Introduction
Esophageal squamous cell carcinoma (ESCC), which is
one of the most common types of cancer, was the sixth leading cause
of cancer-associated mortality in China in 2016 (1). In 2012, an estimated 455,800 new
esophageal cancer cases and 400,200 deaths were reported (2). Although surgery and radiation therapy
are effective therapeutic strategies for certain tumors diagnosed
at an early stage, a number of patients eventually progress to
advanced stages of cancer (3). Based
on previous epidemiologic studies, it is widely accepted that
genetics serve a crucial role in the development and progression of
ESCC (4,5); however, the underlying mechanisms
remain unclear. It is therefore important to fully understand the
development and progression of ESCC, in order to identify novel
therapeutic targets.
The proline-rich protein 11 (PRR11) gene is located
on chromosome 17q22 and contains a bivalent nuclear localization
signal, two proline-rich regions and a zinc finger domains
(6). In recent years, PRR11 has been
reported to serve as a candidate oncogene in various types of
cancer, including pancreatic cancer, breast cancer, non-small cell
lung cancer, hepatocellular carcinoma and ovarian carcinoma
(7–9). PRR11 and its upstream adjacent gene,
spindle and kinetochore associated 2 (SKA2), are a classic
head-to-head gene pair, driven by a prototypical and bidirectional
promoter (10). SKA2, which is
involved in the formation of the Ska complex, is involved in the
maintenance of the mitotic mid-plateau and shut-down of the spindle
checkpoint (11–14). Currently, studies on PRR11 and SKA2
mainly focus on lung and breast cancer (7,8,10); however, the role of PRR11 and SKA2 in
ESCC remains unclear.
The present study evaluated the importance of PRR11
and SKA2 in the progression of ESCC, and demonstrated that the
expression level of these genes was highly expressed in human ESCC
tissues. Furthermore, the present study revealed that PRR11 and
SKA2 serve important roles in the proliferation and migratory and
invasive capacities of ESCC cells in vitro. In addition, the
underlying mechanism of PRR11 and SKA2 in the progression of ESCC
was investigated.
Materials and methods
ESCC tissue samples
A total of 30 pairs of ESCC and adjacent normal
tissues were obtained from patients following resection surgery at
the Second Affiliated Hospital of Zhengzhou University between
January 2014 and December 2017. The adjacent normal tissue <1-cm
away from the tumor tissue. The median age was 52 (41–61) years
old, and there were 22 males and 8 females. Some clinical
parameters, such as sex, age, history of drinking and smoking,
tumor site, TNM staging (the 8th edition) (15), and tumor differentiation. All
patients provided informed signed consent. The specimens were
immediately stored at −80°C until RNA extraction. The present study
was approved by the Ethics Committee of the Second Affiliated
Hospital of Zhengzhou University.
RNA preparation and reverse
transcription-quantitative polymerase chain reaction analysis
Total RNA was extracted from the 30 paired ESCC
tumor tissues and adjacent normal tissues using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions, and 2 µg RNA was reverse
transcribed to cDNA using the Reverse Transcription System (A3500,
Promega Corporation) according to the manufacturers' instructions.
The mRNA level was detected using a PikoReal 96 Real-Time PCR
system (Thermo Fisher Scientific, Inc.) and SYBR® Premix
Ex Taq (Takara Bio, Inc.). The thermal cycling conditions were as
follows: 40 cycles at 95°C for 20 sec, 60°C for 30 sec, and 72°C
for 30 sec, and one cycle of 72°C for 10 min. The sequences of the
primers were as follows: PRR11, forward
5′-GAAGCTGGCTAACATCATCCTG-3′, reverse 5′-CTCTGGGTTATGCAGTTCTGG-3′;
SKA2, forward 5′-GGAACTGATGTTCCAGAAAGCTG-3′, reverse
5′-AGCTCCAGGTCTGTTTGCTT-3′; and GAPDH, forward
5′-ATGACCCCTTCATTGACCTCA-3′ and reverse
5′-GAGATGATGACCCTTTTGGCT-3′. The relative mRNA expression level in
each sample was calculated using the comparative expression level
2−∆∆Cq method (16).
Cell culture
The esophageal squamous cell carcinoma cell lines
EC9706 and EC109, and 293T cell line were purchased from The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM)
supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin, and
10% (v/v) fetal bovine serum (FBS; Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified atmosphere containing 5%
CO2.
Plasmids and cell transfection
A PRR11 and SKA2 coding sequence was constructed and
inserted into a pcDNA3.1-Myc plasmid (Invitrogen; Thermo Fisher
Scientific, Inc.) to generate the PRR11 and SKA2 overexpression
vector. Lentiviral supernatants were produced using the Lenti-X HTX
packaging system (Clontech Laboratories, Inc.) according to the
manufacturer's protocol and used for transfection of EC109 cells.
For negative controls, EC109 cell were transfected with
supernatants from empty vectors. A total of 30 µl lentiviral
particles (5×107 UT/ml) were suspended in DMEM, and
incubated with the EC109 cells (1×106) in a 6-well plate
for 12 h until they reached 70% confluence. Next, transfected cells
were screened by green fluorescence using flow cytometry (IX-71;
Olympus Corporation). Lentiviral short hairpin (sh)RNA targeting
PRR11 and SKA2 were designed using software provided by Qiagen,
Inc. and inserted into a pLKO.1-TRC vector (Han Heng Biotechnology
Co., Ltd.). The lentivirus was packaged in 293T cells and collected
following centrifugation at 4°C and 1,000 × g for 2 h. A total of
50 µl lentiviral particles (1×108 UT/ml) were suspended
in DMEM, and incubated with the EC9706 cells (1×106) in
a 6-well plate for 8 h until they reached 60% confluence. Positive
EC9706 cells were selected by puromycin (4 µg/ml) (Han Heng
Biotechnology Co., Ltd.) for at least 3 days or sorted by flow
cytometry. Subsequent experiments were conducted at 48 h
post-transfection. The expression of PRR11 and SKA2 in the
resistant clones was further confirmed by western blotting. The
sequences of the shRNAs were as follows: shPRR11, forward:
5′-CCGGCCAGAGTTTAGAAGTATTGAACTCGAGTTCAATACTTCTAAACTCTGGTTTTTG-3′
and reverse:
5′-AATTCAAAAACCAGAGTTTAGAAGTATTGAACTCGAGTTCAATACTTCTAAACTCTGG-3′;
shSKA2, forward:
5′-CCGGCAAACTTTGTATGCCCGCTTTCTCGAGAAAGCGGGCATACAAAGTTGTTTTTG-3′ and
revere
5′-AATTCAAAAACAAACTTTGTATGCCCGCTTTCTCGAGAAAGCGGGCATACAAAGTTTG-3′;
and shcontrol, forward: 5′-TGACAAGTGGAACCAGAT-3′ and reverse:
5′-TGTCCCCACTCACGAAGG-3′.
Western blotting
EC9706 and EC109 cells were lysed in RIPA lysis
buffer (Thermo Fisher Scientific, Inc.) for at least 30 min on ice.
The lysates were centrifuged at 10,000 × g for 20 min at 4°C.
Protein concentration was determined using Bradford reagent
(Sigma-Aldrich; Merck KGaA) according to the manufacturer's
instructions. Proteins (10 µg/lane) were separated by 10% SDS-PAGE
and transferred onto polyvinylidene fluoride membranes. Then,
membranes were blocked with 5% fat-free milk for 1 h at room
temperature. Next, membranes were incubated with primary antibodies
at 4°C overnight. The primary antibodies were as follows: PRR11
(1:500; cat. no. 3220; Cell Signaling Technology, Inc.), SKA2
(1:500; cat. no. 2419; Cell Signaling Technology, Inc.),
phosphorylated (p) and total Akt (1:1,000; cat. no. 10176-2-AP;
ProteinTech Group, Inc.), proliferating cell nuclear antigen (PCNA;
1:1,000; cat. no. 10205-2-AP; ProteinTech Group, Inc.), Snail
(1:1,000; cat. no. 13099-1-AP; ProteinTech Group, Inc.), Cyclin D1
(1:1,000; cat. no. 60186-1-Ig; ProteinTech Group, Inc.), N-cadherin
(1:1,000; cat. no. 22018-1-AP; ProteinTech Group, Inc.) and GAPDH
(1:1,000; cat. no. 60004-1-Ig; ProteinTech Group, Inc.). Snail and
N-cadherin are markers of EMT, and cyclin D1 is a marker of the
cell cycle (17,18). Membranes were then incubated with
anti-rabbit horseradish peroxidase-conjugated secondary antibody
(cat. no. 7074; 1:2,000; Sangon Biotech Co., Ltd.) for 2 h at room
temperature. The immunoreactive protein bands were visualized using
an enhanced chemiluminescence kit (Pierce; Thermo Fisher
Scientific, Inc.) and a Gel Dox XR system (Bio-Rad Laboratories,
Inc.).
Crystal violet assay
Control and transfected cells were seeded into
6-well plates at a density of 1,000 cells/well. Cells were cultured
in DMEM with 10% FBS and medium was changed every three days. After
two weeks of culture, the culture medium was removed and cells were
stained with crystal violet (1 ml 0.5% crystal violet solution in
20% methanol) for 10 min at room temperature. Cells were washed
with PBS and images were captured using a digital camera. The
optical density (OD) value was measured at 600 nm with a microplate
reader.
Cell proliferation assay
EC9706 and EC109 cells were seeded in 96-well plates
at a density of 1,000 per well in triplicate. MTT solution (20 µl;
5 mg/ml) (APExBIO Technology LLC) was added to the medium and
incubated at 37°C for 4 h. The medium was removed, and 200 µl DMSO
was added to dissolve the formazan crystals. The absorbance was
measured using an automatic microplate reader at a wavelength of
490 nm.
Boyden chamber assay
A total of 2×105 EC9706 and EC109 cells
were loaded into the upper chamber of a 10-well Boyden Chamber
(Neuro Probe, Gaithersburg, MD, USA) with a polycarbonate membrane
of 8 µm pore sizein 200 µl of medium containing 1% FBS. Medium (250
µl) containing 10% FBS was added to the bottom well of the chamber.
Cells were incubated for at 37°C for 6 h. Cells that have migrated
through the pores into the lower chamber were stained with
hematoxylin and eosin for 10 min at room temperature. Cells were
then then photographed using an inverted microscope (OLYMPUS) at
magnification, ×400. Cells were counted from four randomly selected
fields by ImageJ software (version 1.8.0, National Institutes of
Health). Experiments were repeated in triplicate.
Transwell assay
ESCC cell invasion was examined using a Transwell
assay with polyethylene terephthalate membranes (24-well inserts;
8.0 µm; Corning Inc.). Cell suspension (150 µl) containing
2×105 cells was loaded into the upper well that was
precoated with 20% Matrigel (BD Biosciences) at 37°C for 3 h. DMEM
(500 µl) containing 10% FBS was added to the bottom of the well.
After 48 h, cells that have invaded the bottom of the membrane were
stained with 0.1% crystal violet for 10 min at room temperature and
then photographed using an inverted light microscope (Olympus
Corporation) at 400× magnification Cells were counted from four
randomly selected fields by ImageJ software (version 1.8.0,
National Institutes of Health). Experiments were conducted in three
independent experiments.
Statistical analysis
Data are presented as the mean ± standard deviation
and statistical analyses were performed using GraphPad Prism
software (version 5; GraphPad Software, Inc.). PRR11 and SKA2 mRNA
levels between tumor and adjacent normal tissue groups were
analyzed using the paired Student's t-test. PRR11 and SKA2 mRNA
levels in the cell lines were determined using an unpaired
Student's t-test. Comparisons of multiple groups were analyzed
using the ANOVA followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
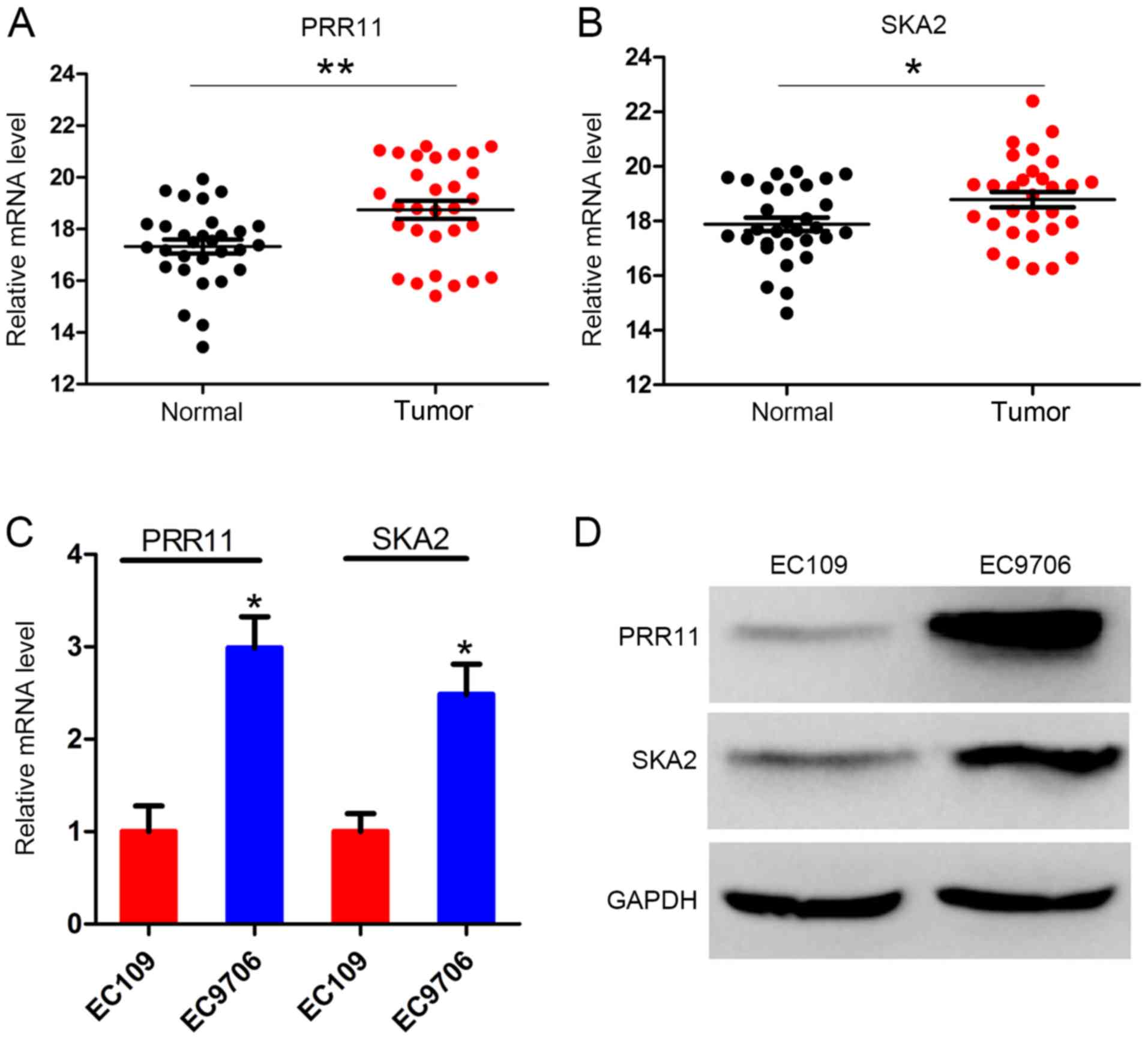
PRR11 and SKA2 are upregulated in ESCC
tissues
The primary focus of the present study was the
expression level of PRR11 and SKA2 in the 30 pairs of ESCC and
adjacent normal tissues. The clinicopathological characteristics of
the 30 patients are presented in Table
I. The results demonstrated that PRR11 expression level was
significantly increased in ESCC tissues compared with the adjacent
normal tissues (P<0.001; Fig.
1A). Similarly, SKA2 expression level was significantly
increased in tumor tissues compared with normal tissues (P<0.05;
Fig. 1B). Furthermore, the mRNA and
protein levels of PRR11 and SKA2 in the two ESCC cell lines EC109
and EC9706 were assessed. The results demonstrated that the mRNA
level of both PRR11 and SKA2 were significantly higher in EC9706
cells compared with EC109 cells. The protein expression of both
PRR11 and SKA2 was also higher in EC9706 cells (Fig. 1C and D).
 | Table I.Clinicopathological characteristics of
the 30 patients included in the present study. |
Table I.
Clinicopathological characteristics of
the 30 patients included in the present study.
| Variables | Number (%) |
|---|
| Sex |
|
| Male | 23 (76.7) |
|
Female | 7 (23.3) |
| Age, years |
|
|
<60 | 19 (63.3) |
| ≥60 | 11 (36.7) |
| Smoking |
|
| Yes | 17 (56.7) |
| No | 13 (43.3) |
| Drinking |
|
| Yes | 12 (40.0) |
| No | 18 (60.0) |
| Tumor site |
|
|
Cervical | 2 (6.7) |
|
Upper | 9 (30.0) |
|
Middle | 10 (33.3) |
|
Lower | 9 (30.0) |
| TNM stage |
|
| IA | 5 (16.7) |
| IB | 9 (30.0) |
| IIA | 7 (23.3) |
| IIB | 5 (16.7) |
| IIIA | 2 (6.7) |
| IIIB | 2 (6.7) |
| Differentiation |
|
| Well | 6 (20.0) |
|
Moderate | 17 (56.7) |
| Poor | 7 (23.3) |
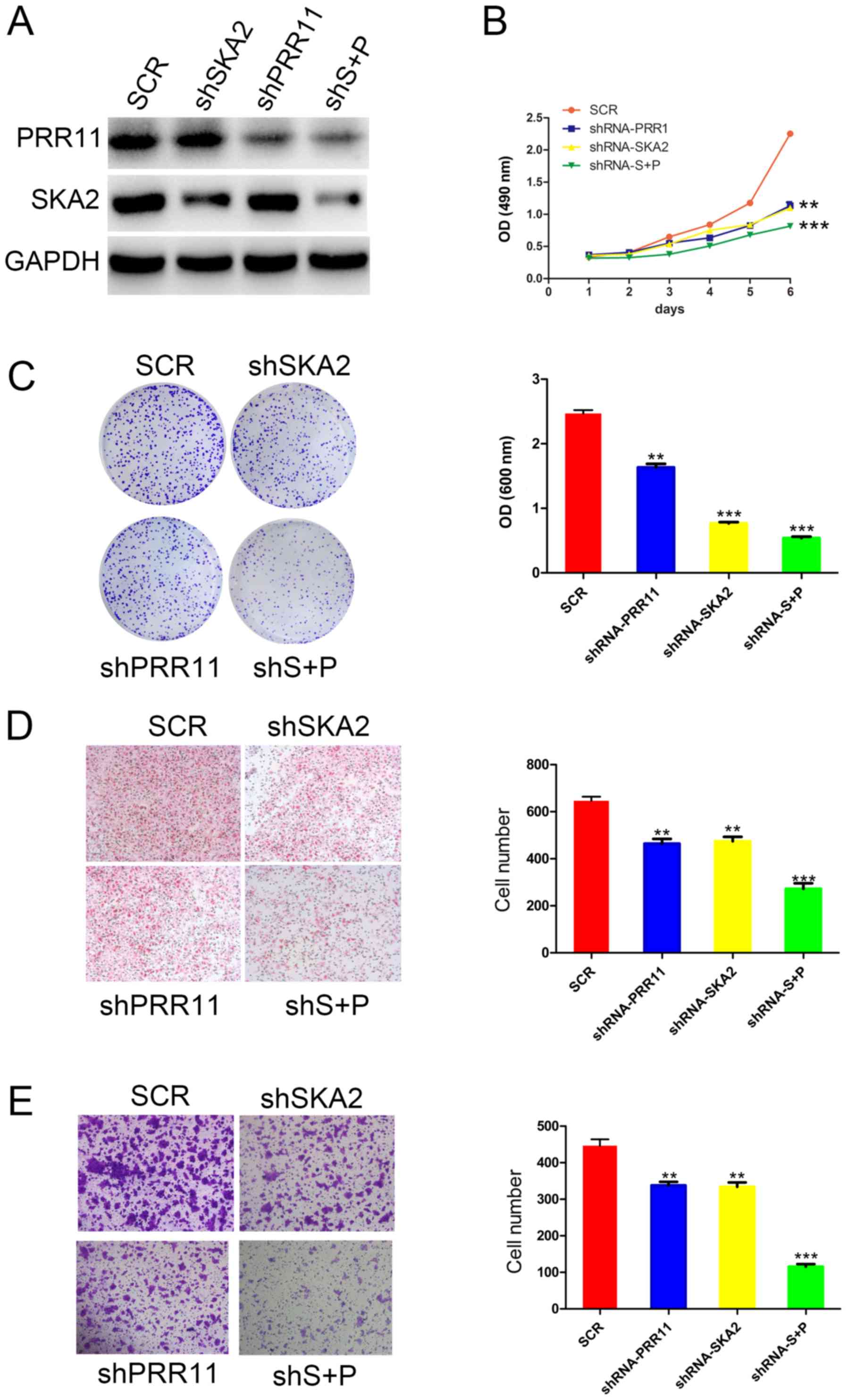
PRR11 and/or SKA2 knockdown inhibits
the proliferation, migration, and invasion of ESCC
To identify the function of the PRR11 and SKA2 gene
pair in ESCC cells, specific shRNA targeting PRR11 and/or SKA2 were
used in EC9706 cells. The efficiency of these knockdowns was
confirmed by western blotting (Fig.
2A). Furthermore, the results from MTT and crystal assays
demonstrated that transfection with shPRR11 or SKA2 significantly
reduced EC9706 cell proliferation compared with untransfected
cells. In addition, double transfection induced a more significant
decrease in cell proliferation compared with single transfections
(P<0.01 and P<0.001; Fig. 2B and
C). In addition, as presented in Fig. 2D, the results from Boyden chamber
assay demonstrated that the migratory ability of EC9706 cells was
significantly decreased following PRR11 or SKA2 knockdown compared
with untransfected cells (P<0.01 and P<0.001, respectively;
Fig. 2D). Cell migratory capacity
was also significantly more decreased following double knockdown of
PRR11 and SKA2 compared with single transfections (P<0.001;
Fig. 2C). Furthermore, the results
from transwell assay indicated that the invasive capacity of EC9706
cells was significantly reduced following PRR11 and/or SKA2
knockdown (P<0.01 and P<0.001; Fig. 2E).
PRR11 and/or SKA2 overexpression
promotes the proliferation, migration, and invasion of ESCC
PRR11 and/or SKA2 were overexpressed in EC109 cells
by transfection. The successful overexpression of PRR11 and/or SKA2
was confirmed by western blotting (Fig.
3A). The results from MTT and crystal violet assays
demonstrated a significantly increased cell proliferation following
PRR11 or SKA2 overexpression compared with untransfected cells. In
addition, the double transfection induced a more significant
increase in cell proliferation compared with single transfections
(P<0.01 and P<0.001; Fig. 3B and
C). Furthermore, as presented in Fig. 3D, the results from Boyden chamber
assay demonstrated that the migratory ability of EC109 cells was
significantly increased following PRR11 or SKA2 overexpression
compared with untransfected cells (P<0.01; Fig. 3D). Cell migratory capacity was more
significantly increased following overexpression of PRR11 and SKA2
compared with single transfections (P<0.001; Fig. 3D). In addition, the results from
transwell assay reported that overexpression of the gene pair
significantly promoted the invasive ability of EC109 cells
(P<0.01 and P<0.001; Fig.
3E).
PRR11 and SKA2 gene pair activates AKT
and epithelial- mesenchymal transition (EMT) signaling
pathways
In order to explore the underlying mechanism of the
gene pair PRR11 and SKA2 on ESCC progression, the protein
expression of p-AKT, total AKT, Snial and N-cadherin of EMT, and
Cyclin D1 of cell cycle signal in EC9706 and EC109 cells by western
blotting. The results demonstrated that expression levels of p-AKT,
Snail and N-cadherin were reduced following PRR11 and SKA2
knockdown in EC9706 cells, and double transfection induced a more
notable decrease in the expressions of these proteins compared with
single transfections (Fig. 4A).
Opposite effects were observed in EC109 cells overexpressing PRR11
and SKA2 (Fig. 4B). In addition,
PRR11 and SKA2 overexpression or knockdown had no effect on Cyclin
D1 expression. Furthermore, PRR11 and SKA2 overexpression increased
the expression of the proliferation marker PCNA, whereas their
knockdown decreased PCNA expression (Fig. 4A and B).
Discussion
Over the past few decades, the prognosis of ESCC has
been relatively poor in China, with the 5-year overall survival
rate <20% and most patients died within 1 year of diagnosis
(2). In addition, this disease lacks
sensitive and specific early diagnosis method. Surgical and
radiation therapies are the main treatments for ESCC; however,
numerous patients eventually develop advanced stages of ESCC
(4). The present study aimed
therefore to identify potential intervention targets for ESCC.
Recent studies reported that the gene pair of PRR11
and SKA2 is involved in tumorigenesis and cancer progression. Zhu
et al (9) reported that PRR11
overexpression facilitates ovarian carcinoma cell proliferation,
migration, and invasion through activation of the
PI3K/AKT/β-Catenin pathway. Furthermore, it was reported that the
gene pair PRR11 and SKA2 is negatively regulated by p53 through
nuclear factor Y in lung cancer cells (10). Also, PRR11 silencing leads to cell
cycle arrest, suppresses colony formation, decreases cell
proliferation and inhibits tumorigenic potential of lung cancer
cells (19). In addition, the PRR11
and SKA2 gene pair has been hypothesized as a potential new target
for the diagnosis and treatment of lung cancer (20). It has reported that overexpression of
PRR11 could promote breast cancer progression by activating EMT
(7). Qiao et al (6) demonstrated that proline-rich protein 11
silencing inhibits hepatocellular carcinoma growth and
epithelial-mesenchymal transition through β-catenin signaling.
The present study demonstrated that PRR11 and SKA2
mRNA levels were significantly increased in ESCC tissues compared
with adjacent normal tissues. Furthermore, cell proliferation,
migratory and invasive capacities were significantly increased
following PRR11 and SKA2 overexpression. In addition, PRR11 and
SKA2 knockdown inhibited the proliferation, invasive and migratory
capacities of ESCC cells. Subsequently, in order to investigate the
underlying mechanism, this study demonstrated that PRR11 and SKA2
overexpression significantly activated the AKT signaling pathway
and certain markers, including Snail and N-cadherin of EMT. AKT
signaling pathway activation is implicated in the development of a
numerous human cancers, including ESCC (21–23).
Furthermore, EMT represents a critical event in the transition from
early to invasive carcinomas, and it was demonstrated that
N-cadherin upregulation is associated with poor prognosis and lower
survival in patients with cancer (24–26).
These findings are consistent with the results from the present
study. To the best of our knowledge, this study was the first to
demonstrate the involvement of PRRl1 and SKA2 in the progression of
ESCC.
In summary, this study demonstrated that the gene
pair PRRl1 and SKA2 may serve a crucial role in the proliferation,
migratory and invasive abilities of ESCC cells. These results
provided a better understanding of the underlying mechanisms of
PRRl1 and SKA2 in ESCC tumor development, and PRRl1 and SKA2 may be
considered as potential targets for the diagnosis and/or treatment
of ESCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Young Core
Instructor Foundation from the Education Commission of Henan
Province (grant no. 2016GGJS-261) and the Science and Technology
Project of Henan Province (grant no. 192102310103).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
JC and HY conducted the experiments. ZN conducted
the experiments and analyzed the data. CK designed the research. HZ
and RP analyzed the data. JC, RP and CK drafted the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Second Affiliated Hospital of Zhengzhou
University. All patients and provided signed informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng YF, Chen HS, Wu SC, Chen HC, Hung
WH, Lin CH and Wang BY: Esophageal squamous cell carcinoma and
prognosis in Taiwan. Cancer Med. 7:4193–4201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ajani JA, D'Amico TA, Almhanna K, Bentrem
DJ, Chao J, Das P, Denlinger CS, Fanta P, Farjah F, Fuchs CS, et
al: Gastric cancer, version 3.2016, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 14:1286–1312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen J, Kwong DL, Cao T, Hu Q, Zhang L,
Ming X, Chen J, Fu L and Guan X: Esophageal squamous cell carcinoma
(ESCC): Advance in genomics and molecular genetics. Dis Esophagus.
28:84–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Walker RC and Underwood TJ: Molecular
pathways in the development and treatment of oesophageal cancer.
Best Pract Res Clin Gastroenterol 36–37. 9–15. 2018. View Article : Google Scholar
|
|
6
|
Qiao W, Wang H, Zhang X and Luo K:
Proline-rich protein 11 silencing inhibits hepatocellular carcinoma
growth and epithelial-mesenchymal transition through β-catenin
signaling. Gene. 681:7–14. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Zhang C, Mai L, Niu Y, Wang Y and
Bu Y: PRR11 and SKA2 gene pair is overexpressed and regulated by
p53 in breast cancer. BMB Rep. 52:157–162. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang L, Lei Y, Zhang Y, Li Y, Bu Y, Song
F and Zhang C: Silencing of PRR11 suppresses cell proliferation and
induces autophagy in NSCLC cells. Genes Dis. 5:158–166. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu J, Hu H, Wang J, Yang Y and Yi P:
PRR11 overexpression facilitates ovarian carcinoma cell
proliferation, migration, and invasion through activation of the
PI3K/AKT/β-catenin pathway. Cell Physiol Biochem. 49:696–705. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Zhang Y, Zhang C, Weng H, Li Y,
Cai W, Xie M, Long Y, Ai Q, Liu Z, et al: The gene pair PRR11 and
SKA2 shares a NF-Y-regulated bidirectional promoter and contributes
to lung cancer development. Biochim Biophys Acta. 1849:1133–1144.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chan YW, Jeyaprakash AA, Nigg EA and
Santamaria A: Aurora B controls kinetochore-microtubule attachments
by inhibiting Ska complex-KMN network interaction. J Cell Biol.
196:563–571. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Q, Sivakumar S, Chen Y, Gao H, Yang
L, Yuan Z, Yu H and Liu H: Ska3 phosphorylated by Cdk1 binds Ndc80
and recruits Ska to kinetochores to promote mitotic progression.
Curr Biol. 27:1477–1484.e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeyaprakash AA, Conti E, Santamaria A,
Jayachandran U, Chan YW, Benda C and Nigg EA: Structural and
functional organization of the Ska complex, a key component of the
kinetochore-microtubule interface. Mol Cell. 46:274–286. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hanisch A, Silljé HH and Nigg EA: Timely
anaphase onset requires a novel spindle and kinetochore complex
comprising Ska1 and Ska2. EMBO J. 25:5504–5515. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vaidya T, Desai S and Mahajan A: (8th).
AJCC and imaging TNM: Time to break-in and assert in the staging
process! Indian J Cancer. 56:271–273. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang J, Antin P, Berx G, Blanpain C,
Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori
G, et al: Guidelines and definitions for research on
epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. Apr
16–2020.(Epub ahead of print). View Article : Google Scholar
|
|
18
|
Leal-Esteban LC and Fajas L: Cell cycle
regulators in cancer cell metabolism. Biochim Biophys Acta Mol
Basis Dis. 1866:1657152020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ji Y, Xie M, Lan H, Zhang Y, Long Y, Weng
H, Li D, Cai W, Zhu H, Niu Y, et al: PRR11 is a novel gene
implicated in cell cycle progression and lung cancer. Int J Biochem
Cell Biol. 45:645–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang C, Zhang Y, Li Y, Zhu H, Wang Y, Cai
W, Zhu J, Ozaki T and Bu Y: PRR11 regulates late-S to G2/M phase
progression and induces premature chromatin condensation (PCC).
Biochem Biophys Res Commun. 458:501–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Song M, Wang P, Zhao R, Chen H,
Zhang M, Shi Y, Liu K, Liu F, Yang R, et al: Targeted therapy of
the AKT kinase inhibits esophageal squamous cell carcinoma growth
in vitro and in vivo. Int J Cancer. 145:1007–1019. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Chen Y, Zhan C, Zhu J, Weng S, Dong
L, Liu T and Shen X: Glypican-1 promotes tumorigenesis by
regulating the PTEN/Akt/β-catenin signaling pathway in esophageal
squamous cell carcinoma. Dig Dis Sci. 64:1493–1502. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Fa X and Zhang Q: MicroRNA-206
exerts anti-oncogenic functions in esophageal squamous cell
carcinoma by suppressing the c-Met/AKT/mTOR pathway. Mol Med Rep.
19:1491–1500. 2019.PubMed/NCBI
|
|
24
|
Rosanò L, Spinella F, Di Castro V, Nicotra
MR, Dedhar S, de Herreros AG, Natali PG and Bagnato A: Endothelin-1
promotes epithelial-to-mesenchymal transition in human ovarian
cancer cells. Cancer Res. 65:11649–11657. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barberà MJ, Puig I, Dominguez D,
Julien-Grille S, Guaita- Esteruelas S, Peiro S, Baulida J, Francí
C, Dedhar S, Larue L and García de Herreros A: Regulation of Snail
transcription during epithelial to mesenchymal transition of tumor
cells. Oncogene. 23:7345–7354. 2004. View Article : Google Scholar : PubMed/NCBI
|