×10−6 | 120 |
| VEGF signaling
pathway |
4.11×10−2 | 35 |
| B cell
activation |
4.11×10−2 | 33 |
| Notch signaling
pathway |
2.6×10−6 | 38 |
 | Table III.Top downregulated pathways identified
in tumours using the Reactome and PANTHER databases following RNA
sequencing. |
Table III.
Top downregulated pathways identified
in tumours using the Reactome and PANTHER databases following RNA
sequencing.
| Pathways | Genes involved | P-value |
|---|
| Proton/oligopeptide
co-transportation (Drug transportation) | SLC15A1, SLC15A2,
SLC15A4 |
1.21×10−7 |
| Organelle
trafficking | KIFC, GALNT1,
KIF5A, MANIA1 |
1.12×10−3 |
| Membrane
trafficking | SEC23B, KIF5C,
GALNT1, KIF5A, DVL2, MANIA1 |
2.82×10−3 |
| MHC Class II
antigen presentation (Immune activation) | SEC23B, KIF5C,
KIF5A |
3.87×10−3 |
Upregulated proteins and their
signalling pathways
Protein expression profiling of PDAC and normal
tissue identified nineteen significantly upregulated proteins and
no downregulated proteins (Fig. 2A
and Table SXI). The adherens
junction protein, nectin-4, was observed to be the most
significantly overexpressed (P=4.85×10−6), followed by
p27Kip1 (P=5.966×10−3), mucin-1 (MUC-1;
P=8.420×10−3) and gelsolin-like actin-capping protein
(CapG; P=1.021×10−2; Fig.
2B). Further analysis of these proteins demonstrated the
enrichment of several pathways (Fig.
S1 and Table IV).
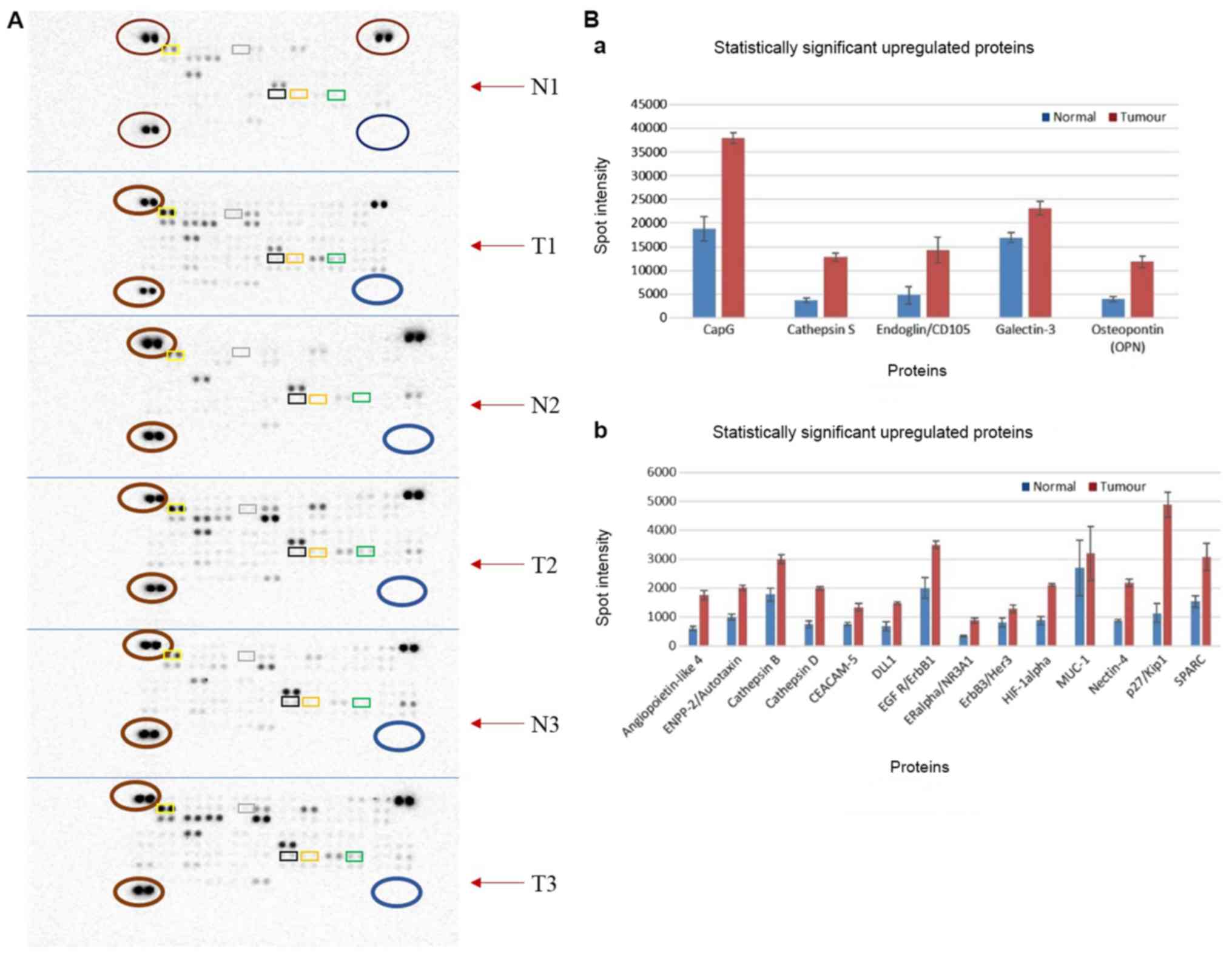 | Figure 2.Differential protein expression
levels using Human Oncology Proteome Profiler arrays. (A) Arrays
demonstrating the protein expression profile for PDAC tumour tissue
and normal tissue from 3 patients (patients 7, 8 and 9). Each
paired spot represents a protein and protein expression increases
as spot intensity increases. Paired spots at the top left, bottom
left and top right (all presented as red circles across the arrays)
served as positive controls, whilst paired spots at the bottom
right (shown in blue circles across all arrays) served as negative
controls. Top 5 significantly overexpressed proteins are indicated
in rectangles: Nectin (orange), p27 (green), MUC-1 (black), CapG
(yellow) and Cathepsin D (grey). (B) Bar charts of upregulated
proteins. Proteins in the top chart have a maximum spot intensity
of 40,000, while those in the bottom chart have a maximum spot
intensity of ~6,000. The blue bar indicates normal expression and
red indicates tumour tissue expression. N1, normal tissue for
patient 7; T1, tumour for patient 7; N2, normal tissue for patient
8; T1, tumour tissue for patient 8; N3, normal tissue for patient
9; T3, tumour for patient 9; MUC-1, mucin 1; CapG, gelsolin-like
actin-capping protein. |
 | Table IV.Top upregulated pathways identified
in tumours from proteome profiler arrays. Generated using
Reactome. |
Table IV.
Top upregulated pathways identified
in tumours from proteome profiler arrays. Generated using
Reactome.
| Upregulated
pathway | P-value | Candidate
proteins |
|---|
| ErbB signaling
pathway |
4.9×10−5 | EGFR, ERBB3,
CDKN1B |
| MHC class II
antigen presentation |
5.74×10−5 | CTSS, CTSB,
CTSD |
| Immune system |
8.14×10−5 | CTSD, CDKN1B,
ERBB3, CTSB, CTSS, EGFR |
| Signalling pathway
in endosomal TLR |
1.68×10−4 | EGFR, CDKN1B,
ERBB3 |
| P13/AKT signalling
in cancer |
1.8×10−4 | ERBB3, EGFR,
CDKN1B |
Discussion
In the present study, using data produced by
transcriptome sequencing and a focused proteome microarray,
plakoglobin was found to be significantly upregulated in early
stage PDAC tumours compared with normal pancreatic tissue. In
addition, analysis of dysregulated pathways within the PDAC tissue
revealed that the IGF-1 and the EGF receptors (IGF1R and EGFR,
respectively), as well as components of the K-RAS signalling
pathway were also upregulated. Plakoglobin, also known as
γ-catenin, is encoded by the junction plakoglobin (JUP) gene
and belongs to the catenin protein family (8). It serves a critical role in cell
adhesion as an important component of desmosomes and adherens
junctions; through interacting as a complex with α- or β-catenin,
plakoglobin associates with the cytoplasmic domains of E-cadherin
or N-cadherin to facilitate the linkage between adherens junctions
and the cytoskeleton (30,31). Recently, plakoglobin was identified
as a transcriptional factor involved in the modulation of WNT
signalling (8). It was also
demonstrated to act as a global regulator of gene expression; it
was found to interact with p53 transcription regulatory elements to
transcriptionally modulate the TCF/LEF family in an APC-dependent
manner (11,32), and upon addition of exogenous
plakoglobin to a mesothelioma cell line, plakoglobin accumulated in
the nucleus and exhibited transcriptional activity (11). Overall, this suggests that
plakoglobin may serve as a transcription factor.
The insulin/IGF-1R, which may signal through the rat
sarcoma (RAS) signalling pathway, is important for transducing
signals that promote the development of pancreatic cancer; the
crosstalk between insulin/IGF and G protein-coupled receptor
signaling was observed to depend on mTOR complex 1 in pancreatic
cell lines (33). The findings from
the present study suggested that the MAPK and PI3K/AKT signalling
pathways may be upregulated in the early PDAC tumour
microenvironment; however, the mechanism behind this dual signaling
remains largely unknown. Alterations in single or groups of genes
involved in one pathway often trigger tumorigenic cellular
behaviour. It has been reported that ~95% of PDAC tumours harbour
activating K-RAS mutations (34) and RAS is a common signaling component
of both the MAPK and PI3K/AKT signaling pathways; however, although
RAS isoforms are structurally similar, they demonstrate distinct
roles in different subcellular environments to mediate different
signalling pathways (35). A
comprehensive analysis of active signaling pathways in pancreatic
cancer demonstrated that 12 of these pathways were often associated
with single or multiple gene alterations that arose from different
signaling pathways (20). Notably,
in the present study, N-RAS was the only isoform identified
as being upregulated in PDAC through transcriptome sequencing
(Table SIII); N-RAS
mutations are frequently observed in melanoma (35).
Receptor tyrosine kinases such as the IGF-IR and
EGFR, which were both found to be upregulated in PDAC in the
present study, transduce signals via the insulin receptor
substrate-1 (IRS) to either the MAPK or PI3K/AKT signaling pathway
to control cell proliferation and survival (33). EGFR expression levels are upregulated
in ~90% of pancreatic tumours, which contributes to chemoresistance
(36–38). Thus, the combination of molecules
found upregulated in the PDAC tumour microenvironment in the
present study further suggested that the PI3K/AKT and MAPK
signaling pathways may be upregulated in PDAC compared to within
normal pancreatic tissue.
The precise signalling role plakoglobin serves
during pancreatic carcinogenesis is largely unclear; however, one
study reported that plakoglobin promoted the phosphorylation of AKT
and ERK in an EGFR-dependent manner, and that its overexpression
promoted cell mobility and migration (39). The data from the present study
demonstrated that both EGFR and insulin/IGF-1R were upregulated in
the PDAC tumour microenvironment. In normal muscle tissue,
plakoglobin bound to the insulin receptor and the p85 regulatory
subunit of PI3K to promote the activation of the PI3K-AKT-FoxO
pathway and muscle growth; in this context, plakoglobin was tightly
regulated by the E3 ligase, Trim-32; however, it is also important
to note that changes in plakoglobin expression alone were
insufficient to influence the signal transduction and muscle growth
(40). In invasive micropapillary
carcinoma of the breast, plakoglobin was observed to be
overexpressed in metastatic tissue, and it contributed to
metastasis through activating the PI3K/AKT/Bcl-2 signalling pathway
(9). This suggested a critical role
for plakoglobin in PI3K/AKT signalling. It has also been reported
that both the MAPK and PI3K/AKT signalling pathways exist in
pancreatic cancer cell lines and that the inhibition of both
pathways leads to cell cycle arrest and apoptosis (41). In addition, plakoglobin was
demonstrated to have an important role in maintaining cell adhesion
in keratinocytes through inhibiting p38 MAPK, hence, the loss of
function of plakoglobin resulted in the activation of p38 MAPK in
these cells (42). However, the
activation of p38 is context-dependent; for example, insulin
stimulated p38 in cultured mouse adipocyte cells but downregulated
it in chick neuronal cells (43,44).
Overall, the findings from the present study, considered alongside
the existing research, suggest that plakoglobin may be an important
context-dependent factor in carcinogenesis. A conceptual schematic
diagram to illustrate the signalling networks upregulated in early
PDAC as well as the hypothesised mechanism of action exerted by
plakoglobin is presented in Fig.
3.
Plakoglobin may also influence cellular outcomes
through its nuclear functions. A candidate nuclear molecule
identified in the present study was MUC1, whose
overexpression was reported in the proteomic study. A previous
study observed that MUC1 was transported in complex with
plakoglobin to the nucleolus in HER2-positive primary invasive
ductal breast carcinomas following treatment with
heregulin/neuregulin-1 (HRG) (39).
This HRG-induced nucleolar translocation of the plakoglobin-MUC1
complex was dependent on MUC1, because a mutation in its
cytoplasmic RRK motif prevented its nuclear transfer (45). Notably, both MUC1 and
HER2 were upregulated in the proteomic study and the
transcriptome sequence data demonstrated that HER1 was also
one of the genes representative of EGFR signalling (Table SV). In addition, both the
transcriptomic and proteomic data analyses revealed further
proteins directly or indirectly associated with the insulin/IGF-1
and EGFR signalling pathways (Fig.
2); this included the adherens junction protein nectin-4, which
has previously been identified as a tumour-associated marker for
breast cancer and as a potential serum biomarker for ovarian cancer
(46,47). Other important proteins included
CapG, a potential oncogene associated with tumour invasion and
metastasis (48) and the cathepsins,
which are proteinases involved in tumour invasion (49). Inflammatory stimuli transcriptionally
driven by NF-κB, which is most likely derived from stromal cells,
reportedly served an important role in the early development of
cancer (21). In conclusion, the
present study suggested that components of both the MAPK and
PI3K/AKT signalling pathways may be dysregulated in the PDAC tumour
microenvironment. These findings proposed an association between
the overexpression of plakoglobin and the upregulation of the
insulin/IGF and MAPK pathways in early resectable PDAC. The
identification of plakoglobin as an important protein in PDAC may
help to understand the mechanisms that mediate the initiation of
tumorigenesis, with the prospect of modulating plakoglobin to
prevent carcinogenesis providing a possible target for future
therapeutic intervention; for example, targeting the negative
regulator of plakoglobin, Trim-32, may be one approach. The
potential involvement of N-RAS in early PDAC has not been
reported before and requires further investigation.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Z Khan for
assistance with some of the sample collections.
Funding
The present study was funded by The South African
National Research Foundation (grant no. 91508; reference no.
CSUR13091741850) and The South African Medical Research Council
through a grant awarded to the Wits Common Epithelial Cancer
Research Centre. MN was funded by The National Research Foundation
(grant no. GUN: 105567).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
EN contributed to project concept and design, all
experimental work, and data acquisition, analysis and
interpretation. EN drafted the article. MN contributed to the
design of the project, data analysis and interpretation, writing of
the manuscript and obtained funding. MB and JD contributed in data
acquisition. MS contributed to the project concept. GC contributed
to the project concept and design, and data interpretation. All
authors revised the work for intellectual content. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The Human Research Ethics Committee of The
University of Witwatersrand approved the collection of biological
samples and data from patients with pancreatic adenocarcinoma
(approval no. M150778) at Chris Hani Baragwanath Academic Hospital
in Johannesburg. All patients gave informed voluntary consent.
Patient consent for publication
All patients provided written consent for sample and
data to be used for the current study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McCrea PD, Maher MT and Gottardi CJ:
Nuclear signaling from cadherin adhesion complexes. Curr Top Dev
Biol. 112:129–196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Broussard JA, Getsios S and Green KJ:
Desmosome regulation and signaling in disease. Cell Tissue Res.
360:501–512. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aktary Z, Kulak S, Mackey J, Jahroudi N
and Pasdar M: Plakoglobin interacts with the transcription factor
p53 and regulates the expression of 14-3-3σ. J Cell Sci.
126:3031–3042. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chidgey M and Dawson C: Desmosomes: A role
in cancer? Br J Cancer. 96:1783–1787. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Simcha I, Geiger B, Yehuda-Levenberg S,
Salomon D and Ben-Ze'ev A: Suppression of tumorigenicity by
plakoglobin: An augmenting effect of N-cadherin. J Cell Biol.
133:199–209. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aktary Z, Alaee M and Pasdar M: Beyond
cell-cell adhesion: Plakoglobin and the regulation of tumorigenesis
and metastasis. Oncotarget. 8:32270–32291. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alaee M, Nool K and Pasdar M: Plakoglobin
restores tumor suppressor activity of p53R175H mutant by
sequestering the oncogenic potential of β-catenin. Cancer Sci.
109:1876–1888. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang L, Ji H, Yin L, Niu X, Wang Y, Liu
Y, Xuan Q, Li L, Zhang H, Zhou X, et al: High expression of
plakoglobin promotes metastasis in invasive micropapillary
carcinoma of the breast via tumor cluster formation. J Cancer.
10:2800–2810. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aktary Z, Chapman K, Lam L, Lo A, Ji C,
Graham K, Cook L, Li L, Mackey JR and Pasdar M: Plakoglobin
interacts with and increases the protein levels of metastasis
suppressor Nm23-H2 and regulates the expression of Nm23-H1.
Oncogene. 29:2118–2129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barker N and Clevers H: Catenins, Wnt
signaling and cancer. Bioessays. 22:961–965. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Besnier LS, Cardot P, Da Rocha B, Simon A,
Loew D, Klein C, Riveau B, Lacasa M, Clair C, Rousset M and Thenet
S: The cellular prion protein PrPc is a partner of the Wnt pathway
in intestinal epithelial cells. Mol Biol Cell. 26:3313–3328. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miravet S, Piedra J, Miró F, Itarte E,
García de Herreros A and Duñach M: The transcriptional factor Tcf-4
contains different binding sites for β-catenin and plakoglobin. J
Biol Chem. 277:1884–1891. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Winn RA, Bremnes RM, Bemis L, Franklin WA,
Miller YE, Cool C and Heasley LE: Gamma-Catenin expression is
reduced or absent in a subset of human lung cancers and
re-expression inhibits transformed cell growth. Oncogene.
21:7497–7506. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Korc M: Role of growth factors in
pancreatic cancer. Surg Oncol Clin N Am. 7:25–41. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Polireddy K and Chen Q: Cancer of the
pancreas: Molecular pathways and current advancement in treatment.
J Cancer. 7:1497–1514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ji B, Tsou L, Wang H, Gaiser S, Chang DZ,
Daniluk J, Bi Y, Grote T, Longnecker DS and Logsdon CD: Ras
activity levels control the development of pancreatic diseases.
Gastroenterology. 137:1072–1082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krah NM, De La O JP, Swift GH, Hoang CQ,
Willet SG, Chen Pan F, Cash GM, Bronner MP, Wright CV, MacDonald RJ
and Murtaugh LC: The acinar differentiation determinant PTF1A
inhibits initiation of pancreatic ductal adenocarcinoma. Elife. Jul
7–2015.(Epub ahead of print). doi: 10.7554/eLife.07125. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee AYL, Dubois CL, Sarai K, Zarei S,
Schaeffer DF, Sander M and Kopp JL: Cell of origin affects tumour
development and phenotype in pancreatic ductal adenocarcinoma. Gut.
Jan 23–2018.(Epub ahead of print). doi:
10.1136/gutjnl-2017-314426.
|
|
20
|
Jones S, Zhang X, Parsons DW, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et
al: Core signaling pathways in human pancreatic cancers revealed by
global genomic analyses. Science. 321:1801–1806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karanikas M, Esempidis A, Chasan ZT,
Deftereou T, Antonopoulou M, Bozali F, Amarantidis K and Man YG:
Pancreatic cancer from molecular pathways to treatment opinion. J
Cancer. 7:1328–1339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Long J, Liu Z, Wu X, Xu Y and Ge C: Gene
expression profile analysis of pancreatic cancer based on
microarray data. Mol Med Rep. 13:3913–3919. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McCleary-Wheeler AL, McWilliams R and
Fernandez-Zapico ME: Aberrant signaling pathways in pancreatic
cancer: A two compartment view. Mol Carcinog. 51:25–39. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Preis M and Korc M: Signaling pathways in
pancreatic cancer. Crit Rev Eukaryot Gene Expr. 21:115–129. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mohelnikova-Duchonova B, Oliverius M,
Honsova E and Soucek P: Evaluation of reference genes and
normalization strategy for quantitative real-time PCR in human
pancreatic carcinoma. Dis Markers. 32:203–210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative Real-Time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pfaffl MW, Horgan GW and Dempfle L:
Relative expression software tool (REST) for group-wise comparison
and statistical analysis of relative expression results in
real-time PCR. Nucleic Acids Res. 30:e362002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Croft D, O'Kelly G, Wu G, Haw R, Gillespie
M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al:
Reactome: A database of reactions, pathways and biological
processes. Nucleic Acids Res. 39:D691–D697. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mi H, Guo N, Kejariwal A and Thomas PD:
PANTHER version 6: Protein sequence and function evolution data
with expanded representation of biological pathways. Nucleic Acids
Res. 35:D247–D252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Knudsen KA and Wheelock MJ: Plakoglobin,
or an 83-kD homologue distinct from beta-catenin, interacts with
E-cadherin and N-cadherin. J Cell Biol. 118:671–679. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rubinfeld B, Souza B, Albert I, Munemitsu
S and Polakis P: The APC protein and E-cadherin form similar but
independent complexes with alpha-catenin, beta-catenin, and
plakoglobin. J Biol Chem. 270:5549–5555. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aktary Z and Pasdar M: Plakoglobin
represses SATB1 expression and decreases in vitro proliferation,
migration and invasion. PLoS One. 8:e783882013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rozengurt E, Sinnett-Smith J and Kisfalvi
K: Crosstalk between insulin/insulin-like growth factor-1 receptors
and G protein-coupled receptor signaling systems: A novel target
for the antidiabetic drug metformin in pancreatic cancer.
ClinCancer Res. 16:2505–2511. 2010.
|
|
34
|
Bryant KL, Mancias JD, Kimmelman AC and
Der CJ: KRAS: Feeding pancreatic cancer proliferation. Trends
Biochem Sci. 39:91–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Posch C and Ortiz-Urda S: NRAS mutant
melanoma-undrugable? Oncotarget. 4:494–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Troiani T, Martinelli E, Capasso A,
Morgillo F, Orditura M, De Vita F and Ciardiello F: Targeting EGFR
in pancreatic cancer treatment. Curr Drug Targets. 13:802–810.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bruns CJ, Solorzano CC, Harbison MT, Ozawa
S, Tsan R, Fan D, Abbruzzese J, Traxler P, Buchdunger E, Radinsky R
and Fidler IJ: Blockade of the epidermal growth factor receptor
signaling by a novel tyrosine kinase inhibitor leads to apoptosis
of endothelial cells and therapy of human pancreatic carcinoma.
Cancer Res. 60:2926–2935. 2000.PubMed/NCBI
|
|
38
|
Morgan MA, Parsels LA, Kollar LE, Normolle
DP, Maybaum J and Lawrence TS: The combination of epidermal growth
factor receptor inhibitors with gemcitabine and radiation in
pancreatic cancer. Clin Cancer Res. 14:5142–5149. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pan H, Gao F, Papageorgis P, Abdolmaleky
HM, Faller DV and Thiagalingam S: Aberrant activation of
gamma-catenin promotes genomic instability and oncogenic effects
during tumor progression. Cancer Biol Ther. 6:1638–1643. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cohen S, Lee D, Zhai B, Gygi SP and
Goldberg AL: Trim32 reduces PI3K-Akt-FoxO signaling in muscle
atrophy by promoting plakoglobin-PI3K dissociation. J Cell Biol.
204:747–758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Roy SK, Srivastava RK and Shankar S:
Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of
FOXO transcription factor, leading to cell cycle arrest and
apoptosis in pancreatic cancer. J Mol Signal. 5:102010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Spindler V, Dehner C, Hübner S and Waschke
J: Plakoglobin but not desmoplakin regulates keratinocyte cohesion
via modulation of p38MAPK signaling. J Invest Dermatol.
134:1655–1664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heidenreich KA and Kummer JL: Inhibition
of p38 mitogen-activated protein kinase by insulin in cultured
fetal neurons. J Biol Chem. 271:9891–9894. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sweeney G, Somwar R, Ramlal T, Volchuk A,
Ueyama A and Klip A: An inhibitor of p38 mitogen-activated protein
kinase prevents insulin-stimulated glucose transport but not
glucose transporter translocation in 3T3-L1 adipocytes and L6
myotubes. J Biol Chem. 274:10071–10078. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, Yu WH, Ren J, Chen W, Huang L,
Kharbanda S, Loda M and Kufe D: Heregulin targets gamma-catenin to
the nucleolus by a mechanism dependent on the DF3/MUC1 oncoprotein.
Mol Cancer Res. 1:765–775. 2003.PubMed/NCBI
|
|
46
|
Fabre-Lafay S, Monville F, Garrido-Urbani
S, Berruyer-Pouyet C, Ginestier C, Reymond N, Finetti P, Sauvan R,
Adélaïde J, Geneix J, et al: Nectin-4 is a new histological and
serological tumor associated marker for breast cancer. BMC Cancer.
7:732007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Derycke MS, Pambuccian SE, Gilks CB,
Kalloger SE, Ghidouche A, Lopez M, Bliss RL, Geller MA, Argenta PA,
Harrington KM and Skubitz AP: Nectin 4 overexpression in ovarian
cancer tissues and serum: Potential role as a serum biomarker. Am J
Clin Pathol. 134:835–845. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu WY, Hunag YY, Liu XG, He JY, Chen DD,
Zeng F, Zhou JH and Zhang YK: Prognostic evaluation of CapG,
Gelsolin, P-gp, GSTP1, and Topo-II Proteins in non-small cell lung
cancer. Anat Rec (Hoboken). 295:208–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gocheva V and Joyce JA: Cysteine
cathepsins and the cutting edge of cancer invasion. Cell Cycle.
6:60–64. 2007. View Article : Google Scholar : PubMed/NCBI
|