Introduction
Liver cancer was the second leading cause of
cancer-associated mortality worldwide in 2015 (1). Patients with HCC have no noticeable
symptoms, making an accurate diagnosis challenging; therefore,
effective and efficient treatment of HCC should be available at a
much earlier stage, and novel biomarkers are required to improve
earlier diagnosis of HCC and guide clinical management (2,3).
HCC is associated with increased expression levels
of α-fetoprotein (AFP) (4). AFP is a
serum glycoprotein that has been widely used as a conventional
biomarker for HCC (5). However, the
expression levels of AFP remain normal in >30% of patients with
HCC at the time of diagnosis. In addition, relatively low AFP
expression levels have been identified in approximately 30% of HCC
(5). Although AFP is often used in
combination with ultrasound for an accurate diagnosis of HCC, novel
biomarkers may improve the diagnostic accuracy and detection rates
(6,7).
Guanine nucleotide-binding protein subunit α (GNA)
is a member of the guanine nucleotide-binding (G protein) family
that serve as modulators or transducers in miscellaneous
transmembrane signaling systems (8,9).
Mutations in G protein subunits α14 (GNA14), αQ (GNAQ) and α11, are
involved in ~50% of cherry hemangiomas and cherry-like hemangiomas,
and pathological and molecular studies have demonstrated common
functions of GNA14 mutations in vascular neoplasms (10). In addition, GNA14 silencing has been
demonstrated to impair the proliferation of endometrial carcinoma
cells by enhancing apoptosis and inducing cell cycle arrest
(11). Additionally, somatic
activating mutations in GNA14 induce the activation of the MAPK
signaling pathway and contribute to the occurrence and development
of congenital and sporadic vascular tumors, providing novel insight
into the underlying mechanisms of carcinogenesis (12).
The present study investigated the prognostic value
of GNA14 expression levels in HCC according using data from The
Cancer Genome Atlas (TCGA). Additionally, biological processes and
signaling pathways associated with GNA14 that may be involved in
the molecular pathogenesis of HCC were identified based on the Gene
Set Enrichment Analysis (GSEA) analysis. Overall, the present study
aimed to explore the role of GNA14 in HCC.
Materials and methods
Data sources
RNA-sequencing (RNA-seq) data (workflow type,
htseq-counts) and matching clinical data of 377 patients were
obtained from the Genomic Data Commons data portal (https://portal.gdc.cancer.gov/) in June 2019,
including information from TCGA database and the Therapeutically
Applicable Research to Generate Effective Treatments program. The
clinicopathological characteristics included age, sex, histological
grade, clinical stage, T, N and M staging (13). According to a previous study
(14), the processing of data and
further analysis were conducted to explore the associations between
GNA14 expression levels and clinicopathological characteristics in
HCC. An unpaired t-test was performed to analyze the difference
between tumor and normal samples.
Integrative Molecular Database of
Hepatocellular Carcinoma (HCCDB) analysis
The HCCDB (http://lifeome.net/database/hccdb/) (15) is an
integrated molecular database of HCC with 15 HCC gene expression
datasets including 3,917 samples. The HCCDB includes 13 datasets
from the Gene Expression Omnibus database and 2 RNA-seq datasets
from TCGA liver hepatocellular carcinoma (LIHC) and the
International Cancer Genome Consortium (ICGC) databases to explore
the expression levels of GNA14 mRNA in HCC.
Oncomine database analysis
Oncomine is a cancer microarray database (oncomine.org/) containing 715 datasets and 86,733
cancer samples (16). In the present
study, Oncomine was used to further assess the expression levels of
GNA14 in HCC and adjacent normal tissue. The parameters were set as
fold-change ≥2 and P<0.05, and the top 10% of ranked genes were
exhibited.
DNA methylation level of GNA14
The DNA methylation levels of GNA14 were analyzed
using the Methylation in Human Cancer database (MethHC) (methhc.mbc.nctu.edu.tw/) (17), a comprehensive database of DNA
methylation and gene expression in human cancer. Unpaired t-tests
were performed to analyze the difference between tumor and normal
samples.
Tumor Immune Estimation Resource
(TIMER) database analysis
A systematic assessment of the correlation between
different immune cells, such as B cells, CD4+ T cells,
CD8+ T cells, neutrophils, macrophages, and dendritic
cells, and GNA14 expression levels across different types of cancer
were performed using the TIMER database (https://cistrome.shinyapps.io/timer/) (18).
GSEA
Gene set enrichment analysis (GSEA) was conducted
using c2.cp.kegg.v6.0.symbols.gmt as a reference gene set
(software.broadinstitute.org/gsea/) (19). A list of genes was obtained from GSEA
to account for the survival differences in patients with
differential expression levels of GNA14. Gene set permutations were
analyzed 1,000 times. In addition, the normalized enrichment score,
normalized P-value and false discovery rate (FDR) q value were
applied to filter the correlative pathways.
Statistical analysis
Statistical analysis was performed using R software
version 3.6.0 (http://www.R-project.org/) (20), R studio software
version 1.2.5019 (21) and GraphPad
Prism version 8.0 software (GraphPad Software, Inc.) for data
processing and analysis. All data were represented as mean ±
standard deviation (SD). According to the median value of GNA14
expression levels in HCC tissues, patients were divided into low
and high GNA14 expression groups. Survival analysis was conducted
by the Kaplan-Meier method. Wilcoxon signed-rank test and log
regression were used to analyze the association of
clinicopathological characteristics and GNA14 expression levels.
The univariate and multivariate Cox proportional hazards model was
performed to evaluate the prognostic value of GNA14 expression.
Receiver operating characteristic (ROC) curves were constructed
using log-rank tests to evaluate the diagnostic value of GNA14
expression in HCC. P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient characteristics
Clinical and RNA-seq data of 377 patients with
primary HCC, including 377 HCC and 50 adjacent normal tissues, were
obtained from TCGA database and are listed in Table I. Of the 377 patients, 122 (32.4%)
were female and 255 (67.6%) were male, with a median age of 61
years at diagnosis, ranging between 16 and 81 years. The
distribution of conventional histological grades G1–4 was 15.4,
49.0, 32.4 and 3.2%, respectively. In addition, division of the
patients based on clinical stage resulted in 125 (51.9%) patients
with stage I, 60 (24.9%) patients with stage II, 55 (22.8%)
patients with stage III and one (0.4%) patient with stage IV
disease. There were 128 (50.4%) patients with T1 stage, 65 (25.6%)
with T2 stage disease, 54 (21.3%) with T3 stage disease and 7
(2.8%) with T4 stage disease. Moreover, only one (0.4%) patient had
confirmed N1 status, 177 (69.7%) with N0 status and others (29.9%)
with Nx status. Additionally, one patient (0.4%) had confirmed M1
status, 186 (72.9%) with N0 status and others (26.7%) with Nx
status.
 | Table I.The clinicopathological
characteristics of 377 patients with hepatocellular carcinoma. |
Table I.
The clinicopathological
characteristics of 377 patients with hepatocellular carcinoma.
| Characteristic | n | % |
|---|
| Age at diagnosis,
years (range) | 61 (16–81) |
|
| Sex |
|
|
|
Female | 122 | 32.4 |
| Male | 255 | 67.6 |
| Grade |
|
|
| G1 | 39 | 15.4 |
| G2 | 124 | 49.0 |
| G3 | 82 | 32.4 |
| G4 | 8 | 3.2 |
| Clinical stage |
|
|
| I | 125 | 51.9 |
| II | 60 | 24.9 |
| III | 55 | 22.8 |
| IV | 1 | 0.4 |
| T stage |
|
|
| T1 | 128 | 50.4 |
| T2 | 65 | 25.6 |
| T3 | 54 | 21.3 |
| T4 | 7 | 2.8 |
| N stage |
|
|
| N0 | 177 | 69.7 |
| N1 | 1 | 0.4 |
| Nx | 76 | 29.9 |
| M stage |
|
|
| M0 | 186 | 72.9 |
| M1 | 1 | 0.4 |
| Mx | 68 | 26.7 |
Differential expression of GNA14 in
HCC
As presented in Fig.
1A, GNA14 expression levels were evaluated in multiple types of
the tumor using the TIMER database. GNA14 expression levels were
significantly decreased in bladder urothelial carcinoma, breast
invasive carcinoma, cholangiocarcinoma, colon adenocarcinoma,
esophageal carcinoma, head and neck squamous cell carcinoma, kidney
chromophobe, kidney renal clear cell carcinoma, kidney renal
papillary cell carcinoma, LIHC and other types of cancer. The
downregulation of GNA14 expression levels in HCC tissues was
further verified using HCCDB and Oncomine (Fig. 1B and C). Therefore, the differential
expression of GNA12 was observed in liver cancer tissues compared
with normal liver tissues using multiple databases.
Association between GNA14 expression
levels and clinicopathological characteristics
RNA-seq data and clinical information of 377
patients with HCC from TCGA were used to analyze the association
between GNA14 expression levels and patient clinicopathological
characteristics. Based on TCGA-LIHC data using an unpaired t-test,
the expression of GNA14 was downregulated in HCC tissues compared
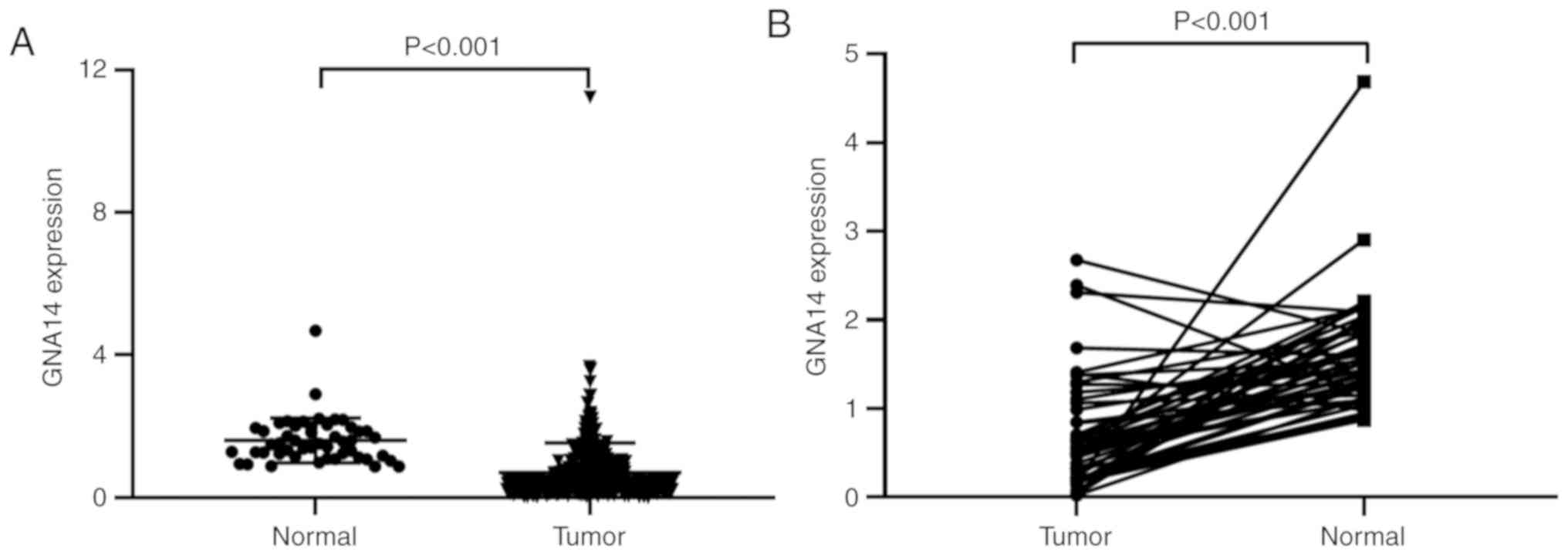
within the normal tissues (P<0.001; Fig. 2A). Moreover, expression of GNA14 in
HCC was lower compared with that in adjacent non-cancerous tissue
(P<0.001; Fig. 2B). In addition,
univariate logistic regression revealed that GNA14 expression
levels were associated with histological grade [odds ratio (OR),
0.114; P=0.009), clinical stage (OR, 0.418; P=0.001) and T stage
(OR, 0.508; P=0.013) in HCC (Table
II). These findings suggested that reduced expression levels of
GNA14 were associated with an advanced histological grade, clinical
stage and T stage in HCC.
 | Table II.GNA14 expression and
clinicopathological characteristics analyzed using log
regression. |
Table II.
GNA14 expression and
clinicopathological characteristics analyzed using log
regression.
| Clinical
characteristics | Total | Odds ratio in GNA14
expression | P-value |
|---|
| Age, years,
continuous | 370 | 1.001
(0.986–1.016) | 0.905 |
| Sex, female vs.
male | 371 | 1.237
(0.801–1.915) | 0.337 |
| Grade, G1 vs.
G3 | 177 | 0.114
(0.016–0.487) | 0.009a |
| M, M0 vs. M1 | 270 | 1.9×10-7
(NA-3.2×1029) | 0.983 |
| N, N0 vs. N1 | 256 | 1.049
(0.124–8.851) | 0.962 |
| Clinical stage, I
vs. III | 256 | 0.418
(0.243–0.710) | 0.001a |
| T, T1 vs. T3 | 261 | 0.508
(0.296–0.864) | 0.013 |
Survival outcomes and multivariate
analysis
According to the median value of GNA14 expression
levels in HCC tissues, patients were divided into low and high
GNA14 expression groups. Kaplan-Meier survival analysis of data
from TCGA-LIHC indicated that low GNA14 expression levels were
associated with a less favorable survival rate compared with that
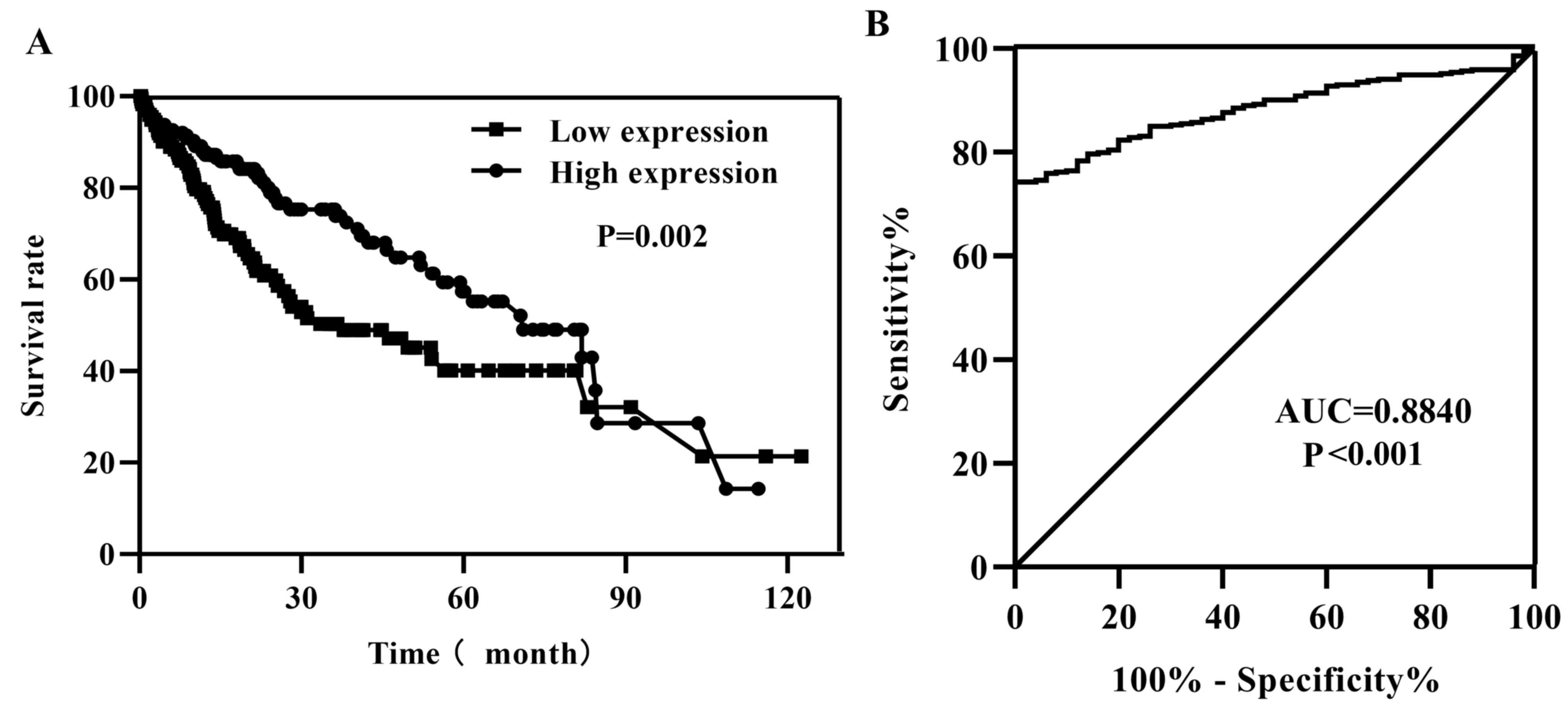
of patients with high GNA14 expression (P=0.002; Fig. 3A). To investigate the characteristics
of GNA14 as a potential tumor marker in HCC, ROC curve analysis was
performed, and the area under the ROC curve was 0.8840 (P<0.001;
Fig. 3B), indicating that GNA14
expression levels were a potential tumor marker for the diagnosis
of HCC. Additionally, univariate Cox regression analysis indicated
that low expression levels of GNA14 were associated with
unfavorable survival [hazard ratio (HR), 0.527; 95% confidence
interval (CI), 0.351–0.791; P=0.002, Table III). Other clinicopathological
features such as clinical stage and T stage were also associated
with less favorable survival (Table
IIIa). In addition, multivariate Cox regression analysis
revealed that low expression levels of GNA14 were associated with
unfavorable survival (HR, 0.636; 95% CI, 0.425–0.953; P=0.028;
Table IIIb. These findings
indicated that GNA14 expression levels may have potential as a
novel predictor for survival in HCC.
 | Table III.Association between overall survival
and clinicopathological features in patients with hepatocellular
carcinoma in The Cancer Genome Atlas Liver Hepatocellular Carcinoma
database using Cox regression and Multivariate Cox regression
analysis. |
Table III.
Association between overall survival
and clinicopathological features in patients with hepatocellular
carcinoma in The Cancer Genome Atlas Liver Hepatocellular Carcinoma
database using Cox regression and Multivariate Cox regression
analysis.
| A, Cox regression
analysis |
|
| Characteristic | HR | 95% CI | P-value |
|---|
| Age, years,
continuous | 1.011 | 0.996–1.026 | 0.154 |
| Sex, female vs.
male | 0.814 | 0.552–1.200 | 0.298 |
| Grade, G1 vs.
G4 | 1.113 | 0.862–1.438 | 0.412 |
| Clinical stage, I
vs. IV | 1.669 | 1.357–2.053 |
<0.001a |
| T, T1 vs. T4 | 1.649 | 1.354–2.009 |
<0.001a |
| M, M0 vs. M1 | 1.180 | 0.950–1.465 | 0.134 |
| N, N0 vs. N1 | 1.076 | 0.863–1.342 | 0.513 |
| GNA14
expression | 0.527 | 0.351–0.791 | 0.002a |
|
| B, Multivariate
Cox regression analysis |
|
|
Characteristic | HR | 95% CI | P-value |
|
| GNA14
expression | 0.636 | 0.425–0.953 | 0.028a |
DNA methylation analysis of GNA14 in
HCC
The methylation levels across the GNA14 gene
regions, including the promoter, TSS1500, TSS200, the 5′
untranslated region (5′UTR), CpG islands and N shores, were
assessed in HCC using the MethHC database. As presented in Fig. 4, the levels of GNA14 DNA methylation
were increased in HCC compared with the normal liver tissues in all
analyzed regions, such as the promoter, TSS1500, TSS200, the 5′
untranslated region (5′UTR), CpG islands and N shores.
Association of tumor-infiltrating
immune cells (TIICs) with GNA14 expression levels in HCC
As presented in Fig.
5A, GNA14 expression levels were partially correlated with the
number of infiltrating B cells (partial correlation, −0.191;
P<0.001) and macrophages (partial correlation, −0.132; P=0.015)
in HCC as determined by the TIMER database. In addition, GNA14
expression levels were partially negatively correlated with catenin
beta 1 (CTNNB1; partial correlation, 0.126; P=0.015), ryanodine
receptor 2 (RYR2; partial correlation, 0.17; P=0.001), filaggrin
(FLG; partial correlation, 0.196; P<0.001), CUB and Sushi
multiple domains 3 (CSMD3; partial correlation, −0.11; P=0.034) and
calcium voltage-gated channel subunit alpha1 E (CACNA1E; partial
correlation, −0.206; P<0.001) expression levels in HCC (Fig. 5B). The results showed that the low
expression of GNA14 might be closely related to immunotherapy.
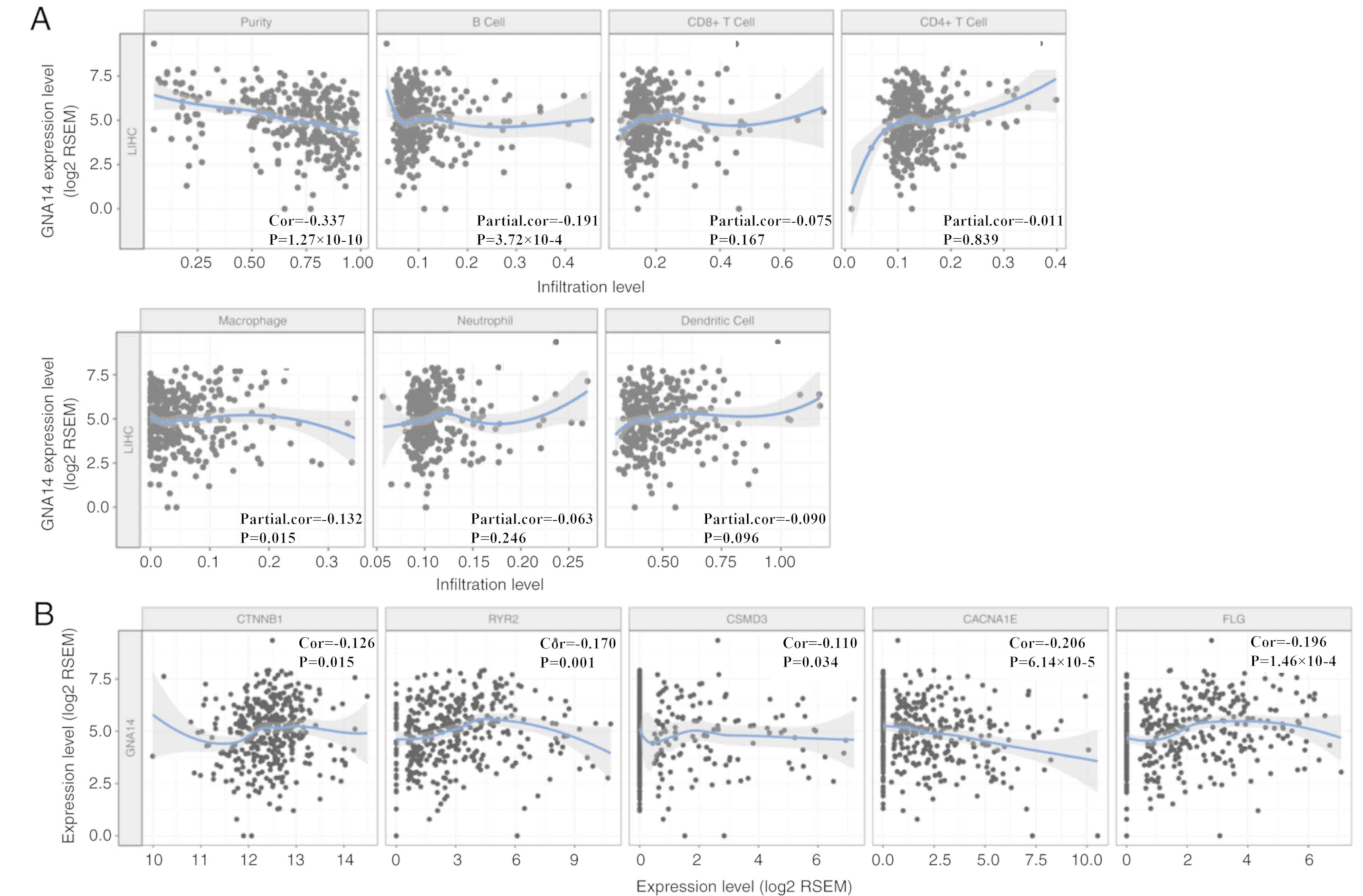 | Figure 5.Correlation of GNA14 expression levels
with TIICs and high-frequency gene expression in HCC using the
TIMER database. (A) Correlation of GNA14 expression levels and
TIICs in HCC, including B cells, CD4+ T cells, CD8+ T cells,
neutrophils, macrophages, and dendritic cells. (B) The correlation
of GNA14 expression and high-frequency gene expression, including
CTNNB1, RYR2, CSMD3, CACNA1E, and FLG, using the ‘correlation’
module of the TIMER database. GNA14, guanine nucleotide-binding
protein subunit α14; HCC, hepatocellular carcinoma; CTNNB1, catenin
beta 1; RYR2, ryanodine receptor 2; CSMD3, CUB and Sushi multiple
domains 3; CACNA1E, calcium voltage-gated channel subunit alpha1 E;
FLG, filaggrin; LIHC, liver hepatocellular carcinoma; TIICs,
tumor-infiltrating immune cells; RSEM, RNA-Seq by Expectation
Maximization. |
Pathway enrichment analysis
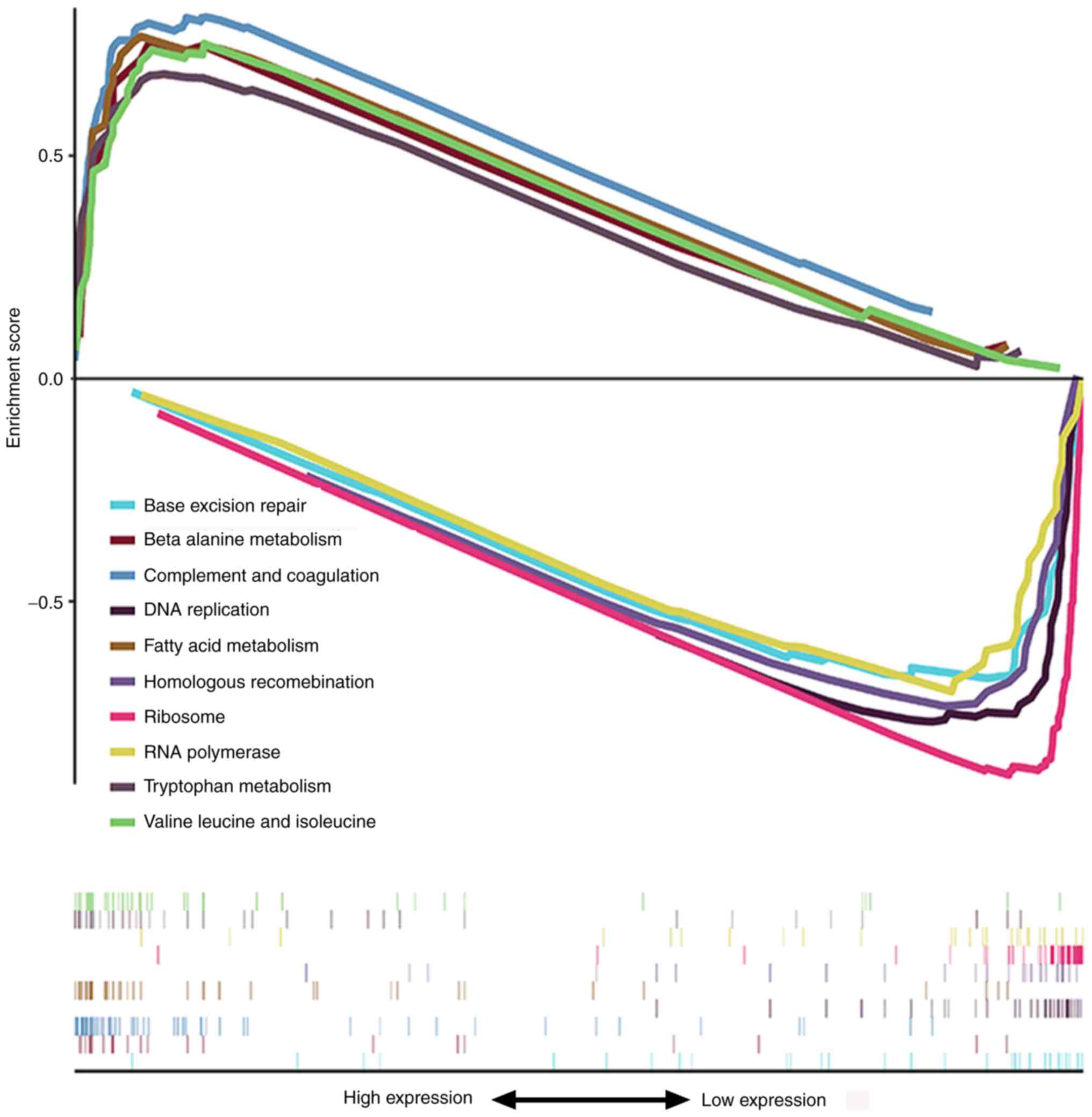
GSEA was used to identify signaling pathways
enriched in high and low GNA14 expression groups. As presented in
Fig. 6 and Table IV, in the low GNA14 expression
group, the significantly enriched signaling pathways were
‘ribosome’, ‘DNA replication’, ‘homologous recombination’, ‘RNA
polymerase’ and ‘base excision repair’. By contrast, ‘complement
and coagulation cascades’, ‘fatty acid metabolism’, ‘valine,
leucine and isoleucine degradation’, ‘beta-alanine metabolism’ and
‘tryptophan metabolism’ were enriched in the high GNA14 expression
group. These results showed that the low expression of GNA14 was
closely related to several tumor-related signaling pathways.
 | Table IV.Pathways were enriched in guanine
nucleotide-binding protein subunit α14 expression differential
phenotype. |
Table IV.
Pathways were enriched in guanine
nucleotide-binding protein subunit α14 expression differential
phenotype.
| Gene set name | NES | NOM P-value | FDR q-value |
|---|
| Complement and
coagulation cascades | 2.290 | <0.001 | <0.001 |
| Fatty acid
metabolism | 1.939 | 0.004 | 0.015 |
| Valine, leucine and
isoleucine degradation | 1.855 | 0.002 | 0.033 |
| Beta alanine
metabolism | 1.933 | <0.001 | 0.014 |
| Tryptophan
metabolism | 1.973 | 0.002 | 0.012 |
| Ribosome | −1.850 | 0.002 | 0.026 |
| DNA
replication | −1.785 | <0.001 | 0.044 |
| Homologous
recombination | −1.888 | <0.001 | 0.027 |
| RNA polymerase | −1.948 | <0.001 | 0.016 |
| Base excision
repair | −1.869 | <0.001 | 0.026 |
Discussion
Recent studies have suggested a potential role for
GNA14 in HCC. For example, activating mutations in GNAQ and GNA14
are frequently presented in hepatic small vessel neoplasms
(22). A recent study on aberrant
DNA methylation of differentially expressed genes in HCC has also
revealed that GNAQ serves as a hub gene (23). Additionally, a previous study
revealed that GNA14 exhibited enhanced methylation in HCC compared
with adjacent non-cancerous tissue (24). To best of our knowledge, the
association between GNA14 expression levels and HCC prognosis has
not been elucidated to date. In the present study, the role of
GNA14 in HCC was investigated using bioinformatics analysis.
A previous study of the GSE6764 gene expression
dataset in the Gene Expression Omnibus database revealed that the
expression levels of GNA14 were reduced in HCC tissues compared
with normal liver tissue (25). In
addition, a CpG island methylation phenotype-associated prognostic
model involved in the expression of PLEKHB1, ESR1, SLCO2A1, and
GNA14 was trained and validated in HCC, serving as an independent
prognostic factor for HCC (26).
However, the role of GNA14 expression levels in diagnosis and
prognosis of hepatocellular carcinoma is still unclear.
In the present study, the RNA-seq data were analyzed
to explore the role of GNA14 in HCC. GNA14 was demonstrated to be
downregulated in HCC compared with normal liver tissue, and these
results were verified using the HCCDB and Oncomine databases. In
the present study, GNA14 expression levels and their potential
prognostic value were investigated. It was demonstrated that GNA14
is abnormally expressed in HCC, and the prognostic value of GNA14
in HCC was also investigated. The results showed that the
downregulation of GNA14 in HCC was associated with an unfavorable
prognosis, indicating that GNA14 might exert a crucial part in HCC.
The results of the present study revealed that low GNA14 expression
levels were associated with advanced clinicopathological features,
including histological grade, clinical stage and T stage, poor
survival outcomes and an unfavorable prognosis. To the best of our
knowledge, the present study was the first to assess the prognostic
value of GNA14 in HCC using bioinformatics analysis. To the best of
our knowledge, the present study is the first to provide novel
insight into the role of GNA14 expression levels in HCC.
DNA methylation is a major epigenetic modification
that regulates gene expression and is crucial for tumor occurrence
and development (27). In the
present study, the methylation levels across the GNA14 gene
regions, including the promoter, TSS1500, TSS200, 5′UTR, CpG island
regions, and N shores, were assessed in HCC. The results indicated
that downregulation of GNA14 was correlated with its
hypermethylation in the promoter region and gene body region.
Furthermore, the correlation between GNA14 expression levels and
the numbers of TIICs was evaluated. Of note, GNA14 expression
levels were partially correlated with the infiltration level of B
cells and macrophages. Additionally, GNA14 expression levels were
partially correlated with CTNNB1, RYR2, FLG, CSMD3 and CACNA1E
expression levels in HCC.
GSEA analysis was performed in the present study
using TCGA-LIHC data to explore the potential function of GNA14 in
HCC. The ‘ribosome’, ‘DNA replication’, ‘homologous recombination’,
‘RNA polymerase’ and ‘base excision repair’ pathways were enriched
in the low GNA14 expression group. ‘Complement and coagulation
cascades’, ‘fatty acid metabolism’, ‘valine, leucine and isoleucine
degradation’, ‘beta-alanine metabolism’ and ‘tryptophan metabolism’
were enriched in the high GNA14 expression group. These signaling
pathways were regulated by GNA14 expression and may be crucial in
the carcinogenesis of HCC. The results indicated that GNA14
expression might have potential prognostic value in HCC and might
be an underlying biomarker for patients with HCC.
There were certain limitations to the present study.
Firstly, the present study lacked experimental validation and
functional research is needed to confirm the role of GNA14 in HCC
in future research. Secondly, adjuvant chemotherapy may have
effects on GNA14 expression and the information on patients who
received adjuvant chemotherapy was not obtained. Lastly,
cancer-specific survival was not analyzed in the present study.
Unlike overall survival, cancer-specific survival excludes death
due to causes unrelated to cancer. It would be more appropriate to
use different survival terms to describe prognosis in future
research.
In conclusion, GNA14 may have the potential to guide
the diagnosis and treatment of HCC. Further experimental studies
should be explored to verify the biological function of GNA14 in
HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in The Cancer Genome Atlas Research
Network repository (http://cancergenome.nih.gov/).
Authors' contributions
TY, SL and WX designed the study. TY collected the
data. SL contributed to data analysis and interpretation. TY and SL
drafted and revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GNA14
|
guanine nucleotide-binding protein
subunit α14
|
|
TIICs
|
tumor-infiltrating immune cells
|
References
|
1
|
Kulik L and El-Serag HB: epidemiology and
management of hepatocellular carcinoma. Gastroenterology.
156:477–491.e1. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 2:160182016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kanwal F and Singal AG: Surveillance for
hepatocellular carcinoma: Current best practice and future
direction. Gastroenterology. 157:54–64. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X and Wang Q: Alpha-fetoprotein and
hepatocellular carcinoma immunity. Can J Gastroenterol Hepatol.
2018:90492522018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bai DS, Zhang C, Chen P, Jin SJ and Jiang
GQ: The prognostic correlation of AFP level at diagnosis with
pathological grade, progression, and survival of patients with
hepatocellular carcinoma. Sci Rep. 7:128702017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marrero JA and Lok AS: Newer markers for
hepatocellular carcinoma. Gastroenterology. 127 (5 Suppl
1):S113–S119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Daniele B, Bencivenga A, Megna AS and
Tinessa V: Alpha-fetoprotein and ultrasonography screening for
hepatocellular carcinoma. Gastroenterology. 127 (5 Suppl
1):S108–S112. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pierce KL, Premont RT and Lefkowitz RJ:
Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 3:639–650.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bockaert J and Pin JP: Molecular tinkering
of G protein-coupled receptors: An evolutionary success. EMBO J.
18:1723–1729. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liau JY, Lee JC, Tsai JH, Chen CC, Chung
YC and Wang YH: High frequency of GNA14, GNAQ, and GNA11 mutations
in cherry hemangioma: A histopathological and molecular study of 85
cases indicating GNA14 as the most commonly mutated gene in
vascular neoplasms. Mod Pathol. 32:1657–1665. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Lv X, Xu F, Wei M, Liu C and Yang
Y: GNA14 silencing suppresses the proliferation of endometrial
carcinoma cells through inducing apoptosis and G2/M cell
cycle arrest. Biosci Rep. 38:BSR201805742018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lim YH, Bacchiocchi A, Qiu J, Straub R,
Bruckner A, Bercovitch L, Narayan D; Yale Center for Mendelian
Genomics, ; McNiff J, Ko C, et al: GNA14 somatic mutation causes
congenital and sporadic vascular tumors by MAPK activation. Am J
Hum Genet. 99:443–450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vogel A, Cervantes A, Chau I, Daniele B,
Llovet JM, Meyer T, Nault JC, Neumann U, Ricke J, Sangro B, et al:
Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 30:871–873. 2019.
View Article : Google Scholar
|
|
14
|
Wu H and Zhang J: Decreased expression of
TFAP2B in endometrial cancer predicts poor prognosis: A study based
on TCGA data. Gynecol Oncol. 149:592–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lian Q, Wang S, Zhang G, Wang D, Luo G,
Tang J, Chen L and Gu J: HCCDB: A Database of hepatocellular
carcinoma expression atlas. Genomics Proteomics Bioinformatics.
16:269–275. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang WY, Hsu SD, Huang HY, Sun YM, Chou
CH, Weng SL and Huang HD: MethHC: A database of DNA methylation and
gene expression in human cancer. Nucleic Acids Res. 43:(Database
Issue). D856–D861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna: 2012, http://www.R-project.org/
|
|
21
|
RStudio Team, . RStudio: Integrated
Development for R. RStudio, Inc.; Boston, MA: 2015, http://www.rstudio.com/
|
|
22
|
Joseph NM, Brunt EM, Marginean C,
Nalbantoglu I, Snover DC, Thung SN, Yeh MM, Umetsu SE, Ferrell LD
and Gill RM: Frequent GNAQ and GNA14 mutations in hepatic small
vessel neoplasm. Am J Surg Pathol. 42:1201–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cai C, Wang W and Tu Z: Aberrantly DNA
Methylated-differentially expressed genes and pathways in
hepatocellular carcinoma. J Cancer. 10:355–366. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao W, Kondo Y, Shen L, Shimizu Y, Sano T,
Yamao K, Natsume A, Goto Y, Ito M, Murakami H, et al: Variable DNA
methylation patterns associated with progression of disease in
hepatocellular carcinomas. Carcinogenesis. 29:1901–1910. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li G, Xu W, Zhang L, Liu T, Jin G, Song J,
Wu J, Wang Y, Chen W, Zhang C, et al: Development and validation of
a CIMP-associated prognostic model for hepatocellular carcinoma.
EBioMedicine. 47:128–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanwal R and Gupta S: Epigenetic
modifications in cancer. Clin Genet. 81:303–311. 2012. View Article : Google Scholar : PubMed/NCBI
|