Introduction
Colorectal cancer (CRC) was the third most commonly
diagnosed malignancy and the second leading cause of
cancer-associated mortality worldwide in 2012 (1). This could be effectively reduced by
early screening. It is estimated that there were 3,763,000 newly
diagnosed cases of CRC and 191,000 deaths attributable to CRC in
2015 (2). There has recently been a
growing interest in identifying the causes of CRC development and
novel approaches to prevent CRC and improve early diagnosis and
treatments. Although researchers have made great progress in
revealing the underlying mechanisms of CRC development and
metastasis, certain aspects remain unclear. Additionally, early
diagnosis of CRC remains unsatisfactory due to metastasis, genetic
heterogeneity and living habits (3).
Further investigations are required to identify novel and useful
biomarkers for the early diagnosis and treatment of CRC.
Long non-coding RNAs (lncRNAs) are transcripts of
>200 nucleotides in length with limited or no coding potential
(4). LncRNAs exhibit an mRNA-like
structure with a poly-A tail and a promoter region. They not only
assist in the intermediary delivery of genetic information, but
they also serve a number of regulatory functions, including dynamic
expression and differential splicing (5). They have attracted a lot of attention
due to their involvement in a number of physiological processes
(6). LncRNAs are involved in
multiple regulatory processes, including X-chromosome silencing,
genomic imprinting, chromatin modification, transcriptional
activation, transcriptional interference and intranuclear transport
(7).
Several studies have revealed an association between
aberrant lncRNA expression and human diseases such as cancer,
neurodegenerative and cardiovascular diseases (8,9). LncRNAs
influence almost all of the hallmarks of cancer, including
sustaining proliferative signaling, evading growth suppressors,
resisting cell death, enabling replicative immortality, inducing
angiogenesis, promoting invasion/metastasis, genome instability,
inflammation, reprogramming of energy metabolism and evading immune
destruction (10). Aberrant lncRNA
expression is associated with the progression of various types of
cancer, such as breast, liver, bladder, prostate, lung, gastric and
CRC (11,12).
Increasing evidence has demonstrated that specific
lncRNAs are abnormally expressed in numerous types of cancer,
including CRC, and function as tumor suppressor genes, oncogenes or
both (13). Increasing studies have
highlighted that lncRNA dysregulation serves a pivotal role in
proliferation, angiogenesis, metastasis, invasion, apoptosis and
genome instability in CRC, which are consequently associated with
clinical implications (14–17). Colon cancer-associated transcript
(CCAT) 2 was identified as a novel lncRNA that is upregulated in
microsatellite-stable CRC, contributing to CRC pathogenesis such as
tumor growth, metastasis and chromosomal instability (18). The long intergenic non-protein coding
RNA 2598 is upregulated in CRC in a stage-dependent manner and is
able to promote the migration, invasion and
epithelial-to-mesenchymal transition (EMT) of CRC cells (19). Master factors of EMT, such as Snail,
play important role in CRC (20,21).
Snail is a conserved zinc-finger transcription factor, which can
induce EMT in a variety of tissues, and different EMT programs
might exist (22,23). Lnc-GNAT1-1 is expressed at low levels
in CRC and functions as a tumor suppressor by regulating RKIP (Raf
kinase inhibitor protein)-NF-κB-Snail circuit (24). The expression of
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is
associated with CRC metastasis, while decreased expression of
MALAT1 inhibits nuclear translocation of β-catenin and attenuates
Wnt/β-catenin signaling, resulting in decreased CRC invasion and
metastasis (25–27). Furthermore, lncRNAs with altered
expression levels in CRC tissues may serve as potential prognostic
biomarkers and therapeutic targets (28,29).
Although a total of 556 upregulated and 1,040 downregulated lncRNAs
have been identified in CRC tissues, the underlying mechanisms of
each lncRNA in CRC remain unclear (30).
Therefore, the present study investigated the
expression profile of lncRNAs in CRC and paired normal tissues
using a PCR array to assess the association between abnormally
expressed lncRNAs and CRC, and to identify potential prognostic
biomarkers for CRC.
Materials and methods
Clinical samples
Three patients (two females aged 50 and 47 years and
one male aged 48 years) with CRC were randomly recruited from the
Qilu Hospital of Shandong University (Jinan, China) for
RT2 lncRNA PCR array Human Cancer PathwayFinder (Qiagen,
GmbH). A total of 130 CRC and paired adjacent normal tissues were
collected from patients. The patient cohort consisted of 96 males
and 34 females; 72 people were >60 years old and 58 people were
≤60 years old who underwent surgical resection at the Department of
General Surgery at Qilu Hospital of Shandong University between
April 2013 and December 2014 in order to validate the results of
the PCR array. All patients had pathologically confirmed colorectal
adenocarcinoma. Tumors were staged according to the TNM staging
system of the American Joint Committee on Cancer (7th edition)
(31). None of the patients had
received radiotherapy or chemotherapy prior to surgical resection.
All isolated samples were frozen in liquid nitrogen and then stored
at −80°C prior to RNA extraction. The experimental protocol of the
present study was approved by the Ethics Committee of Qilu Hospital
of Shandong University. All tissues were removed by the same method
and the same inclusion criteria were used as in the bigger patient
cohort. The distance between normal tissue and cancerous tissue
>5 cm, and the normal mucosa had no visible deformation. Written
informed consent was provided by all participants.
PCR array
Gene expression was measured using the
RT2 lncRNA PCR array Human Cancer PathwayFinder (Qiagen,
GmbH) (32). Mature RNA was isolated
from tissues using an RNA extraction kit (cat. no. 74106; Qiagen
GmbH) according to the manufacturer's protocol. The RNA quality was
determined using a spectrophotometer and the RNA was subsequently
reverse transcribed using RT2 First Stand kit (cat no.
330401; Qiagen GmbH) according to the manufacturer's protocol with
steps at 42°C for 5 min to remove genomic DNA, ice bath for at
least 1 min, 42°C for 15 min to perform reverse transcription and
95°C for 5 min to interrupt the reaction. The cDNA was used for the
RT2 lncRNA PCR array (Qiagen, GmbH) in combination with
RT2 SYBR® Green qPCR Mastermix (Qiagen,
GmbH). A total of 89 lncRNA genes associated with CRC were assessed
on three paired samples using the RT2 lncRNA PCR array.
After qPCR, CT values were exported to an Excel file to create a
table of CT values. This table was then uploaded on to
the data analysis web portal at http://www.qiagen.com/geneglobe. Samples were assigned
to control and test groups. CT values were normalized
based on a/an Automatic selection from full panel of reference
genes. The data analysis web portal calculates fold
change/regulation using ∆∆Ct method, in which ∆Ct is
calculated between gene of interest (GOI) and an average of
reference genes (HKG), followed by ∆∆ Ct calculations
[∆Ct (Test Group)-∆Ct (Control Group)]. Fold
Change is then calculated using 2−∆∆Ct formula. The data
analysis web portal also plots scatter plot, volcano plot,
clustergram, and heat map. This data analysis report was exported
from the QIAGEN web portal at GeneGlobe. Fold change was calculated
as the normalized lncRNA expression in the cancer tissue samples
divided by the normalized expression in the paired normal tissue
samples. P<0.05 (two-tailed) was considered to indicate a
statistically significant difference.
Reverse transcription-quantitative
(RT-q)PCR analysis
Total RNA was extracted from 130 patient tissues
using an RNeasy Mini kit (Qiagen, GmbH). Reverse transcription was
performed using the First Strand kit (Qiagen, GmbH) with 1 µg of
RNA per sample according to the manufacturer's protocol. qPCR
reactions were run on a LC96 thermal cycler (Roche Diagnostics)
with an initial activation step at 95°C for 15 sec, followed by 40
cycles at 95°C for 15 sec and 60°C for 1 min. The primers for
TRERNA1 PCR were purchased from Qiagen, GmbH. Detection was
repeated in triplicates. Primer sequences were as follows: Snail
forward, 5′-CCTCGCTGCCAATGCTCATCTG-3′ and reverse,
5′-GCTCTGCCACCCTGGGACTC-3′; and GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. The fluorophore was RT2
SYBR Green qPCR Mastermix (Qiagen, GmbH). Comparative
quantification was assessed using the 2−ΔΔCT method
using GAPDH as the endogenous control (33).
Cell culture
The normal human colorectal FHC cell line and the
human CRC HCT-8, DLD-1, SW480, HCT-116 and SW620 cell lines were
purchased from the Cell Resource Center at the Shanghai Institute
of Biochemistry and Cell Biology at the Chinese Academy of
Sciences. HCT-8, DLD-1 and FHC cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% (v/v) FBS (Gibco; Thermo Fisher Scientific, Inc.). HCT-116 and
SW480 were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS. All cell lines were incubated in a
humidified atmosphere at 37°C with 5% CO2.
Small interfering (si)RNA-mediated
knockdown of TRERNA1
SiRNAs were purchased from Suzhou Ribo Life Science
Co., Ltd. HCT-8 and HCT-116 cells were transfected with 5 nmol
siRNAs with jetPRIME® transfection reagent
(Polyplus-transfection SA) when the cell density was
1×106 cells in a 6-well plate according to the
manufacturer's protocol. Subsequent experiments were performed 48h
after transfection. The siRNAs sequences were as follows:
Si-negative control (Si-NC),
5′-UUCUUCGAAACGUGUCACGUT-3′; and Si-TRERNA1,
5′-GAAGGGAACCAGUGCUAAAUU-3′.
Transwell assay
Matrigel (cat. no. 356234; Corning Life Sciences)
and serum-free medium were mixed in a 1:4 ratio (v/v) to create a
mixed liquid. A total of 20 µl mixed liquid was added to Transwell
chambers (cat. no. 3422; Corning Life Sciences) inserted in 24-well
plates, which were incubated for 3 h at 37°C to speed up the
solidification of Matrigel. SiRNA-transfected HCT-8 or HCT-116
cells were counted, and 5×104 cells/well were plated
into the upper chambers in serum-free medium (HCT-8 cells were
incubated in RPMI-1640 medium; HCT-116 cells were incubated in DMEM
medium). A total of 600 µl medium (HCT-8 cells were incubated in
RPMI-1640 medium; HCT-116 were incubated in DMEM medium)
supplemented with 20% (v/v) FBS was added in the lower chambers.
After 18 h of incubation at 37°C, the cells and Matrigel in the
upper chambers were removed using cotton swabs, and the cells in
the lower chambers were fixed with anhydrous methanol at room
temperature for 15 min and stained with 0.05% crystal violet
staining solution (Beijing Solarbio Science & Technology Co.,
Ltd.) at room temperature for 15 min. Finally, the cells were
imaged using a light microscope (magnification, ×100) and counted
to compare cell invasion between the control and transfected
groups. The same steps without the addition of Matrigel were
followed to analyze cell migration.
Western blot analysis
HCT-8 and HCT-116 cells were lysed using the high
RIPA lysis buffer with proteinase inhibitor PMSF (Beijing Solarbio
Science & Technology Co., Ltd.) at a ratio of 100:1 (v/v),
after transfection for 48 h. The protein concentration was
determined using the bicinchoninic acid reagent kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. A total
of 30 µg of protein was loaded per lane and separated using a 10%
SDS-PAGE gel and subsequently transferred onto PVDF membranes (EMD
Millipore). The membranes were blocked using 5% skimmed milk for 1
h at room temperature, followed by overnight incubation with
primary antibodies against Snail (1:1,000; cat. no. 3879; Cell
Signaling Technology, Inc.), E-cadherin (1:1,000; cat no. 3195;
Cell Signaling Technology, Inc.) and β-actin (1:1,000; cat no.
3195; Cell Signaling Technology, Inc. x) at 4°C. The membranes were
subsequently incubated with anti-rabbit horseradish
peroxidase-labeled secondary antibodies (1:5,000; cat. no.
SA00001-2; Wuhan Sanying Biotechnology) for 1 h at room
temperature. Finally, the PVDF membranes were developed using the
enhanced chemiluminescence method using Luminata™ Western HRP
Substrates (EMD Millipore) and detected using a chemiluminometer
(Tanon Science and Technology Co., Ltd.).
Prediction of lncRNA/mRNA and miRNA
interactions
MiRanda (v1.0b) was used to predicted the miRNAs
binding both TRERNA1 and Snail according to Anton's study (34). A hit between any predicted miRNA and
a target lncRNA/mRNA was considered with a score of 100 or higher,
corresponding to at least a perfect seed match.
Statistical analysis
Statistical analysis was conducted using SPSS v25.0
(IBM Corp.). All data was presented as mean ± SD. Comparisons
between two groups were performed by Student's unpaired t-test, and
categorical data were analyzed using Fisher's exact test. A paired
t-test was used to analyze the differential expression of TRERNA1
in cancer tissues compared with adjacent normal tissues. If
variance tested by one-way ANOVA was statistically significant,
Tukey's multiple comparisons test was used to make comparisons
among multiple groups. Overall survival between experimental groups
were compared by a log-rank (Mantel-Cox) test in the Kaplan-Meier
analysis. Spearman's correlation coefficient analysis was used to
assess the correlation between TRERNA1 and Snail levels in CRC
tissues. P<0.05 (two-tailed) was considered to indicate a
statistically significant difference.
Results
Differentially expressed lncRNAs in
CRC tissues
To test the effect of lncRNAs in CRC tissues, three
pairs of CRC and non-tumorous adjacent tissues were selected.
RT2 lncRNA PCR array Human Cancer PathwayFinder was used
to simultaneously monitor the expression levels of 89 lncRNAs that
resulted associated with tumorigenesis and CRC by relative RT-qPCR.
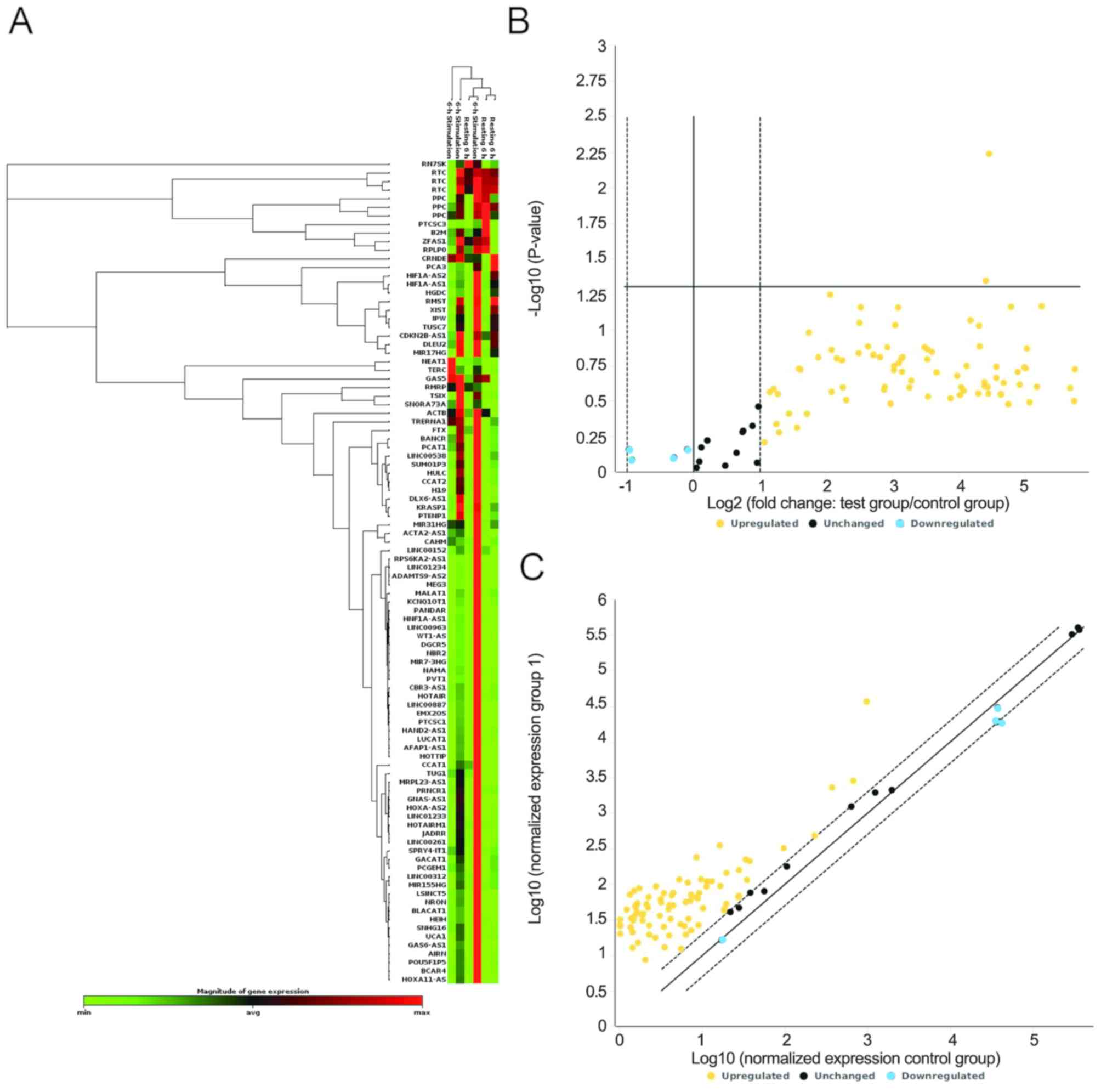
More information regarding lncRNA genes is listed in Table SI. The differential expression of
the 89 lncRNAs in CRC tissues compared with non-tumorous adjacent
tissues is displayed in Fig. 1A. The
volcano plots in Fig. 1B indicate
significant changes in gene expression. The expression levels of 4
lncRNAs were downregulated in CRC tissues (fold change >2),
while 75 lncRNAs were upregulated in CRC tissues compared with
non-tumorous adjacent tissues (fold change >2). A total of 10
lncRNAs exhibited no significant changes (fold change <2). The
scatter plot analysis compared the normalized expression levels of
each gene on the array between the control group and test group by
plotting them against one another, to visualize large gene
expression changes. Changes in lncRNA expression were observed
between tumor and non-tumorous adjacent tissues (Fig. 1C). Additionally, 2/89 lncRNAs were
significantly upregulated (P<0.05), namely TRERNA1 and
BRAF-activated non-protein coding RNA (BANCR), with fold changes of
21.88 and 21.14, respectively (Table
I). It has been demonstrated that TRERNA1 is upregulated in
gastric carcinoma and hepatic carcinoma, and that it promotes cell
migration and invasion in gastric cancer and hepatic carcinoma
(35–37). However, little is known about the
role of TRERNA1 in CRC. Therefore, considering the results of PCR
array TRERNA1 was selected for follow-up experiments.
 | Table I.Fold-change of long non-coding RNAs
detected via PCR array. |
Table I.
Fold-change of long non-coding RNAs
detected via PCR array.
| Position | Gene symbol | Fold change | P-value |
|---|
| A05 | BANCR | 21.14 | 0.045 |
| G05 | TRERNA1 | 21.88 | 0.005 |
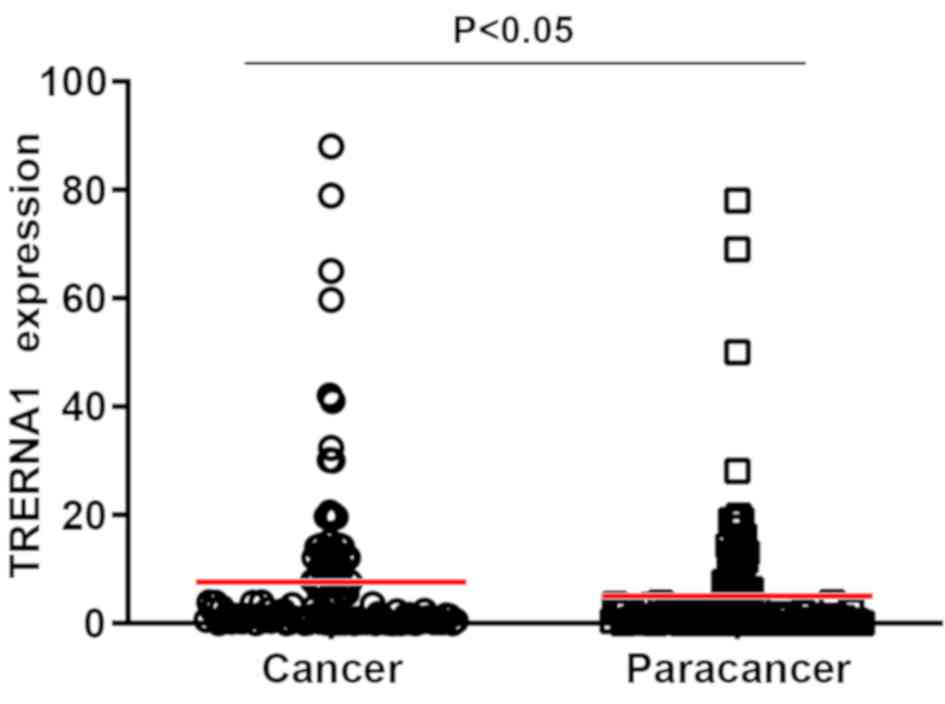
TRERNA1 expression is upregulated in
CRC tissues
To assess the effect of TRERNA1 in CRC, TRERNA1
expression was analyzed in 130 CRC and non-tumorous adjacent
tissues by RT-qPCR. The results revealed that TRERNA1 expression
was upregulated in CRC tissues compared with non-tumorous adjacent
tissues (Fig. 2). In addition, the
association between TRERNA1 expression and the clinicopathological
parameters of patients with CRC was analyzed. The results suggested
that TRERNA1 expression was associated with distant metastasis,
perineural invasion, TNM stage, node metastasis stage and tumor
diameter, but not with age, sex, tumor location and differentiation
(Fig. 3A-D; Table II). For example, the expression
levels of TRERNA1 in patients with M1 stage were significantly
higher than in those with M0 stage (Table II). According to pathological stage
(pTNM), patients with stage III–IV exhibited higher expression
levels of TRERNA1 compared with patients with stage I–II (Fig. 3D). Next, the effect of TRERNA1 on
pathological node metastasis stage was analyzed (pN), which
revealed that patients with stage I–II exhibited higher expression
levels of TRERNA1 compared with patients with stage 0 (Fig. 3E).
 | Table II.Association between TRERNA1
expression and the clinicopathological parameters of patients with
colorectal cancer (n=130). |
Table II.
Association between TRERNA1
expression and the clinicopathological parameters of patients with
colorectal cancer (n=130).
| Variable | Patients, n | TRERNA1 expression
level, median (95% CI) | Z-value | P-value |
|---|
| Sex |
|
| −0.32 | 0.753 |
|
Male | 96 | 1.61
(0.35–9.63) |
|
|
|
Female | 34 | 1.38
(0.49–7.59) |
|
|
| Age, years |
|
| 1.18 | 0.239 |
|
≤60 | 58 | 2.43
(0.99–7.76) |
|
|
|
>60 | 72 | 1.38
(0.31–9.63) |
|
|
| Tumor location |
|
| 1.68 | 0.093 |
|
Colon | 62 | 1.475
(0.25–6.93) |
|
|
|
Rectum | 68 | 2.43
(0.5–12.94) |
|
|
| Tumor diameter,
cm |
|
| 2.20 | 0.028a |
| ≤5 | 84 | 1.02
(0.19–7.76) |
|
|
|
>5 | 46 | 2.89
(0.74–10.42) |
|
|
| Tumor
differentiation |
|
| −0.56 | 0.575 |
|
Poor | 30 | 1.35
(0.21–10.16) |
|
|
|
Well/moderate | 100 | 1.95
(0.39–9.4) |
|
|
| pT stage |
|
| 0.97 | 0.332 |
|
T1-3 | 98 | 1.54
(0.37–7.76) |
|
|
| T4 | 32 | 1.89
(0.44–13.56) |
|
|
| pN stage |
|
| −2.92 | 0.004a |
| N0 | 62 | 1.38
(0.21–3.83) |
|
|
|
N1-2 | 68 | 3.18
(0.86–13.50) |
|
|
| Distant metastasis
(M stage) |
|
| 4.33 |
<0.001a |
| M0 | 114 | 1.36
(0.37–6.27) |
|
|
| M1 | 16 | 19.83
(11.38–30.03) |
|
|
| pTNM stage |
|
| 3.06 | 0.002a |
|
I–II | 66 | 1.38
(0.21–3.81) |
|
|
|
III–IV | 64 | 5.83
(0.63–14.31) |
|
|
| Lymphovascular
invasion |
|
| −0.57 | 0.567 |
|
Yes | 10 | 1.36
(0.39–5.57) |
|
|
| No | 120 | 1.61
(0.38–9.63) |
|
|
| Perineural
invasion |
|
| 2.08 | 0.038a |
|
Yes | 56 | 3.15
(0.57–12.52) |
|
|
| No | 74 | 1.26
(0.33–7.59) |
|
|
Prognostic potential of TRERNA1 in
CRC
All 130 patients included in the present study were
classified into two sub-groups based on the expression levels of
TRERNA1. Patients with TRERNA1 expression levels higher than the
median value 1.55 were assigned to the high expression group, while
patients with TRERNA1 expression levels lower than the median value
were assigned to the low expression group. The term overall
survival (OS) time was used to describe the chance of survival. The
association between TRERNA1 expression and OS time in patients with
CRC was investigated via Kaplan-Meier analysis. The results of the
survival curve revealed that patients with CRC with high TRERNA1
expression had shorter OS times than those with low TRERNA1
expression (Fig. 4). In addition,
the multivariate analysis revealed that high TRERNA1 expression,
distant metastasis and patient node metastasis were associated with
a less favorable prognosis in patients with CRC (Table III). The results of the present
study revealed that TRERNA1 may serve as a prognostic marker for
patients with CRC.
 | Table III.Univariate and multivariate overall
survival analysis of clinicopathological factors. |
Table III.
Univariate and multivariate overall
survival analysis of clinicopathological factors.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex (male vs.
female) | 0.86
(0.37–1.99) | 0.727 |
|
|
| Age, years (≤60 vs.
>60) | 0.77
(0.38–1.55) | 0.461 |
|
|
| Tumor diameter, cm
(≤5 vs. >5) | 1.01
(0.51–2.03) | 0.968 |
|
|
| Tumor location
(colon vs. rectum) | 2.31
(1.12–4.73) | 0.023 |
|
|
| Differentiation
(well/moderate vs. poor) | 0.71
(0.34–1.51) | 0.374 |
|
|
| pT stage (T1-3 vs.
T4) | 2.34
(1.16–4.72) | 0.017 |
|
|
| pN stage (N0 vs.
N1-2) | 3.85
(1.58–9.37) | 0.003 |
|
|
| Distant metastasis
(M0 vs. M1) | 3.97
(1.90–8.28) | <0.001 | 2.51
(1.10–5.71) | 0.029a |
| pTNM stage (I–II
vs. III–IV) | 7.99
(2.80–22.82) | <0.001 | 6.24
(2.10–18.58) | 0.001a |
| Lymphovascular
invasion (no vs. yes) | 1.43
(0.54–3.84) | 0.474 |
|
|
| Perineural invasion
(no vs. yes) | 1.20
(0.59–2.41) | 0.616 |
|
|
| TRERNA1 expression
(low vs. high) | 2.76
(1.27–5.97) | 0.010 | 2.68
(1.25–5.73) | 0.011a |
siRNA-mediated knockdown of TRERNA1
inhibits invasion and migration of CRC cells
Based on results from clinical pathology analyses
and previous studies (35,36), it was hypothesized that TRERNA1 may
affect the invasion and migration abilities of CRC cells.
Therefore, TRERNA1 expression in the normal human colorectal FHC
cell line and five CRC cell lines was detected. The results
revealed that TRERNA1 expression was upregulated in HCT-8, DLD-1,
SW480 and HCT-116 cell lines compared with in FHC cells (Fig. 5A). Subsequently, the human CRC HCT-8
and HCT-116 cell lines were used to detect the effect of TRERNA1 on
the invasion and migration abilities of CRC cells in vitro.
TRERNA1 expression was knocked down with specific siRNA sequences,
and the results are presented in Fig.
5B. Following TRERNA1 knockdown, the number of HCT-8 and
HCT-116 cells that migrated or invaded to the other side of the
membrane was significantly decreased in the experimental groups
compared with the control groups (Fig.
5C and D). This suggests that the downregulation of TRERNA1
expression inhibits the invasion and migration abilities of CRC
cells.
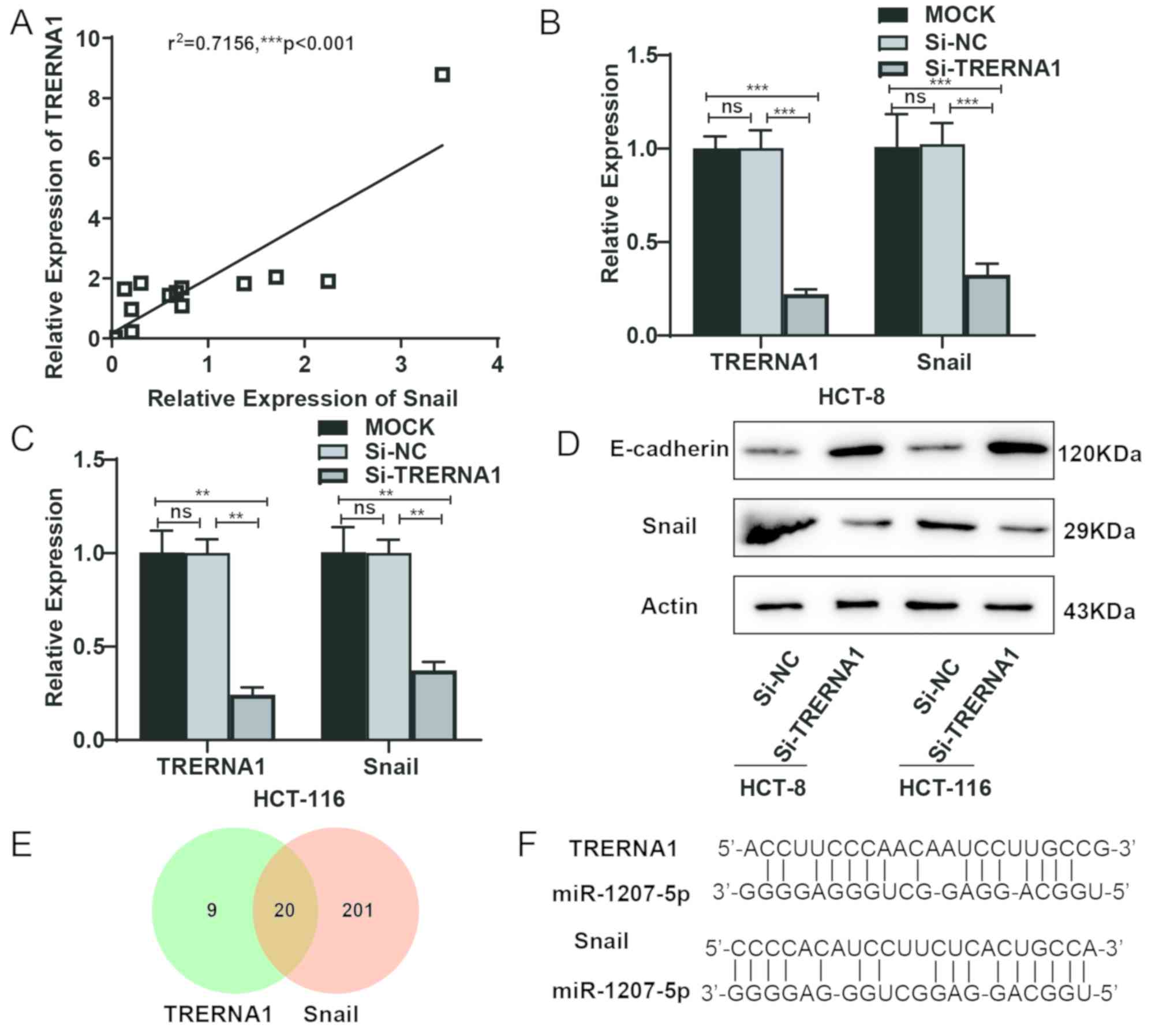
Knockdown of TRERNA1 suppresses Snail
expression
The present study revealed that high TRERNA1
expression was associated with CRC metastasis and that the invasion
and migratory abilities of CRC cells were inhibited following
TRERNA1 knockdown. Additionally, previous studies demonstrated that
TRERNA1 regulates Snail expression as an enhancer of SNAI1 promoter
(35). Results from the present
study revealed that TRERNA1 expression was positively correlated
with Snail expression in CRC tissues (Fig. 6A). TRERNA1 knockdown suppressed the
expression levels of Snail mRNA (Fig. 6B
and C). Western blot results revealed that following TRERNA1
knockdown, Snail expression was downregulated, while E-cadherin
expression was upregulated (Fig.
6D). These results suggested that TRERNA1 may affect EMT in CRC
metastasis via regulation of Snail expression. A total of 20
microRNAs (miRs) were predicted to bind both TRERNA1 and Snail,
using miRanda v1.0b (energy <-100; Fig. 6E; Tables
SII–SIV). MiR-1207-5p was of
particular interest, since studies have demonstrated that
miR-1207-5p is expressed at low levels in CRC (38,39). It
was therefore speculated that TRERNA1 may regulate miR-1207-5p and
Snail expression via endogenous competition (Fig. 6F).
Discussion
Numerous genetic factors influence the pathogenesis,
development, metastasis and prognosis of CRC, including lncRNAs
(29). The present study revealed
two differentially expressed lncRNAs, TRERNA1 and BANCR, in CRC
samples compared with adjacent normal tissues, via PCR array. It
has been previously reported that BANCR regulates the microRNA
(miR)-203/chromosome segregation 1 like axis and increases
chemosensitization of CRC cells to Adriamycin, and BANCR
upregulation is associated with lymph node metastasis and a poor
prognosis in patients with CRC (28,40,41). It
has been previously demonstrated that upregulation of BANCR is
associated with lymph node metastasis and a poor prognosis in
patients with CRC (17). However,
there are a few studies on the role of TRERNA1 in CRC. The findings
from the present study revealed that high TRERNA1 expression in CRC
tissues was negatively associated with survival time and acted as
an independent prognostic factor for patients with CRC.
LncRNAs, which are >200 nucleotides in length,
have different functions in various cell processes and diseases.
Numerous lncRNAs are abnormally expressed in CRC (42). CCAT1 was the first identified lncRNA
with potential as a diagnostic marker in CRC and tumor-associated
tissues (43). A total of >2,300
lncRNAs were dysregulated among 33,045 lncRNAs tested at the
genome-wide level in CRC (44),
among which TRERNA1was not included.
TRERNA1 is an EMT master regulator transcription
factor that has been demonstrated to serve an important role in the
invasion and metastasis of gastric cancer (35,36).
Additionally, TRERNA1 is associated with invasion and a poor
response to chemotherapy in chronic lymphocytic leukemia (45). The RT2 lncRNA PCR array is
a highly sensitive gene expression profiling tool used for
analyzing focused panels of cancer-associated genes (46). From the PCR array results, the top
candidates were selected according to the established criteria
(>15-fold change and P<0.05) and RT-qPCR was performed to
investigate the expression levels of TRERNA1 in CRC tissues. The
results revealed that TRERNA1 expression was significantly
increased in the remaining 130 cancerous tissues compared with
paired adjacent normal tissues, in accordance with the study
conducted by Kim et al (44),
which suggested that elevated TRERNA1 expression may be a common
feature in tumor progression and metastasis.
Snail is a zinc finger transcription factor that
inhibits E-cadherin transcription. E-cadherin downregulation is
associated with EMT, a process that occurs during embryonic
development and in invasive cancer cells (47–49). In
the present study, TRERNA1 regulated Snail expression and further
affected E-cadherin expression. It was speculated that TRENRNA1 may
affect CRC metastasis by regulating Snail, which is consistent with
the in vitro results of functional experiments conducted in
the present study. Bioinformatics analysis revealed that TRERNA1
may regulate Snail expression via endogenous competitive effects in
combination with miR-1207-5p. However, it is necessary to further
investigate how TRERNA1 regulates Snail expression via binding to
miR-1270-5p in subsequent experiments.
At present, only a few studies have evaluated the
prognostic value of TRERNA1 expression and suggested that TRERNA1
may act as a prognostic factor in gastric cancer (35,36).
TRERNA1 has been demonstrated to serve as an onco-lncRNA and to
promote the metastasis and invasion of gastric cancer cells
(35). In the present study, TRERNA1
expression was associated with the invasion and metastasis of CRC
cells. Multivariate Cox regression model analysis was conducted to
further assess the association between TRERNA1 expression and OS
time in patients with CRC. The current results indicate that high
TRERNA1 expression may serve as an independent predictor of poor
prognosis in patients with CRC, and that TRERNA1 may therefore be
used as a prognostic marker in patients with CRC.
In conclusion, the present study was, to the best of
our knowledge, the first to provide evidence that TRERNA1 was
upregulated in CRC tissues compared with paired normal tissues.
High TRERNA1 expression was positively associated with distant
metastasis, perineural invasion, node metastasis stage, tumor
diameter and OS time in patients with CRC. Knockdown of TRERNA1
inhibited the invasion and metastasis of CRC cells. However,
extensive functional studies and additional well-designed studies
with different ethnic groups are required to confirm the role of
TRERNA1 in CRC prognosis.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Key Research and Development Foundation of Shandong Province (grant
nos. 2017GSF218034, 2016GSF201010 and 2019GSF108016), the Science
Foundation of Qilu Hospital of Shandong University (grant no.
2017QLQN16) and the China Postdoctoral Science Foundation (grant
no. 2019M652393).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HQ and QH conceived the study; HQ and WW designed
the experiments; WW performed the experiments; WW, XT and HQ
analyzed the data and wrote the paper. All authors have read and
approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Qilu Hospital of Shandong University (Jinan, China).
Written informed consent was provided by all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jeun M, Lee HJ, Park S, Do EJ, Choi J,
Sung YN, Hong SM, Kim SY, Kim DH, Kang JY, et al: A novel
blood-based colorectal cancer diagnostic technology using
electrical detection of colon cancer secreted protein-2. Adv Sci
(Weinh). 6:18021152019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marisa L, Svrcek M, Collura A, Becht E,
Cervera P, Wanherdrick K, Buhard O, Goloudina A, Jonchère V, Selves
J, et al: The balance between cytotoxic T-cell lymphocytes and
immune checkpoint expression in the prognosis of colon tumors. J
Natl Cancer Inst; 110. 2018, PubMed/NCBI
|
|
4
|
van Bakel H, Nislow C, Blencowe BJ and
Hughes TR: Most ‘dark matter’ transcripts are associated with known
genes. PLoS Biol. 8:e10003712010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perkel JM: Visiting ‘noncodarnia’.
Biotechniques. 54:301, 303–304. 2013. View Article : Google Scholar
|
|
6
|
Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A,
Egranov SD, Zhang Y, Xia W, Gong J, et al: Oncogenic lncRNA
downregulates cancer cell antigen presentation and intrinsic tumor
suppression. Nat Immunol. 20:835–851. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Oliveira JC, Oliveira LC, Mathias C,
Pedroso GA, Lemos DS, Salviano-Silva A, Jucoski TS, Lobo-Alves SC,
Zambalde EP, Cipolla GA and Gradia DF: Long non-coding RNAs in
cancer: Another layer of complexity. J Gene Med.
21:e30652019.PubMed/NCBI
|
|
11
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q,
Zhang P, Yang Z, Liu W, Zhang H, Chen N, et al: Long non-coding RNA
CCAL regulates colorectal cancer progression by activating
Wnt/β-catenin signalling pathway via suppression of activator
protein 2α. Gut. 65:1494–1504. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Neve B, Jonckheere N, Vincent A and Van
Seuningen I: Epigenetic regulation by lncRNAs: An overview focused
on UCA1 in colorectal cancer. Cancers (Basel). 10(pii): E4402018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Y, Zhao L, Lei L, Lau WB, Lau B, Yang
Q, Le X, Yang H, Wang C, Luo Z, et al: LncRNAs: The bridge linking
RNA and colorectal cancer. Oncotarget. 8:12517–12532.
2017.PubMed/NCBI
|
|
15
|
Chen DL, Lu YX, Zhang JX, Wei XL, Wang F,
Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, et al: Long
non-coding RNA UICLM promotes colorectal cancer liver metastasis by
acting as a ceRNA for microRNA-215 to regulate ZEB2 expression.
Theranostics. 7:4836–4849. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu
XY, Yu ZW, Jia YH, Bai XF, Li L, et al: The lncRNA CRNDE promotes
colorectal cancer cell proliferation and chemoresistance via
miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol
Cancer. 16:92017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu T, Han Z, Li H, Zhu Y, Sun Z and Zhu
A: LncRNA DLEU1 contributes to colorectal cancer progression via
activation of KPNA3. Mol Cancer. 17:1182018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Y, Yang X, Chen Z, Tian L, Jiang G,
Chen F, Li J, An P, Lu L, Luo N, et al: m6A-induced
lncRNA RP11 triggers the dissemination of colorectal cancer cells
via upregulation of Zeb1. Mol Cancer. 18:872019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Beyes S, Andrieux G, Schrempp M, Aicher D,
Wenzel J, Antón-García P, Boerries M and Hecht A: Genome-wide
mapping of DNA-binding sites identifies stemness-related genes as
directly repressed targets of SNAIL1 in colorectal cancer cells.
Oncogene. 38:6647–6661. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang H, Li JM, Wei W, Yang R, Chen D, Ma
XD, Jiang GM and Wang BL: Regulation of ATP-binding cassette
subfamily B member 1 by Snail contributes to chemoresistance in
colorectal cancer. Cancer Sci. 111:84–97. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ye X, Tam WL, Shibue T, Kaygusuz Y,
Reinhardt F, Ng Eaton E and Weinberg RA: Distinct EMT programs
control normal mammary stem cells and tumour-initiating cells.
Nature. 525:256–260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ye C, Shen Z, Wang B, Li Y, Li T, Yang Y,
Jiang K, Ye Y and Wang S: A novel long non-coding RNA lnc-GNAT1-1
is low expressed in colorectal cancer and acts as a tumor
suppressor through regulating RKIP-NF-κB-Snail circuit. J Exp Clin
Cancer Res. 35:1872016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY,
Chen SY and Li ZG: MALAT1 promotes colorectal cancer cell
proliferation/migration/invasion via PRKA kinase anchor protein 9.
Biochim Biophys Acta. 1852:166–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT1
promotes tumour growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L,
Sun J, Cai J, Qin J, Ren J and Li Q: Resveratrol inhibits invasion
and metastasis of colorectal cancer cells via MALAT1 mediated
Wnt/β-catenin signal pathway. PLoS One. 8:e787002013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen X, Bai Y, Luo B and Zhou X:
Upregulation of lncRNA BANCR associated with the lymph node
metastasis and poor prognosis in colorectal cancer. Biol Res.
50:322017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ragusa M, Barbagallo C, Statello L,
Condorelli AG, Battaglia R, Tamburello L, Barbagallo D, Di Pietro C
and Purrello M: Non-coding landscapes of colorectal cancer. World J
Gastroenterol. 21:11709–11739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Z, Jia H, Gu T, Hu Q, Yu J, Zang D,
Song N and Wang H: RNA sequencing and bioinformatics analysis of
the long noncoding RNA-mRNA network in colorectal cancer. J Cell
Biochem. 119:9957–9966. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Patel KR, Andreadi C, Britton RG,
Horner-Glister E, Karmokar A, Sale S, Brown VA, Brenner DE, Singh
R, Steward WP, et al: Sulfate metabolites provide an intracellular
pool for resveratrol generation and induce autophagy with
senescence. Sci Transl Med. 5:205ra1332013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu H, Hu Y, Liu X, Song W, Gong P, Zhang
K, Chen Z, Zhou M, Shen X, Qian Y and Fan H: LncRNA TRERNA1
function as an enhancer of SNAI1 promotes gastric cancer metastasis
by regulating epithelial-mesenchymal transition. Mol Ther Nucleic
Acids. 8:291–299. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu H, Liu X, Gong P, Song W, Zhou M, Li Y,
Zhao Z and Fan H: Elevated TFAP4 regulates lncRNA TRERNA1 to
promote cell migration and invasion in gastric cancer. Oncol Rep.
40:923–931. 2018.PubMed/NCBI
|
|
37
|
Song W, Gu Y, Lu S, Wu H, Cheng Z, Hu J,
Qian Y, Zheng Y and Fan H: LncRNA TRERNA1 facilitates
hepatocellular carcinoma metastasis by dimethylating H3K9 in the
CDH1 promoter region via the recruitment of the EHMT2/SNAI1
complex. Cell Prolif. 52:e126212019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X and Wu X: The role of
MicroRNA-1207-5p in colorectal cancer. Clin Lab. 63:1875–1882.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yan Y, Su M and Qin B: CircHIPK3 promotes
colorectal cancer cells proliferation and metastasis via modulating
of miR-1207-5p/FMNL2 signal. Biochem Biophys Res Commun.
524:839–846. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma S, Yang D, Liu Y, Wang Y, Lin T, Li Y,
Yang S, Zhang W and Zhang R: LncRNA BANCR promotes tumorigenesis
and enhances adriamycin resistance in colorectal cancer. Aging
(Albany NY). 10:2062–2078. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li AX, Xin WQ and Ma CG: Fentanyl inhibits
the invasion and migration of colorectal cancer cells via
inhibiting the negative regulation of Ets-1 on BANCR. Biochem
Biophys Res Commun. 465:594–600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong
H, Zhou JY and Yang SM: Long non-coding RNAs in colorectal cancer.
Oncotarget. 7:5226–5239. 2016.PubMed/NCBI
|
|
43
|
Nissan A, Stojadinovic A,
Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A,
Dayanc BE, Ritter G, Gomceli I, et al: Colon cancer associated
transcript-1: A novel RNA expressed in malignant and pre-malignant
human tissues. Int J Cancer. 130:1598–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim T, Jeon YJ, Cui R, Lee JH, Peng Y, Kim
SH, Tili E, Alder H and Croce CM: Role of MYC-regulated long
noncoding RNAs in cell cycle regulation and tumorigenesis. J Natl
Cancer Inst. 107(pii): dju5052015.PubMed/NCBI
|
|
45
|
Miller CR, Ruppert AS, Fobare S, Chen TL,
Liu C, Lehman A, Blachly JS, Zhang X, Lucas DM, Grever MR, et al:
The long noncoding RNA, treRNA, decreases DNA damage and is
associated with poor response to chemotherapy in chronic
lymphocytic leukemia. Oncotarget. 8:25942–25954. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang YR, Jang HJ, Yoon S, Lee YH, Nam D,
Kim IS, Lee H, Kim H, Choi JH, Kang BH, et al: OGA heterozygosity
suppresses intestinal tumorigenesis in Apc(min/+) mice.
Oncogenesis. 3:e1092014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Barrallo-Gimeno A and Nieto MA: The Snail
genes as inducers of cell movement and survival: Implications in
development and cancer. Development. 132:3151–3161. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Batlle E, Sancho E, Francí C, Domínguez D,
Monfar M, Baulida J and García De Herreros A: The transcription
factor snail is a repressor of E-cadherin gene expression in
epithelial tumour cells. Nat Cell Biol. 2:84–89. 2000. View Article : Google Scholar : PubMed/NCBI
|