Introduction
Colon cancer is one of the most common malignant
tumors of the digestive system and its incidence rate is increasing
every year in China (1,2). The incidence rate in China was
27.08/100,000, and the mortality rate was 13.13/100,00 in 2014
(3). Patients with colon cancer
typically suffer recurrence and metastasis, occasionally between
1–2 years after surgery (4).
It has been previously determined that chemokine
receptors are involved in the development and metastasis of breast
cancer, prostate cancer, colon cancer, liver cancer (5–8).
Recently, there is increasing evidence that chemokines and their
receptors play a key role in the development and progression of
cancer (9–11). CXCL14 is expressed by islet δ-cells
where it may exert paracrine effects to inhibit insulin secretion
in a CXCR4/CXCR7-independent manner through reductions in β-cell
ATP levels (10). Zhang et al
(11) revealed that angiogenesis was
enhanced with increased SDF1 and that angiogenesis was weakened
with the inhibition of CXCR7. They demonstrated that PI3K/AKT was
involved in the downstream pathway in the coculture. VEC
angiogenesis induction by NPCs was enhanced with an increase in
pAKT or a decrease in PTEN. The chemokine receptor investigated in
the current study is chemokine receptor 7 (CXCR7), which is a new
receptor for C-X-C motif chemokine ligand 12 [CXCL12; also known as
stromal cell-derived factor-1 (SDF-1)], after the discovery of the
CXCR4 receptor, and its binding affinity for CXCL12 is up to 10
times higher compared with that of the CXCR4 receptor (12,13).
Studies have shown that CXCR7 can inhibit tumor cell
growth and proliferation in prostate cancer and neuroblastoma by
binding to CXCL12 (14,15). Stacer et al (16) found that high expression of CXCR7 in
endothelial cells can regulate the metastasis of breast cancer
cells. A previous study revealed that CXCR7 enhances PC3 and C4-2B
prostate cancer cell invasion and metastasis by regulating the
expression levels of cell adhesion molecules, such as fibronectin,
cadherin-11, CD44, and matrix metalloproteinases (14). A previous report demonstrated that
CXCR7 is highly expressed in human colon cancer cells (17). Over the last 8 years, a number of
studies have confirmed that CXCR7 is also expressed in other types
of cancer, such as pancreatic cancer, thyroid cancer, prostate
cancer, breast cancer, esophageal cancer, liver cancer, and bladder
cancer, and it has been shown to promote tumor growth and
metastasis (5,18–23). The
results of our previous study (24)
indicated that the protein and mRNA expression of CXCR7 in Caco-2
cells was low compared with that in RKO, SW480, and HCT116 colon
cancer cells. However, whether CXCR7 has similar functions in
Caco-2 and HCT116 cells remains to be elucidated.
Therefore, the primary aim of the current study is
to assess the protein and mRNA expression levels of CXCR7 in Caco-2
and HCT116 cells, and secondly to inhibit the expression level of
CXCR7 in Caco-2 and HCT116 cells and investigate the subsequent
biological activity of these cells.
Materials and methods
Cell culture
Caco-2 and HCT116 cells were purchased from the Cell
Bank of the Chinese Academy of Sciences and cultured in Dulbecco's
modified Eagle's medium (HyClone; GE Healthcare Life Sciences)
containing 10% fetal bovine serum (FBS), and 100 U/ml penicillin
and 100 µg/ml streptomycin. The cells were cultured in an incubator
at 37°C in a humidified incubator with 5% CO2.
Cell transfection
Caco-2 and HCT116 cells were seeded at a density of
4×105 cells/well in a 6-well plate overnight, and the
medium was replaced with fresh medium without FBS. Cy5
fluorescence-labeled siRNA (Guangzhou RiboBio Co., Ltd.) was
transfected into Caco-2 and HCT116 cells by Lipofectamine 3000.
After 6 h, the siRNA transfection efficiency was observed under the
inverted fluorescence microscope. The three CXCR7 interfering
segments were as follows: siRNA1, 5′-CGUCCAACAAUGAGACCUAdTdT-3′;
siRNA2, 5′-CGUCCAACAAUGAGACCUAdTdT-3′; and siRNA3,
5′-GCUAUGACACGCACUGCUAdTdT-3′. siRNAs [siRNA1, siRNA2, siRNA3, and
siRNA Negative Control (NC); Guangzhou RiboBio Co., Ltd.] were
transfected into Caco-2 and HCT116 cells using
Lipofectamine® 3000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), in accordance with the
manufacturer's instructions. Follow-up subsequent experimentation 6
h after transfection.
Reverse transcription-quantitative PCR
(qRT-PCR) after transfection
The total RNA of the cells was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), Reverse transcription of total RNA was carried out by using
a Prime Script RT Master mix (Takara Biotechnology Co., Ltd.). The
following primers were used: CXCR7 forward,
5′-TCTGCATCTCTTCGACTACTCA-3′ and reverse
5′-GTAGAGCAGGACGCTTTTGTT-3′; GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse 5′-GAAGATGGTGATGGGATTTC-3′.
GAPDH was used as the internal control. Subsequently, 1 µl cDNA was
amplified using 10 µl SYBR Premix Ex TaqII kit (Takara
Biotechnology Co., Ltd.) and 0.8 µl primers in a Light Cycler 480
instrument (Roche Diagnostics). The following thermal cycling
conditions were used: Initial denaturation at 95°C for 30 sec, 45
cycles of 95°C for 5 sec, 60°C for 30 sec, melting curve at 95°C
for 5 sec, 60°C for 60 sec, 95°C for 5 sec, and cooling at 40°C for
30 sec. Relative mRNA expressions was calculated using the
2−ΔΔCT method: 2−ΔΔCT (ΔΔCt=[Ct (CXCR7)-Ct
(GAPDH)] target- [Ct (CXCR7)-Ct (GAPDH)]xinternal standard
(25).
Western blot analysis
Transfected cells were incubated in
radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of
Biotechnology) for 15 min. The protein concentration of the samples
was calculated using a bicinchoninic acid (BCA) protein
concentration assay kit. Each sample (20 mg/lane) was mixed with
loading buffer (Beyotime Institute of Biotechnology) and boiled for
5 min at 95°C in the heating module. Then, 20 mg per lane of
proteins from each sample were loaded and separated by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE),
later transferred to polyvinylidene difluoride membranes (PVDF;
Bio-Rad Laboratories, Inc.). The membranes were blocked at room
temperature for 2 h with 3% bovine serum albumin (BSA, Hyclone; GE
Healthcare Life Sciences) and washed them three times with TBST.
The dilution ratio for the anti-CXCR7 antibody (cat. no. ab72100)
was 1:1,000 and for the anti-GAPDH antibody (cat. no. ab181602) was
1:2,000 (both Abcam). The primary antibody was incubated overnight
at 4°C. After several washings, membranes were incubated with
horseradish peroxidase-conjugated secondary antibody (anti-rabbit)
(1:2,000 dilution) for 2 h at 37°C. The protein bands were
developed by adding ECL solution (Bio-Rad Laboratories, Inc.). The
levels of protein expression were evaluated by Image Pro Plus 6.0
(IPP) software (Media Cybernetics, Inc.).
Immunofluorescence assay after
transfection
The cells were washed three times with
phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for
20 min at RT, and incubated with 0.1% Triton X-100 penetrant for 25
min. After washing the well plates, the samples were blocked in
goat serum (HyClone; GE Healthcare Life Sciences) for 30 min at RT,
and incubated with CXCR7 antibody (cat. no. ab72100; dilution
1:100; Abcam) overnight at 4°C. Subsequently, the cells were
incubated with fluorescent secondary antibody (1:200) for 1 h at
37°C, washed three times with PBS, incubated with
4′,6-diamidino-2-phenylindole for 30 min at RT, and images were
obtained using a fluorescence microscope (magnification, ×200). The
levels of fluorescence intensity were evaluated by Image Pro Plus
6.0 (IPP) software.
Cell Counting Kit-8 (CCK-8) and
5-ethynyl-2′-deoxyuridine (EdU) assays to detect cell proliferation
after transfection
Cells were seeded in 96-well plates at 5,000 cells
per well. After 24, 48 and 72 h of culture, 100 µl of Dulbecco's
Modified Eagle Medium (DMEM) medium (HyClone; GE Healthcare Life
Sciences) containing 10% CCK-8 reagent (Dojindo Molecular
Technologies) was added to each well, in accordance with the
manufacturer's instructions. The plates were incubated at 37°C for
1 and 2 h, and then, the absorbance was measured at 450 nm using a
microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Next, 50 µM EdU medium was prepared, and 200 µl of the medium was
added to each well for 2 h. The wells were then subjected to Apollo
staining for 30 min at RT, in accordance with the manufacturer's
instructions, and images were obtained using a fluorescence
microscope (magnification, ×200).
Cell migration and invasion assay
after transfection
The transfected cells were seeded into the upper
chamber of a Transwell insert at a density of 4×105
cells/well without FBS, and 500 µl of DMEM (HyClone; GE Healthcare
Life Sciences) containing 20% FBS was added to the lower chamber;
the chambers were then incubated at 37°C for 6 h. The migrant cells
that were adhered to the lower surface of the membrane were fixed
with 4% paraformaldehyde for 20 min at room temperature, and then
stained with 0.1% crystal violet for 30 min at RT. The number of
cells under the membrane surface was counted in five different
fields using a light microscope at a magnification of ×200.
Transfected Caco-2 and HCT116 cells were seeded into 6-well plates
at a density of 4×105 cells/well prior to a wound
healing assay. When the confluence of cells was >90%, a scratch
was created using a 10 µl pipette tip in the middle of the well.
Cells were cultured with serum-free medium for a further 24, 48 and
72 h. A light microscope was used to observe wound healing
(magnification, ×200). Matrigel (100 µl; 1:6 dilution; BD
Biosciences) was added to the upper chamber to coagulate for 30 min
at 37°C, and cells were cultured for 24 h for invasion experiments.
The additional steps were the same as aforementioned.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Statistical differences between the groups were assessed
using SPSS version 19.0 software (IBM Corp.). Differences between
groups were calculated using a one-way analysis of variance
(ANOVA), and P<0.05 was considered to indicate a statistically
significant difference. The experiment above was performed in
triplicate.
Results
Expression level of CXCR7 in Caco-2
and HCT116 cells after transfection
siRNA1, siRNA2, siRNA3, and siRNANC were transfected
into Caco-2 and HCT116 cells, and fluorescence imaging confirmed
that the transfection was effective (Fig. 1A). In the present study, RT-qPCR and
western blot analysis showed that the expression level of CXCR7 in
the transfected group (siRNA3) was significantly lower compared
with that in the NC group (qPCR: Caco-2, 0.87±0.11 and HCT116,
1.05±0.10; WB: Caco-2, 0.69±0.03 and HCT116, 0.79±0.03), and the
effect of siRNA3 (PCR: Caco-2, 0.34±0.15 and HCT116, 0.31±0.03; WB:
Caco-2, 0.06±0.02, and HCT116, 0.16±0.05) was the most significant
in reducing CXCR7 expression. Consequently, siRNA3 was used in
subsequent experiments (Fig. 1B and
C).
Immunofluorescence detection following
transfection
siRNA3 and siRNANC were transfected into Caco-2 and
HCT116 cells, and the expression of CXCR7 was detected. The results
showed that the fluorescence intensity of Caco-2 and HCT116 cells
in the siRNA3 transfected group (Caco-2, 2.12±0.14 and HCT116,
3.45±0.95) was significantly lower compared with that in the NC
group (Caco-2, 7.88±0.55 and HCT116, 8.78±0.41, P<0.01), and the
difference between the control and the NC group was not significant
(Fig. 1D).
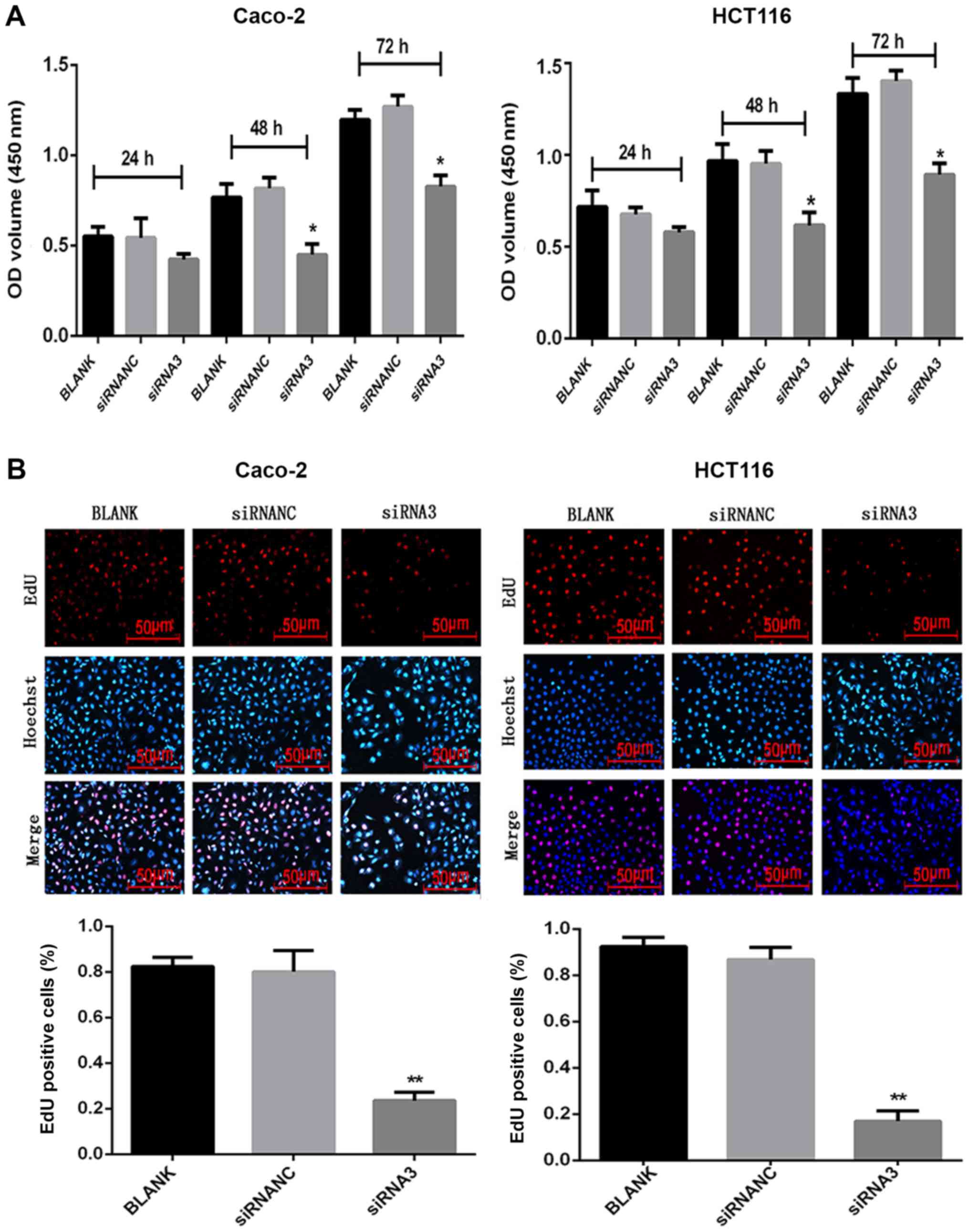
Effect of CXCR7 on the proliferation
of Caco-2 and HCT116 cells
Following transfection of siRNA3 into Caco-2 and
HCT116 cells, their proliferation was measured using CCK-8 and EdU
assays. After 48 and 72 h, 10 µl CCK-8 solution was added to each
well and incubated for 1 h. The CCK-8 assay demonstrated that the
proliferation of the siRNA3-transfected cells (48 h: Caco-2,
0.31±0.02 and HCT116, 0.58±0.04; 72 h: Caco-2, 0.67±0.06 and
HCT116, 0.73±0.06) was reduced compared with that in the NC group
(48 h: Caco-2, 0.65±0.03 and HCT116, 0.75±0.08; 72 h: Caco-2,
0.89±0.09 and HCT116, 0.93±0.06) (Fig.
S1). After 48 and 72 h, 10 µl CCK-8 solution was added to each
well and incubated for 2 h. The CCK-8 assay demonstrated that the
proliferation of the siRNA3-transfected cells (48 h: Caco-2,
0.45±0.05 and HCT116, 0.62±0.07; 72 h: Caco-2, 0.83±0.05 and
HCT116, 0.89±0.06) was reduced compared with that in the NC group
(48 h: Caco-2, 0.82±0.06 and HCT116, 0.95±0.07; 72 h: Caco-2,
1.27±0.06 and HCT116, 1.40±0.06) (Fig.
2A). The CCK-8 assay results were not statistically significant
at 24 h, for either cell line. The EdU results revealed that the
number of proliferating cells was significantly reduced (P<0.05)
after siRNA3 transfection (Caco-2, 0.24±0.04 and HCT116, 0.17±0.04)
compared with the NC group (Caco-2, 0.80±0.09 and HCT116,
0.87±0.05) (Fig. 2B).
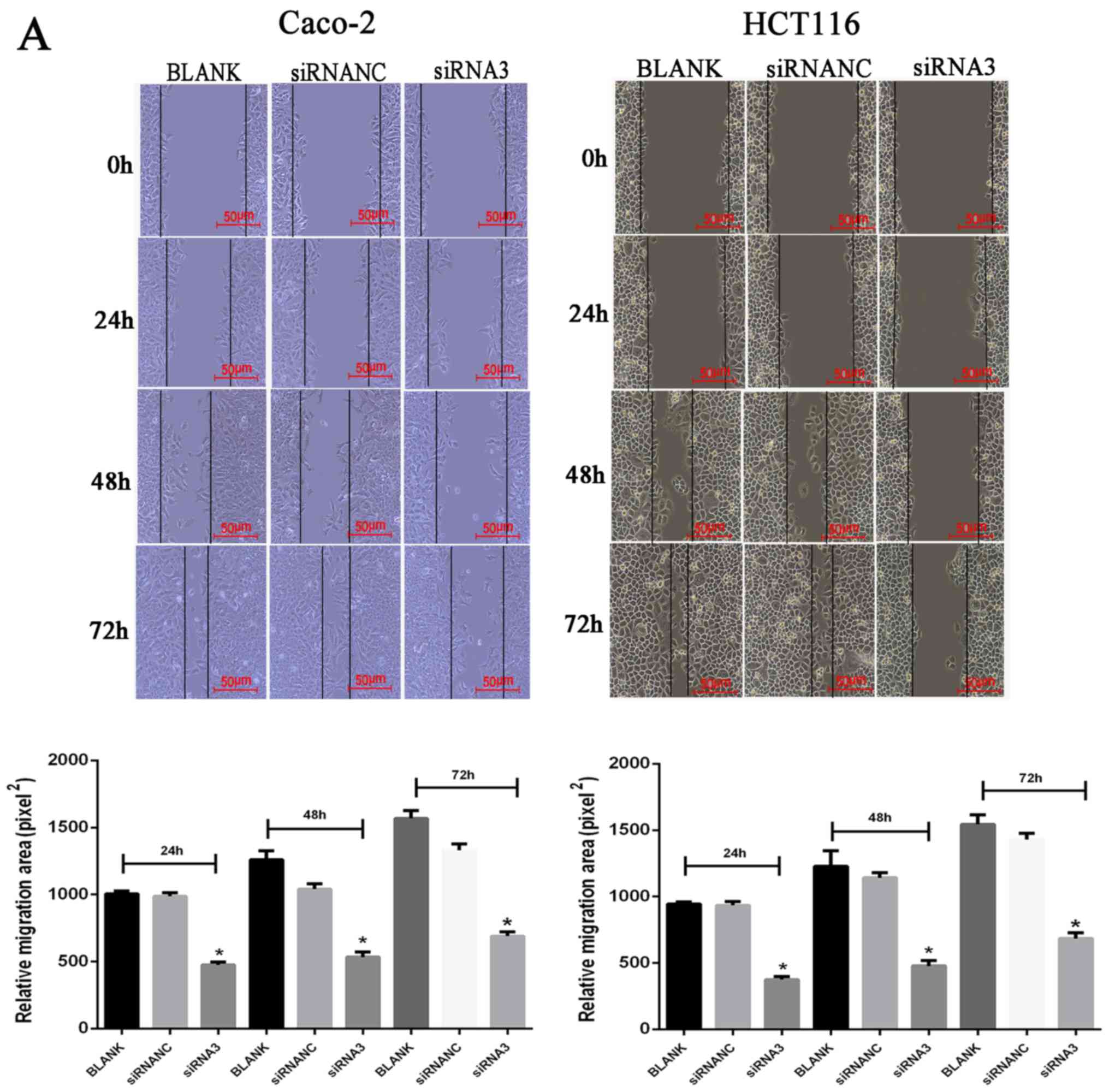
Effect of CXCR7 on the migration of
Caco-2 and HCT116 cells
Following transfection of siRNA3 into Caco-2 and
HCT116 cells, their migration ability was determined using a wound
healing and Transwell assays. The wound healing assay results
revealed that the cell migration area was significantly decreased
(P<0.05) after Caco-2 and HCT116 transfection of siRNA3 (24 h:
Caco-2, 460.58±46.88 and HCT116, 368.91±38.19; 48 h: Caco-2,
548.00±12.83 and HCT116, 471.33±33.37; 72 h: Caco-2, 713.33±19.55
and HCT116, 666.67±42.92) compared with that in the NC group (24 h:
Caco-2, 944.65±53.11 and HCT116, 931.31±23.39; 48 h: Caco-2,
1,060.67±38.06 and HCT116, 1,194.00±75.45; 72 h: Caco-2,
1,372.67±61.63 and HCT116, 1,449.33±56.05) (Fig. 3A). The Transwell assay results
revealed that the number of transmembrane cells was significantly
decreased (P<0.01) after Caco-2 and HCT116 transfection of
siRNA3 Caco-2, 195±6.02 and HCT116, 128.33±11.37) compared with
that in the NC group (Caco-2, 389.67±22.15 and HCT116,
411.43±12.53), indicating that downregulation of CXCR7 expression
significantly inhibited cell migration (Fig. 3B).
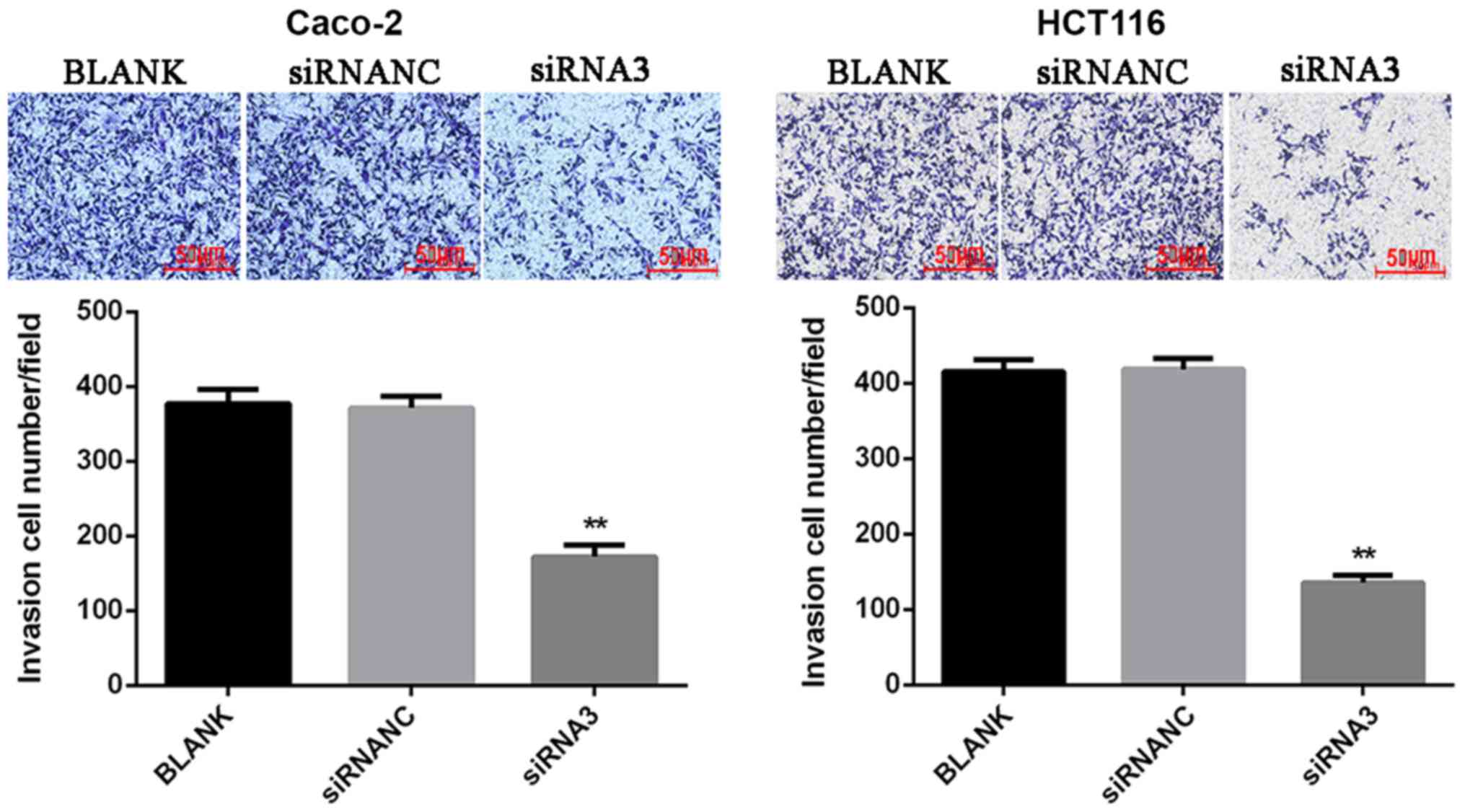
Effect of CXCR7 on Caco-2 and HCT116
cell invasion
To determine the cell invasion ability, a Matrigel
assay was used to detect the number of transmembrane cells. The
results showed that the number of invasions of Caco-2 and HCT116
cells transfected with siRNA3 (Caco-2, 172.33±14.32 and HCT116,
135.67±10.02) was significantly reduced (P<0.01) compared with
that in the NC group (Caco-2, 371.33±15.82 and HCT116
418.33±14.19). This suggests that downregulation of CXCR7 may
inhibit cell invasion (Fig. 4).
Discussion
The development of cancer is a complex process,
including proliferation, migration, and invasion (26). Previous studies have found that
chemokines and chemokine receptors play an important role in tumor
invasion, growth, and metastasis (27,28).
Romain et al (29) found that
the mRNA or protein expression of CXCR4 and CXCR7 was similar to
that of the normal mucosa in the polyps and early-stage carcinomas
but significantly increased in late stage carcinomas.
Recent studies have reported that another receptor
for SDF-1, CXCR7, is overexpressed in various human malignancies,
such as ovarian cancer, colorectal cancer, and breast cancer
(7,30,31).
Experimental studies showed that mice treated with anti-SDF-1
showed higher expression of CXCR7 compared with that in controls,
indicating that CXCR7 regulates colon cancer angiogenesis and tumor
growth independently of SDF-1 (32).
Moreover, a previous study revealed that the chemokine receptor
CXCR7 demonstrates increased expression in colorectal cancer tumors
(33). Guillemot et al
(34) suggested that the activation
of CXCR7 on tumor blood vessels by its ligands may facilitate the
progression of CRC within lung cancer but not within liver cancer.
Wang et al (35) found that
CXCR7-knockdown negatively affected cell survival and migration
in vitro, suggesting that CXCR7 functions in tumor
aggravation. The experiments performed in the present study also
demonstrated the proliferation, invasion and migration ability of
Caco-2 and HCT116 cells was significantly reduced following
transfection with CXCR7 siRNA, inhibiting the tumor-like behavior
of these cells, which supports the previous studies.
In the present study, downregulation of CXCR7
expression has been shown to inhibit tumor cell proliferation,
migration, and invasion. The CCK-8 assay and EdU results revealed
that cell proliferation was reduced after transfection compared
with that in the NC group. The wound healing assay and Transwell
assay results revealed that the migration area and the number of
migrating cells were significantly reduced in the siRNA3
transfection group compared with that in the NC group. In our
previous study (24), SW480 and
Caco-2 cells were inoculated subcutaneously into the right lower
limb tissue of mice. CXCR7 was upregulated or downregulated prior
to inoculation in mice. The expression of CXCR7 was measured using
RT-qPCR, western blot analysis, and immunohistochemistry. Firstly,
RT-qPCR and western blot analysis revealed that knockdown of CXCR7
in SW480 and Caco-2 cells decreased the phosphorylation of AKT and
ERK and expression of VEGF. In contrast, overexpression of CXCR7 in
SW480 and Caco-2 cells increased the phosphorylation of AKT and ERK
and expression of VEGF. Secondly, the immunohistochemical staining
was higher in CXCR7-overexpressing Caco-2 cells compared with that
in the NC group. In contrast, the staining was lower in the
CXCR7-silenced SW480 cells compared with that in the NC group.
These results indicate that CXCR7 simultaneously regulates the
ERK/AKT signaling pathway and expression of VEGF in colon cancer
in vitro and in vivo. Our previous study revealed
that CXCR7 is a key factor in tumorigenesis by promoting cell
proliferation and angiogenesis (24). The results of the present study
support the current conclusions and also indicate that changes in
the expression of CXCR7 are key factors in the development and
progression of colon cancer. The effects of CXCR7 on colon cancer
migration and invasion was further investigated. The results showed
that silencing CXCR7 also inhibits colon cancer migration and
invasion. Wang et al (36)
found that CXCR7 is involved in CXCL12 cell cycle and proliferation
regulation in mouse neural progenitor cells, and CXCL12
pretreatment leads to shortening of the G0/G1
phase and prolongation of S phase, and increases cyclin D1 and
β-catenin expression. Notably, in vitro experiments
performed by Kim et al (37)
on U937 cell migration revealed that the silencing of CXCR7 did not
change the proliferation or apoptosis of U937 cells. This indicates
that the effect of dysregulation of CXCR7 expression is specific to
different cancer types.
A previous study has shown that a CXCR7 antagonist
(CCX771) inhibits tumor growth and metastasis in a mouse breast
cancer model (22). Lin et al
(38) also found that CCX771 can
inhibit the proliferation of HepG2 hepatoma cells overexpressing
CXCR7 by blocking the activated mitogen activated protein kinase
signaling pathway. The results of the current study indicate that
downregulation of CXCR7 expression inhibits cell proliferation,
invasion, and migration. Gebauer et al (39) found that the high expression of CXCR7
in colorectal cancer is associated with poor prognosis in patients.
Therefore, inhibition of CXCL12/CXCR7 may represent a new approach
for tumor targeted therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Heilongjiang
Provincial Science Fund Project (grant no. H2017078).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL contributed to collecting data, data analysis and
interpretation, and drafting the manuscript. XMW contributed to the
concept and design of the study and for the final approved version,
and supervise the study. ZTL analyzed the data and drafted the
manuscript. YJL performed cell culture experiments and helped
revise the manuscript. LS analyzed the data and organized the data.
ZZ contributed to the concept and design of the study. YXZ
critically revised the article and acquired the data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Sun K, Zheng R, Zeng H, Zhang S,
Xia C, Yang Z, Li H, Zou X and He J: Cancer incidence and mortality
in China, 2014. Chin J Cancer Res. 30:1–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lai HW, Wei JC, Hung HC and Lin CC: Tumor
sidedness influences prognostic impact of lymph node metastasis in
colon cancer patients undergoing curative surgery. Sci Rep.
9:198922019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Singh RK and Lokeshwar BL: The
IL-8-regulated chemokine receptor CXCR7 stimulates EGFR signaling
to promote prostate cancer growth. Cancer Res. 71:3268–3277. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xue TC, Han D, Chen RX, Zou JH, Wang Y,
Tang ZY and Ye SL: High expression of CXCR7 combined with alpha
fetoprotein in hepatocellular carcinoma correlates with
extra-hepatic metastasis to lung after hepatectomy. Asian Pac J
Cancer Prev. 12:657–663. 2011.PubMed/NCBI
|
|
7
|
Li XX, Zheng HT, Huang LY, Shi DB, Peng
JJ, Liang L and Cai SJ: Silencing of CXCR7 gene represses growth
and invasion and induces apoptosis in colorectal cancer through ERK
and β-arrestin pathways. Int J Oncol. 45:1649–1657. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miao Z, Luker KE, Summers BC, Berahovich
R, Bhojani MS, Rehemtulla A, Kleer CG, Essner JJ, Nasevicius A,
Luker GD, et al: CXCR7 (RDC1) promotes breast and lung tumor growth
in vivo and is expressed on tumor-associated vasculature. Proc Natl
Acad Sci USA. 104:15735–15740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Floranović MP and Veličković LJ: Effect of
CXCL12 and its receptors on unpredictable renal cell carcinoma.
Clin Genitourin Cancer. Dec 4–2019.(Epub ahead of print).
View Article : Google Scholar
|
|
10
|
Atanes P, Hawkes RG, Olaniru OE,
Ruz-Maldonado I, Amisten S and Persaud SJ: CXCL14 inhibits insulin
secretion independently of CXCR4 or CXCR7 receptor activation or
cAMP inhibition. Cell Physiol Biochem. 52:879–892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H, Wang P, Zhang X, Zhao W, Ren H
and Hu Z: SDF1/CXCR7 signaling axis participates in angiogenesis in
degenerated discs via the PI3K/AKT pathway. DNA Cell Biol.
38:457–467. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Balabanian K, Lagane B, Infantino S, Chow
KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M and
Bachelerie F: The chemokine SDF-1/CXCL12 binds to and signals
through the orphan receptor RDC1 in T lymphocytes. J Biol Chem.
280:35760–35766. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Daniel SK, Seo YD and Pillarisetty VG: The
CXCL12- CXCR4/CXCR7 axis as a mechanism of immune resistance in
gastrointestinal malignancies. Semin Cancer Biol. Dec 23–2019.(Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Shiozawa Y, Wang J, Wang Y, Jung
Y, Pienta KJ, Mehra R, Loberg R and Taichman RS: The role of
CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate
cancer. J Biol Chem. 283:4283–4294. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liberman J, Sartelet H, Flahaut M,
Mühlethaler-Mottet A, Coulon A, Nyalendo C, Vassal G, Joseph JM and
Gross N: Involvement of the CXCR7/CXCR4/CXCL12 axis in the
malignant progression of human neuroblastoma. PLoS One.
7:e436652012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stacer AC, Fenner J, Cavnar SP, Xiao A,
Zhao S, Chang SL, Salomonnson A, Luker KE and Luker GD: Endothelial
CXCR7 regulates breast cancer metastasis. Oncogene. 35:1716–1724.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang HX, Tao LY, Qi KE, Zhang HY, Feng D,
Wei WJ, Kong H, Chen TW, Lin QS and Chen DJ: Role of CXC chemokine
receptor type 7 in carcinogenesis and lymph node metastasis of
colon cancer. Mol Clin Oncol. 3:1229–1232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hao M, Zheng J, Hou K and Wang J, Chen X,
Lu X, Bo J, Xu C, Shen K and Wang J: Role of chemokine receptor
CXCR7 in bladder cancer progression. Biochem Pharmacol. 84:204–214.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Heinrich EL, Lee W, Lu J, Lowy AM and Kim
J: Chemokine CXCL12 activates dual CXCR4 and CXCR7-mediated
signaling pathways in pancreatic cancer cells. J Transl Med.
10:682012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tachezy M, Zander H, Gebauer F, von Loga
K, Pantel K, Izbicki JR and Bockhorn M: CXCR7 expression in
esophageal cancer. J Transl Med. 11:2382013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Z, Yang L, Teng X, Zhang H and Guan H:
The involvement of CXCR7 in modulating the progression of papillary
thyroid carcinoma. J Surg Res. 191:379–388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wani N, Nasser MW, Ahirwar DK, Zhao H,
Miao Z, Shilo K and Ganju RK: C-X-C motif chemokine 12/C-X-C
chemokine receptor type 7 signaling regulates breast cancer growth
and metastasis by modulating the tumor microenvironment. Breast
Cancer Res. 16:R542014. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao ZW, Fan XX, Song JJ, Xu M, Chen MJ,
Tu JF, Wu FZ, Zhang DK, Liu L, Chen L, et al: ShRNA knock-down of
CXCR7 inhibits tumour invasion and metastasis in hepatocellular
carcinoma after transcatheter arterial chemoembolization. J Cell
Mol Med. 21:1989–1999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Wang X, Li Z, Zhang Z and Zhang Y:
Chemokine receptor 7 targets the vascular endothelial growth factor
via the AKT/ERK pathway to regulate angiogenesis in colon cancer.
Cancer Med. 8:5327–5340. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Balkwill F: Cancer and the chemokine
network. Nat Rev Cancer. 4:540–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ben-Baruch A: Organ selectivity in
metastasis: Regulation by chemokines and their receptors. Clin Exp
Metastasis. 25:345–356. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Romain B, Hachet-Haas M, Rohr S, Brigand
C, Galzi JL, Gaub MP, Pencreach E and Guenot D: Hypoxia
differentially regulated CXCR4 and CXCR7 signaling in colon cancer.
Mol Cancer. 13:582014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu Y, Li H, Xue B, Jiang X, Huang K, Ge J,
Zhang H and Chen B: SDF-1/CXCR7 axis enhances ovarian cancer cell
invasion by MMP-9 expression through p38 MAPK pathway. DNA Cell
Biol. 33:543–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qian T, Liu Y, Dong Y, Zhang L, Dong Y,
Sun Y and Sun D: CXCR7 regulates breast tumor metastasis and
angiogenesis in vivo and in vitro. Mol Med Rep.
17:3633–3639. 2018.PubMed/NCBI
|
|
32
|
Kollmar O, Rupertus K, Scheuer C, Nickels
RM, Haberl GC, Tilton B, Menger MD and Schilling MK: CXCR4 and
CXCR7 regulate angiogenesis and CT26.WT tumor growth independent
from SDF-1. Int J Cancer. 126:1302–1315. 2010.PubMed/NCBI
|
|
33
|
Yang D, Dai T, Xue L, Liu X, Wu B, Geng J,
Mao X, Wang R, Chen L and Chu X: Expression of chemokine receptor
CXCR7 in colorectal carcinoma and its prognostic significance. Int
J Clin Exp Pathol. 8:13051–13058. 2015.PubMed/NCBI
|
|
34
|
Guillemot E, Karimdjee-Soilihi B, Pradelli
E, Benchetrit M, Goguet-Surmenian E, Millet MA, Larbret F, Michiels
JF, Birnbaum D, Alemanno P, et al: CXCR7 receptors facilitate the
progression of colon carcinoma within lung not within liver. Br J
Cancer. 107:1944–1949. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang H, Tao L, Qi KE, Zhang H, Feng D, Wei
W, Kong H, Chen T, Lin Q and Chen D: CXCR7 functions in colon
cancer cell survival and migration. Exp Ther Med. 10:1720–1724.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Xu P, Qiu L, Zhang M, Huang Y and
Zheng JC: CXCR7 participates in CXCL12-mediated cell cycle and
proliferation regulation in mouse neural progenitor cells. Curr Mol
Med. 16:738–746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim HY, Lee SY, Kim DY, Moon JY, Choi YS,
Song IC, Lee HJ, Yun HJ, Kim S and Jo DY: Expression and functional
roles of the chemokine receptor CXCR7 in acute myeloid leukemia
cells. Blood Res. 50:218–226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin L, Han MM, Wang F, Xu LL, Yu HX and
Yang PY: CXCR7 stimulates MAPK signaling to regulate hepatocellular
carcinoma progression. Cell Death Dis. 5:e14882014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gebauer F, Tachezy M, Effenberger K, von
Loga K, Zander H, Marx A, Kaifi JT, Sauter G, Izbicki JR and
Bockhorn M: Prognostic impact of CXCR4 and CXCR7 expression in
pancreatic adenocarcinoma. J Surg Oncol. 104:140–145. 2011.
View Article : Google Scholar : PubMed/NCBI
|