Introduction
Hepatocellular carcinoma (HCC) is the most common
type of liver cancer and a malignant tumor that is detrimental to
human health (1,2). Geographically, the incidence rates in
North and West Africa, and East and Southeast Asia are high. In
China, the annual incidence of HCC is 20.37/100,000, which accounts
for 50% of all reported HCC cases worldwide (3), and its mortality rate (15.2%) was
reported to be the second highest among all cancer-associated
mortalities in 2018 (4,5). Previous studies have reported that the
5-year survival rate of patients with HCC is only 3–5%, and
~600,000 patients die from HCC annually in China (3,6).
Although patients with HCC can be treated using various measures,
including surgical resection, liver transplantation, local
treatment and chemotherapy, the prognosis of HCC remains poor
(7,8). This is due to high recurrence and
metastasis, which are the predominant causes of mortality in
patients with HCC (9). Regarding
patients with severe early disease who undergo liver resection or
liver transplantation, >80% exhibit recurrence following
surgery, which affects their quality of life (10). A number of factors have predictive
prognostic value for HCC, including tumor size, tumor
differentiation, vascular invasion, marginal resection, and novel
immunological and tissue biomarkers (11–15).
Currently, different biomarkers, such as α-fetoprotein (AFP) and
des-γ-carboxyprothrombin, are being implemented as a base to
establish a series of diagnostic and long-term prognostic
guidelines for HCC (16). However,
recent studies have failed to provide sufficient unbiased data to
establish these biomarkers as effective monitoring, diagnostic and
prognostic tools, due to their poor specificity and sensitivity
(16,17). Thus, the development of novel
molecular markers that can be used to accurately determine the
prognosis or recurrence of HCC has critical clinical application
value, and selecting appropriate treatment plans for patients is
beneficial to improve the survival rate of HCC.
KIAA1522 is a newly cloned, large protein-coding
gene of unknown function (18).
Information from the Gene Expression Atlas (http://www.ebi.ac.uk/gxa) and The European
Bioinformatics Institute databases (https://www.ebi.ac.uk) demonstrates that KIAA1522 mRNA
expression is upregulated in various tumor tissues, such as lung
and breast cancer, suggesting that KIAA1522 may play a role in
tumorigenesis and cancer development (18). However, to the best of our knowledge,
KIAA1522 expression in HCC has not yet been fully investigated.
Thus, the present study performed bioinformatics analysis, using
the Oncomine (www.oncomine.org) and OncoLnc
(www.oncolnc.org) databases, in order to analyze
KIAA1522 expression and assess its association with the clinical
prognosis of HCC. KIAA1522 expression in HCC and adjacent normal
tissues was detected via immunohistochemical staining and reverse
transcription-quantitative (RT-q)PCR analysis. Additionally, the
association between KIAA1522 expression and the clinicopathological
characteristics and prognosis of HCC was analyzed by combining
clinical pathology and follow-up data.
Materials and methods
Bioinformatics analysis
The Chen Liver dataset (19) was searched within the Oncomine
database with the following specifications: Gene, KIAA1522;
analysis type, cancer vs. normal analysis; and cancer type, liver
cancer. KIAA1522 expression levels in HCC and adjacent normal liver
tissues were analyzed in the database and plotted using GraphPad
Prism software (version 6; http://www.graphpad.com). Subsequently, the dataset
[which comprised 217 datapoints from The Cancer Genome Atlas (TCGA)
database (https://tcga-data.nci.nih.gov/tcga)] was searched for
within the OncoLnc database with the following parameters:
KIAA1522; liver hepatocellular carcinoma; lower percentile, 30%;
and upper percentile, 30%. A survival curve was generated and
plotted using GraphPad Prism 6.
Patients and samples
A total of 79 paraffin-embedded primary HCC tissue
samples, from 64 males and 15 females (median age, 55 years; age
range, 24–73 years) were surgically resected and collected between
January 2013 and December 2013 at The Affiliated Hospital of
Qingdao University (Qingdao, China). The inclusion criteria were as
follows: Postoperative pathology confirmed as HCC and
Tumor-Node-Metastasis (TNM) staging (20), TnN0M0. The exclusion criteria were as
follows: Received any anticancer treatment prior to surgery;
serious complications or mortality within 30 days following
surgery; non-tumor associated mortalities, and incomplete clinical,
pathological and surgical data.
According to the eighth edition of the 2017 American
Joint Committee on Cancer TNM staging system (www.cancerstaging.org), there were 39 cases in
clinical stage I, 10 cases in clinical stage II and 30 cases in
clinical stage IIIa. The pathological differentiation grade, based
on the World Health Organization tumor histological classification
criteria (21), were as follows: A
total of 3 cases with high differentiation, 50 cases with moderate
differentiation and 26 cases with poor differentiation. Liver
function grading was performed using the Child-Pugh grading
standard (22), which was scored
according to the most recent clinical data prior to surgery, and
included 78 cases of grade A and 1 case of grade B.
RT-qPCR
Total RNA was extracted from surgically resected HCC
tissues using TRIzol® reagent (Invitrogen; Thermo fisher
Scientific, Inc.) and reversed transcribed into first-strand cDNA
using the First-Strand Synthesis System for RT-PCR (Takara
Biotechnology Co., Inc.). qPCR was subsequently performed using the
SYBR Green kit (Takara Biotechnology Co., Inc.). The following
primer sequences were used for qPCR: KIAA1522 forward,
5′-TGGATGAGCACCAGGACAAC-3′ and reverse, 5′-GTCCGGGAGGACTGGATACT-3′;
and β-actin forward, 5′-CCTCTCCCAAGTCCACACAG-3′ and reverse,
5′-GGGCACGAAGGCTCATCATT-3′. The following thermocycling conditions
were used for qPCR: 24 cycles of initial denaturation at 98°C for
30 sec; 30 cycles of 98°C for 15 sec, 72°C for 30 sec; and a final
extension at 72°C for 30 sec, and then cooled at 4°C for 15 sec.
Relative mRNA expression levels were measured using the
2−ΔΔCq method (23). All
experiments were performed in triplicate.
Immunohistochemistry
A total of 79 paraffin-embedded primary HCC tissue
samples were fixed in 4% formaldehyde for 24 h at room temperature
and embedded in paraffin. Paraffin-embedded samples were cut into
4-um-thick sections. Tissue samples were subsequently
deparaffinized in xylene at room temperature for 15 min, and
rehydrated in a descending ethanol series (anhydrous ethanol I, 5
min; anhydrous ethanol II, 5 min; 95% ethanol, 3 min; 90% ethanol,
3 min; 80% ethanol, 2 min and 70% ethanol, 2 min). Following
antigen retrieval with 10 mM citrate buffer (pH 6; Beijing g-clone
Biotechnology Co., Ltd.; http://www.g-clone.com) at 100°C for 10–15 min, tissue
sections were blocked with 3% hydrogen peroxide for 10 min at 37°C
to inhibit endogenous peroxidase activity. Tissue samples were
subsequently incubated with rabbit anti-human KIAA1522 antibody
(1:200; cat. no. PAB22604; Abnova), overnight at 4°C in a
humidified chamber. The PV-9000 Two-step Immunohistochemistry kit
(OriGene Technologies, Inc.) was used to detect primary antibodies.
Tissue sections were dehydrated in a descending ethanol series,
washed twice with running water and mounted. The slides were
subsequently stained with 3,3′-diaminobenzidine for 10 min at room
temperature, and counterstained with haematoxylin at room
temperature for 1 min.
Simultaneously, two pathologists assessed the
pathological sections in a blinded manner, and when the results
varied, a third person assessed the pathological sections to
discuss the differing observations. HCC tissue samples were
observed in 10 randomly selected fields of view under high power
light microscope (magnification, ×200). The degree of staining of
positive cells was scored as follows: 0, no staining; 1, pale
yellow; 2, brownish yellow; and 3, brown. The percentage of
positive cells were scored as follows: 0, <5%; 1, 6–25%; 2,
26–50%; 3, 51–75% and 4, >75%. The two scores were added
together, and all cases were divided into the low and high
expression groups. Total scores <2 were assigned to the low
expression group, while scores ≥2 were assigned to the high
expression group.
Case follow-up
Patients who met the study criteria were closely
followed via outpatient review and telephone. The regular follow-up
plan was set as follows: Every 3 months for 1 year post-surgery,
and every 6 months between 2 and 3 years post-surgery. The review
included liver function, AFP, abdominal ultrasound, chest
radiograph, enhanced CT or MRI and needle biopsy, among other
examinations. Recurrence was defined by imaging studies, or
biopsy-confirmed new lesions in the liver or outside the liver.
Disease-free survival (DFS) time was defined as the time from the
date of surgery to the time of recurrence or follow-up. Overall
survival (OS) time was defined as the time from the date of surgery
to the time of mortality or follow-up. Both DFS and OS were
calculated on a monthly basis, and the follow-up deadline was
December 2017.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 24.0; IBM Corp.). The χ2, continuity
correction χ2 and Fisher's exact probability tests were
used to determine the associations between KIAA1522 expression and
clinicopathological characteristics in HCC. The independent sample
t-test was used to analyze the difference between KIAA1522
expression in adjacent and HCC tissues. Kaplan-Meier survival
analysis and the log-rank test were used to compare the association
between KIAA1522 expression and postoperative recurrence and
survival in patients with HCC. The Cox proportional hazard model
was used to select variables by the forward logistic regression
method for univariate and multivariate analyses of recurrence and
survival. P<0.05 was considered to indicate a statistically
significant difference.
Results
KIAA1522 expression in HCC and
adjacent normal tissues
Analysis of the Chen Liver dataset within the
Oncomine database demonstrated that KIAA1522 mRNA expression was
significantly higher in HCC tissues compared with that in adjacent
normal liver tissues (P<0.01; Fig.
1A). This observation was validated in the patient cohort
assessed in the present study via RT-qPCR analysis of the matched
HCC and adjacent non-tumor tissues, which also demonstrated that
KIAA1522 mRNA expression was significantly higher in HCC tissues
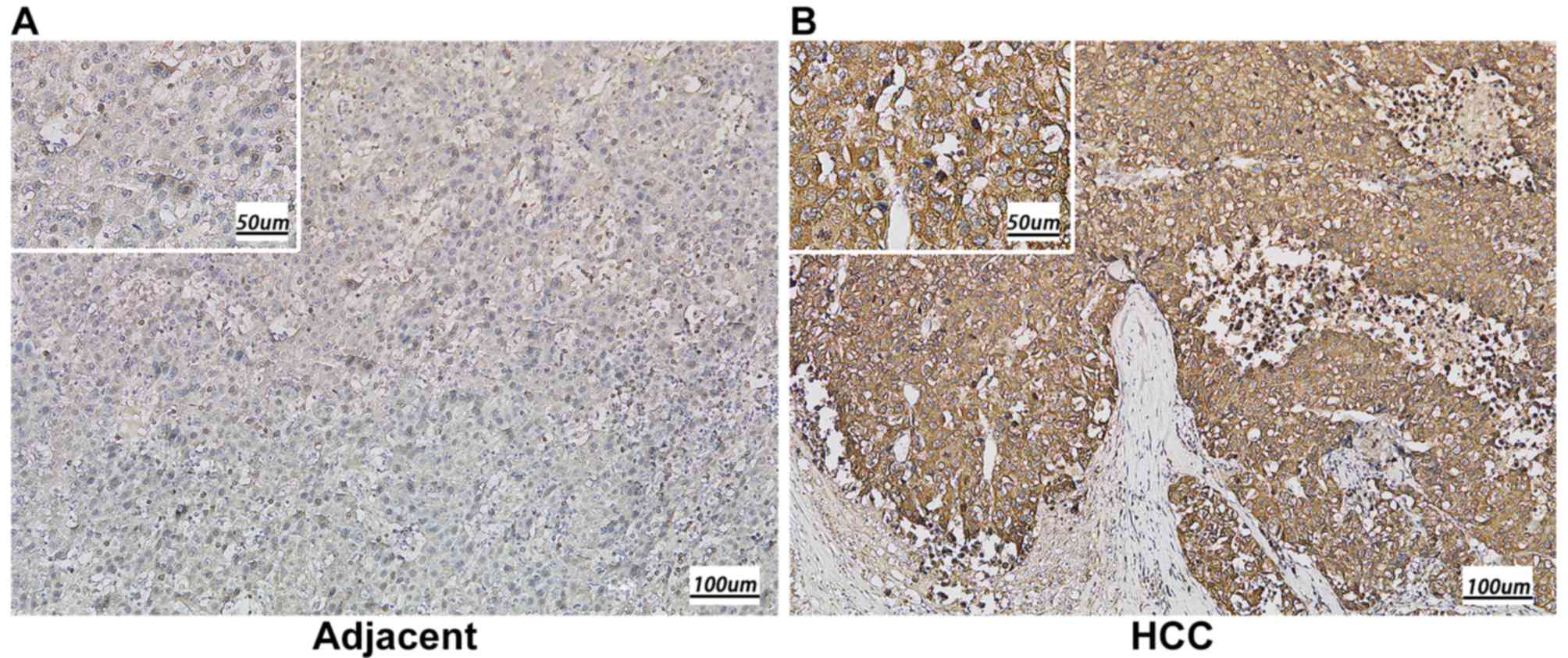
compared with that in adjacent normal tissues (P<0.001; Fig. 1B). Subsequently, immunohistochemistry
was performed to detect KIAA1522 expression in 79 paraffin-embedded
human HCC tissues, including 50 cases of clinical stage I, 19 cases
of clinical stage II and 10 cases of clinical stage IIIa. The
results indicated that KIAA1522 protein expression was
predominantly localized in the cytoplasm, and the high expression
rate of KIAA1522 protein was significantly higher in the HCC
tissues (89.9%) compared with the adjacent normal tissues (15.2%)
(P<0.01; Fig. 2 and Table I).
 | Table I.KIAA1522 protein expression in HCC and
adjacent normal tissues. |
Table I.
KIAA1522 protein expression in HCC and
adjacent normal tissues.
|
| KIAA1522 |
|
|
|---|
|
|
|
|
|
|---|
| Tissue | Negative, n (%) | Positive, n (%) | Total, n | P-value |
|---|
| HCC | 8
(10.1) | 71 (89.9) | 79 | <0.01a |
| Adjacent normal | 67 (84.8) | 12 (15.2) | 79 |
|
Association between KIAA1522
expression and clinicopathological characteristics in HCC
The association between KIAA1522 expression and the
clinicopathological characteristics in HCC are presented in
Table II. Notably, KIAA1522
expression was not demonstrated to be significantly associated with
any of the assessed clinicopathological characteristics in HCC,
including age, sex, Child-Pugh classification, tumor number and
size, degree of differentiation and clinical stage.
 | Table II.Association between KIAA1522
expression and clinicopathological characteristics in patients with
hepatocellular carcinoma. |
Table II.
Association between KIAA1522
expression and clinicopathological characteristics in patients with
hepatocellular carcinoma.
|
|
| KIAA1522, n (%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Patient, n | High | Low | χ2 | P-value |
|---|
| Sex |
|
|
| 0.395 | 0.530a |
|
Male | 64 | 37 (57.8) | 27 (42.2) |
|
|
|
Female | 15 | 5 (33.3) | 10 (66.7) |
|
|
| Age, years |
|
|
| 0.750 | 0.386a |
|
<50 | 16 | 8 (50.0) | 8 (50.0) |
|
|
|
≥50 | 63 | 39 (61.9) | 24 (38.1) |
|
|
| Alcohol
consumption |
|
|
| 0.069 | 0.793a |
|
Yes | 21 | 13 (61.9) | 8 (38.1) |
|
|
| No | 58 | 34 (58.6) | 24 (41.4) |
|
|
| HBV infection |
|
|
| 0.000 |
>0.999b |
|
Yes | 74 | 44 (59.5) | 30 (40.5) |
|
|
| No | 5 | 3 (60.0) | 2 (40.0) |
|
|
| Liver
cirrhosis |
|
|
| 0.002 | 0.965a |
|
Yes | 64 | 38 (59.4) | 26 (40.6) |
|
|
| No | 15 | 9 (60.0) | 6 (40.0) |
|
|
| AFP level,
ng/l |
|
|
| 2.005 | 0.157a |
|
≤400 | 54 | 35 (64.8) | 19 (35.2) |
|
|
|
>400 | 25 | 12 (48.0) | 13 (52.0) |
|
|
| Child-Pugh
grade |
|
|
|
|
>0.999c |
| A | 78 | 46 (59.0) | 32 (41.0) |
|
|
| B | 1 | 0 (0.0) | 1 (100.0) |
|
|
| Tumor number |
|
|
| 0.000 |
>0.999b |
|
Single | 69 | 41 (59.4) | 28 (40.6) |
|
|
|
Multiple | 10 | 6 (60.0) | 4 (40.0) |
|
|
| Tumor size, cm |
|
|
| 1.114 | 0.286a |
| ≤5 | 57 | 36 (63.2) | 21 (36.8) |
|
|
|
>5 | 22 | 11 (50.0) | 11 (50.0) |
|
|
| Pathological
differentiation |
|
|
| 0.000 | 0.242b |
|
High | 3 | 2 (66.7) | 1 (33.3) |
|
|
| Middle
and low | 76 | 45 (59.2) | 31 (40.8) |
|
|
| Microvascular tumor
thrombus |
|
|
| 0.264 | 0.607b |
|
Yes | 27 | 15 (55.6) | 12 (44.4) |
|
|
| No | 52 | 32 (61.5) | 20 (38.5) |
|
|
| Capsule
invasion |
|
|
| 3.174 | 0.075a |
|
Yes | 50 | 26 (52.0) | 24 (48.0) |
|
|
| No | 29 | 21 (72.4) | 8 (27.6) |
|
|
| Surgical resection
range |
|
|
| 2.420 | 0.120a |
| Liver
lobe | 41 | 21 (51.2) | 20 (48.8) |
|
|
| Liver
segment | 38 | 26 (68.4) | 12 (31.6) |
|
|
| TNM stage |
|
|
| 0.005 | 0.943a |
|
I+II | 49 | 29 (59.2) | 20 (40.8) |
|
|
|
IIIa | 30 | 18 (60.0) | 12 (40.0) |
|
|
Association between KIAA1522
expression and postoperative prognosis in HCC
The KIAA1522 dataset, including 217 TCGA primary
liver cancer datapoints, was downloaded from the OncoLnc database
and a survival curve was generated, which demonstrated that high
KIAA522 expression was closely associated with low OS time of
patients with HCC (P<0.01; Fig.
3A). Survival analysis was performed on the 79 clinical cases
from the present study using the Kaplan-Meier method and log-rank
test, in order to determine the association between KIAA1522
expression in HCC and OS time following surgery. The results
demonstrated that the mean OS time was 49.700 months (95% CI,
45.320–53.950). The mean OS time in the low expression group was
59.200 months (95% CI, 57.405–60.907), while the mean OS time in
the high expression group was 36.600 months (95% CI,
27.607–43.636). The mean OS time in the high expression group was
significantly decreased compared with the low expression group
(P<0.001; Fig. 3B).
Database analysis failed to provide a survival curve
associated with recurrence of KIAA1522 expression in HCC. Thus,
KIAA1522 expression in HCC was compared with postoperative
recurrence, as described above. The results demonstrated that the
overall mean DFS time was 41.100 months (95% CI, 35.691–46.480).
The mean DFS time in the low expression group was 43.400 months
(95% CI, 36.994–49.875), while the mean DFS time in the high
expression group was 37.241 months (95% CI, 35.691–46.480). No
significant difference in DFS time was observed between the two
groups (P=0.254 Fig. 3C).
Univariate and multivariate Cox
regression analyses of prognosis
Univariate and multivariate analyses of
clinicopathological characteristics with regard to OS are presented
in Table III. Factors that may
influence the OS time of patients with HCC were included in the
univariate Cox regression analysis. The results demonstrated that
TNM staging, KIAA1522 expression, tumor size, surgical resection,
hepatic capsule invasion and vascular tumor thrombus had a
significant impact on postoperative survival time (all P<0.05).
Conversely, sex, age and AFP levels, among others, were
demonstrated to have no significant effect on postoperative
survival time in patients with HCC (all P>0.05). Subsequently,
multivariate Cox regression analysis was performed on the
clinicopathological characteristics which demonstrated statistical
significance in the univariate analysis. The results demonstrated
that surgical resection range, hepatic capsule invasion and
vascular tumor thrombus did not affect the OS time of patients with
HCC. However, tumor size >5 cm (P=0.031), TNM clinical stage
IIIa (P=0.047) and high KIAA1522 expression (P=0.001) were
demonstrated to be independent risk factors affecting postoperative
OS time (all P<0.05). According to the hazard ratio (HR) values,
these factors were considered to have an impact on the OS time in
the following order: High KIAA1522 expression, TNM clinical stage
IIIa and tumor size >5 cm (Table
III).
 | Table III.Univariate and multivariate analyses
of clinicopathological characteristics with regard to overall
survival. |
Table III.
Univariate and multivariate analyses
of clinicopathological characteristics with regard to overall
survival.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Characteristic | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex (female vs.
male) | 2.478
(0.577–10.644) | 0.222 |
|
|
| Age (<50 vs. ≥50
years) | 0.575
(0.223–1.480) | 0.251 |
|
|
| Alcohol
consumption | 1.492
(0.602–3.699) | 0.387 |
|
|
| HBV infection | 1.356
(0.182–10.103) | 0.655 |
|
|
| TBIL level (≤22 vs.
>22 µmol/l) | 1.067
(0.359–3.172) | 0.907 |
|
|
| ALB level
(<35vs. ≥35 g/l) | 1.454
(0.338–8.102) | 0.615 |
|
|
| ALT level (≤60 vs.
>60 µ/l) | 1.688
(0.681–4.183) | 0.258 |
|
|
| AST level (≤42 vs.
>42 µ/l) | 1.883
(0.760–4.668) | 0.172 |
|
|
| PLT level (<100
vs. ≥100 l) | 5.188
(0.696–30.166) | 0.108 |
|
|
| Liver
cirrhosis | 0.767
(0.281–2.094) | 0.604 |
|
|
| AFP level (≤400 vs.
>400 ng/l) | 1.833
(0.772–4.355) | 0.170 |
|
|
| Child-Pugh grade (A
vs. B) | 0.049
(0.020–5.284) | 0.710 |
|
|
| Tumor number
(single vs. multiple) | 1.704
(0.573–5.068) | 0.338 |
|
|
| Tumor size (≤5 vs.
>5 cm) | 4.603
(1.934–10.953) |
0.001a | 3.070
(1.107–8.512) | 0.031a |
| Differentiation
(high vs. middle and low) | 2.506
(0.526–5.812) | 0.517 |
|
|
| Surgical resection
range (liver lobe vs. segment) | 2.876
(0.967–8.566) |
0.040a |
|
|
| Capsule
invasion | 2.813
(1.090–7.255) |
0.032a |
|
|
| Vascular tumor
thrombus | 6.541
(2.363–18.101) |
0.001a |
|
|
| TNM stage (I+II vs.
IIIa) | 3.354
(1.388–8.102) |
0.007a | 3.116
(0.904–10.740) | 0.047a |
| KIAA1522 (low vs.
high) | 13.294
(3.898–45.344) |
0.001a | 19.413
(5.197–72.500) | 0.001a |
Discussion
Currently, a variety of methods, including
hepatectomy, liver transplantation, radiofrequency ablation,
transarterial chemoembolization and sorafenib are available for the
treatment of patients with HCC at different stages (1,24).
However, the long-term prognosis of HCC remains unsatisfactory.
Thus, elucidating the underlying molecular mechanism of HCC and
identifying essential molecules can help improve the early
prediction of prognosis, and aid in the development of novel
treatment strategies. Examining the expression or the prognostic
value of a specific gene or panel of genes with key biological
functions is one approach to identifying essential molecules.
Additionally, high-throughput technology can identify biomarkers at
the genomic or proteomic level (25,26), and
online expression profile datasets can also be used to search for
potential biomarkers (1). According
to several databases, KIAA1522 expression is elevated in HCC
tissues and is closely associated with the prognosis of HCC, which
prompted its analysis in the present study.
Previous studies have reported that the newly cloned
gene, KIAA1522, is overexpressed in several types of human cancer
(18,27–29).
However, the association between aberrant KIAA1522 expression and
malignant tumor growth remains unclear. Xie et al (18) reported that KIAA1522 is overexpressed
in oesophageal squamous cell carcinoma (ESCC), and the
overexpression of KIAA1522 can enhance the malignant proliferative
capacity and anoikis resistance by activating the ERK signaling
pathway to promote tumor formation and progression. This indicates
that aberrant KIAA1522 expression plays a carcinogenic role in
ESCC. Furthermore, Li et al (28) demonstrated that KIAA1522 is a direct
target of miR-125b-5p in breast cancer and is involved in tumor
cell proliferation, colony formation, cell migration and cell
invasion. Liu et al (29)
indicated that the high KIAA1522 expression can be used as an
independent biomarker for predicting poor survival and platinum
resistance in patients with non-small cell lung cancer. KIAA1522 is
involved in oncogenic KRAS signaling in lung cancer cells and may
be a novel target for lung cancer treatment. These studies
demonstrate that KIAA1522 plays a key role in the proliferation,
metastasis and invasion of various cancer cells. Although KIAA1522
is overexpressed in several tumor tissues, to the best of our
knowledge, its association with HCC remains unknown.
The present study applied bioinformatics technology,
using the Oncomine and OncoLnc databases, to determine the
association between KIAA1522 expression and clinical prognosis. The
results demonstrated that KIAA1522 mRNA expression was
significantly higher in HCC tissues compared with that in adjacent
normal tissues. Furthermore, the high KIAA1522 expression group
exhibited a significantly lower OS time compared with the low
expression group. Subsequently, immunohistochemical staining was
performed to detect KIAA1522 protein expression levels in the 79
HCC and adjacent normal tissue samples, while RT-qPCR was performed
to determine KIAA1522 mRNA expression levels. The results
demonstrated that both KIAA1522 protein and mRNA expression levels
were significantly higher in the HCC tissues compared with those in
the adjacent normal tissues. Taken together, these results indicate
that KIAA1522 is upregulated in HCC at both the molecular and
protein levels.
Clinical data from 79 patients with HCC was analyzed
to determine whether KIAA1522 expression levels were associated
with the relevant clinicopathological characteristics. The results
demonstrated that KIAA1522 protein expression in HCC was not
associated with age, sex, alcoholism, cirrhosis, Child-Pugh
classification, number and size of tumors, degree of
differentiation and clinical stage. However, this may be inaccurate
due to the small sample size used in the present study, thus
further studies with larger sample sizes are required for
verification. The association between KIAA1522 expression and
postoperative prognosis in HCC was assessed during the follow-up
period, which demonstrated that the OS time of patients in the high
KIA1522 expression group was significantly lower compared with that
of patients in the low expression group. No significant difference
was observed for DFS time and KIAA1522 expression, indicating that
KIAA1522 expression was not associated with postoperative
recurrence. This may be due to the small sample size used in the
present study and untimely patient postoperative review, thus
future studies will aim to increase the sample size to verify this
view. The association between KIAA1522 expression and postoperative
prognosis, and the risk factors affecting survival and recurrence
following hepatectomy were also assessed. Univariate and
multivariate Cox regression analyses demonstrated that high
KIAA1522 expression in HCC was significantly and positively
associated with the short-term survival of patients. This suggests
that KIAA1522 may play a key role in the occurrence and development
of HCC and may serve as a potential molecular marker for predicting
the prognosis of patients with HCC. Further investigation on the
association between KIAA1522 expression and postoperative survival
in HCC may provide a novel and effective approach to improve the OS
time of patients with HCC.
Additionally, tumor size and TNM staging were
demonstrated to be independent risk factors affecting OS time. A
previous study combined a variety of liver cancer staging systems
and clinicopathological factors to predict the prognosis of liver
cancer (17). Based on the results
of the present study, it is speculated that late TNM stages lead to
a large tumor burden, which may result in micro-hepatic metastasis
of HCC prior to surgery, thus affecting prognosis. This also
suggests that early detection, diagnosis and surgery can
effectively decrease the clinical stage of preoperative tumors,
which may be an effective means to prevent postoperative recurrence
and improve prognosis. The results of the present study
demonstrated that a tumor size >5 cm may be an independent risk
factor for the OS time of patients with HCC following surgery. This
is consistent with previous finding that indicated that the
prognosis patients with large liver tumors were worse than that of
patients with small liver tumors (30). Zhou et al (31) reported that the survival rate of
patients with HCC, with tumors <5 cm following surgery, is
significantly higher compared with patients with tumors >5 cm.
At the same time, when dissecting the pathological specimens during
liver transplantation, it was demonstrated that the larger the
tumor is, the higher the vascular invasion and the degree of
metastasis. This finding is consistent with the results of the
present study. However, the present study failed to demonstrate a
significant association between KIAA1522 expression levels and TNM
stage and tumor size in the HCC tissues, which may be due to the
small sample size used.
Overall, the results of the present study
demonstrated that KIAA1522 expression was associated with
postoperative prognosis in HCC, and overexpression of KIAA1522 is
associated with a poor prognosis in patients with HCC. The results
of the present study demonstrated that TNM staging, tumor size and
high KIAA1522 expression were all independent risk factors for
postoperative survival, suggesting that KIAA1522 may serve as a
novel molecular marker for predicting the survival of patients with
HCC. Based on bioinformatics analysis, this preliminary small
sample-level study was performed on the postoperative prognosis of
79 patients with HCC in The Affiliated Hospital of Qingdao
University in 2013. A major limitation of the present study is use
of a small sample size, which may have introduced bias. Thus,
future studies will use larger sample sizes, gather more relevant
data and aim to perform an in-depth investigation of the underlying
molecular mechanism of KIAA1522 in HCC.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Key Research and
Development Plan of Shandong Province (grant no. 2018GSF118233),
the Science and Technology Plan of Qingdao City Shinan District
(grant no. 2018-4-018-YY) and the Natural Science Foundation of
Shandong Province (grant no. ZR2012HQ039).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
CS and BH contributed to the study design. MZ, YXi,
JY and ZC acquired the data. YXu, MZ and YXi performed the
experiments. YXu drafted the initial manuscript. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Affiliated Hospital of Qingdao University and
written informed consent was obtained from all patients prior to
the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Llovet JM, Schwartz M and Mazzaferro V:
Resection and liver transplantation for hepatocellular carcinoma.
Semin Liver Dis. 25:181–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grazi GL, Ercolani G, Pierangeli F, Del
Gaudio M, Cescon M, Cavallari A and Mazziotti A: Improved results
of liver resection for hepatocellular carcinoma on cirrhosis give
the procedure added value. Ann Surg. 234:71–78. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng RM, Zong YN, Cao SM and Xu RH:
Current cancer situation in China: Good or bad news from the 2018
Global Cancer Statistics? Cancer Commun (Lond). 39:222019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Desjardins LA: Hepatocellular carcinoma.
Clin J Oncol Nurs. 6:107–108. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Yao M, Dong Z, Zhang Y and Yao D:
Circulating specific biomarkers in diagnosis of hepatocellular
carcinoma and its metastasis monitoring. Tumour Biol. 35:9–20.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Contratto M and Wu J: Targeted therapy or
immunotherapy? Optimal treatment in hepatocellular carcinoma. World
J Gastrointest Oncol. 10:1082018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji F, Fu SJ, Shen SL, Zhang LJ, Cao QH, Li
SQ, Peng BG, Liang LJ and Hua YP: The prognostic value of combined
TGF-β1 and ELF in hepatocellular carcinoma. BMC Cancer. 15:1162015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ng KK, Lo CM, Liu CL, Poon RT, Chan SC and
Fan ST: Survival analysis of patients with transplantable recurrent
hepatocellular carcinoma: Implications for salvage liver
transplant. Arch Surg. 143:68–74. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim DY, Paik YH, Ahn SH, Youn YJ, Choi JW,
Kim JK, Lee KS, Chon CY and Han KH: PIVKA-II is a useful tumor
marker for recurrent hepatocellular carcinoma after surgical
resection. Oncology. 72 (Suppl 1):52–57. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu SJ, Qi CY, Xiao WK, Li SQ, Peng BG and
Liang LJ: Glypican-3 is a potential prognostic biomarker for
hepatocellular carcinoma after curative resection. Surgery.
154:536–544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo Y, Liang X, Lu M, Weng T, Liu Y and Ye
X: Mammalian target of rapamycin as a novel target in the treatment
of hepatocellular carcinoma. Hepatogastroenterology. 57:913–918.
2010.PubMed/NCBI
|
|
16
|
Marrero JA, Feng Z, Wang Y, Nguyen MH,
Befeler AS, Roberts LR, Reddy KR, Harnois D, Llovet JM, Normolle D,
et al: Alpha-fetoprotein, des-gamma carboxyprothrombin, and
lectin-bound alpha-fetoprotein in early hepatocellular carcinoma.
Gastroenterology. 137:110–118. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cristea CG, Gheonea IA, Săndulescu LD,
Gheonea DI, Ciurea T and Purcarea MR: Considerations regarding
current diagnosis and prognosis of hepatocellular carcinoma. J Med
Life. 8:120–128. 2015.PubMed/NCBI
|
|
18
|
Xie ZH, Yu J, Shang L, Zhu YQ, Hao JJ, Cai
Y, Xu X, Zhang Y and Wang MR: KIAA1522 overexpression promotes
tumorigenicity and metastasis of esophageal cancer cells through
potentiating the ERK activity. Onco Targets Ther. 10:3743–3754.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th Edition of theAJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maiolino P, Restucci B, Papparella S,
Paciello O and De Vico G: Correlation of nuclear morphometric
features with animal and Human World Health Organization
international histological classifications of canine spontaneous
seminomas. Vet Pathol. 41:608–611. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Durand FO and Valla D: Assessment of the
prognosis of cirrhosis: Child-Pugh versus MELD. J Hepatol. 42
(Suppl):S100–S107. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Method. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Franssen B, Alshebeeb K, Tabrizian P,
Marti J, Pierobon ES, Lubezky N, Roayaie S, Florman S and Schwartz
ME: Differences in surgical outcomes between hepatitis B- and
hepatitis C-related hepatocellular carcinoma: A retrospective
analysis of a single North American center. Ann Surg. 260:650–658.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu CQ, Ding K, Strumpf D, Weir BA,
Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et
al: Prognostic and predictive gene signature for adjuvant
chemotherapy in resected non-small-cell lung cancer. J Clin Oncol.
28:4417–4424. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu CQ, Pintilie M, John T, Strumpf D,
Shepherd FA, Der SD, Jurisica I and Tsao MS: Understanding
prognostic gene expression signatures in lung cancer. Clin Lung
Cancer. 10:331–340. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagase T, Kikuno R, Ishikawa K, Hirosawa M
and Ohara O: Prediction of the coding sequences of unidentified
human genes. XVII. The Complete Sequences of 100 New cDNA clones
from brain which code for large proteins in vitro. DNA Res.
7:143–150. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Wang Y, Fan H, Zhang Z and Li N:
miR-125b-5p inhibits breast cancer cell proliferation, migration
and invasion by targeting KIAA1522. Biochem Biophys Res Commun.
504:277–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu YZ, Yang H, Cao J, Jiang YY, Hao JJ,
Xu X, Cai Y and Wang MR: KIAA1522 is a novel prognostic biomarker
in patients with non-small cell lung cancer. Sci Rep. 6:247862016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takamoto T, Sano K, Hashimoto T, Ichida A,
Shimada K, Maruyama Y and Makuuchi M: Practical contribution of
virtual hepatectomy for colorectal liver metastases: A
propensity-matched analysis of clinical outcome. J Gastrointest
Surg. 22:2037–2044. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou XD, Tang ZY, Ma ZC, Fan J, Wu ZQ, Qin
LX, Zhou J, Yu Y, Sun HC and Qiu SJ: Twenty-year survivors after
resection for hepatocellular carcinoma-analysis of 53 cases. J
Cancer Res Clin Oncol. 135:1067–1072. View Article : Google Scholar : PubMed/NCBI
|