Introduction
Cervical cancer is one of the most common cancer
types in women worldwide (1). In
developing countries, cervical cancer is a major health problem. In
Thailand, cervical cancer is one of five cancer types leading to
the death of 7 Thai women per day (2). Several studies have identified risk
factors for cervical cancer. Human papillomavirus (HPV) infection
is the most important risk factor for cervical cancer (3). HPV can be divided into low-risk and
high-risk groups. High-risk HPV types, including HPV16, 18 and 56,
are those with high oncogenic potential that can be the cause of
neoplastic transformation (4). The
mechanism by which high-risk HPV leads to cervical cancer is still
under investigation. The role of E6 and E7 in p53 and pRB
degradation, respectively, is well known in cancer transformation
(5). The role of E6 and E7 proteins
in tumorigenesis is still being explored. Recent studies have
illustrated that not only the degradation of p53 and pRB, but also
the function of these two oncoproteins, are involved in the
promoter methylation of tumor suppressor genes, resulting in
tumorigenesis (6,7). Our previous study found that cervical
cancer tissues infected with the integrated form of either HPV 16
or 18 can exhibit methylation at the cyclin A1 (CCNA1)
promoter (8). HPV is composed of 8
genes: L1, L2, E1, E2, E4, E5, E6 and E7. The
integrated form of HPV exhibits overexpression of E6 and
E7 because of E2 disruption. E6 and E7 are
oncoproteins that cause cervical cancer and head and neck cancer
(9).
There is much evidence showing the association
between HPV infection and promoter methylation in cancer. A study
by Sator et al (10) showed a
higher level of promoter methylation and lower gene expression
levels of seventy-five genes including IRS1, GNA11, GNAI2, EREG,
CCNA1, RGS4, and PKIG resulting from HPV infection in
head and neck cancer. A study by Lechner et al (11) showed increased mRNA expression of
both DNA methyltransferase 3α (Dnmt3a) and DNA
methyltransferase 1 (Dnmt1) in HPV-positive head and neck
cancer cell lines. Moreover, they observed that promoter
methylation increased in E6 and E7-transfected head and neck cancer
cell lines (10). A study by
Kitkumthorn et al (12)
demonstrated that CCNA1 promoter methylation was detected in
cervical intraepithelial neoplasia (CIN) grade III and invasive
cancer related to HPV.
Promoter methylation of genes is a good biomarker
for identifying women who are at risk of cervical cancer
development. CCNA1 promoter methylation is an efficient
biomarker that can be diagnosed from precancerous stages to
invasive cervical cancer (13). Our
previous study found that HPV16-E7 can induce CCNA1 promoter
methylation by forming a complex with Dnmt1 at the CCNA1
promoter (14). Therefore, there is
a need to identify genes in which HPV can induce promoter
methylation, in addition to CCNA1. Herein, not only HPV-E7
but also HPV-E6 were included for the assessment of their
methylation ability. The promoter methylation and expression of two
tumor suppressor genes, cell adhesion molecule 1 (CADM1) and
death associated protein kinase 1 (DAPK1), were then
observed in E6 and E7-transfected cervical cancer cell lines: C33a,
which is HPV-negative; and SiHa, which is HPV 16-positive. The
studies of Steenbergen et al and Banzai et al found
CADM1 and DAPK1 promoter methylation in HPV-related
cervical cancer, respectively (15,16).
CADM1 is involved in cell adhesion, while DAPK1 is
involved in apoptosis (17).
Promoter methylation of these genes suppresses their expression,
leading to carcinogenesis. The aim of the present study was to
investigate promoter methylation of CADM1 and DAPK1
in the cells with HPV-E6 or E7 transfection together with the
mechanism by which HPV-E7 induced promoter methylation of the
genes.
Materials and methods
Cell culture
The SiHa (HPV 16-positive) cell line was kindly
provided by Dr.Silvio Gutkind (Moores Cancer Center, UCSD, USA)
with proof that there was no contamination, and the C33a
(HPV-negative) cell line was purchased from the American Type
Culture Collection (HTB-31TM; lot no. 63596879). The cells were
grown and maintained in DMEM (Sigma-Aldrich; Merck KGaA)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 1% antibiotic-antimycotic (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in an atmosphere of 5% CO2.
Recombinant plasmid
The E6 recombinant plasmid was inserted into
a PCDNA3 vector, which was provided by Assoc. Prof. Dr. Andrew
Yeudal (Augusta University, USA). The E7 recombinant plasmid
was inserted into PCDNA3.1 myc-his, which was constructed as per a
previous study (13). Both
recombinant plasmids were sent for sequencing to confirm that the
sequences were correct.
Transfection
E6 and E7 recombinant plasmids were
transfected into C33a cells. C33a cells were seeded into 6-well
plates at 3×105 cells/ml and incubated overnight. Next,
2 µg E6 or E7 recombinant plasmid and pcDNA
3.1/myc-his (PC) empty plasmid (Invitrogen; Thermo Fisher
Scientific, Inc.) were transfected using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. After 72 h, transfected cells were
collected to study E6- and E7-mediated CADM1 and
DAPK1 promoter methylation, and CADM1 and
DAPK1 mRNA expression. Moreover, cells transfected with E7
were harvested for chromatin immunoprecipitation. The transfection
was performed in triplicate for all experiments.
Isolation of DNA
SiHa, C33a and C33a cells transfected with E6
or E7 were subjected to DNA extraction. Briefly, cells were
digested with lysis buffer II containing SDS (Sigma-Aldrich; Merck
KGaA) and proteinase K (USB) at 50°C overnight. Phenol/chloroform
extraction and ethanol precipitation were then carried out as
previously described (14).
Preparation of RNA
SiHa, C33a and C33a cells transfected with E6
or E7 and empty vector were subjected to RNA extraction.
Total RNA was extracted using the TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Then, 5 µg total RNA
from each sample was synthesized to cDNA using the RevertAid
first-strand cDNA synthesis kit, according to the manufacturer's
specifications (Thermo Fisher Scientific, Inc.).
Sodium bisulfite treatment and
methylation-specific PCR
A total of 750 ng DNAsample was subjected to
bisulfite treatment using the EZ DNA Methylation-Gold kit (Zymo
Research Corp.), according to the protocol provided by the
manufacturer. The eluted DNA was subsequently used to carry out
methylation-specific PCR using 0.3 µM methylated and unmethylated
specific primers (Table I). The DNA
was initially denatured for 15 min at 95°C, followed by 35 cycles
of 1 min at 95°C, 1 min at 60°C for CADM1 and 53°C for
DAPK1, and 1 min at 72°C, and 72°C for 7 min. Then, 10 µl
PCR products were observed by gel electrophoresis on 8% acrylamide
gels and stained with SYBR (Lonza Group, Ltd.). The methylated and
unmethylated band intensities of each sample were visualized and
measured using Storm840 and ImageQuant Software (Amersham
Biosciences; GE Healthcare). The experiment was performed in
triplicate.
 | Table I.Primer sequences, amplicon sizes and
annealing temperature and conditions for PCR analysis. |
Table I.
Primer sequences, amplicon sizes and
annealing temperature and conditions for PCR analysis.
| Primers | Sequence
(5′-3′) | Amplicon sizes,
bp | Annealing
temperature, °C |
|---|
| MSP |
| CADM1 |
|
|
|
| CADM1-met (F) |
GCGTCGTCGAACGTTAGCG | 165 | 60 |
|
CADM1-met (R) |
GTTAACTACCTCCGAAACCCG |
|
|
|
CADM1-unmet (F) |
TGAATGTTAGTGTTAGGGGGTG | 72 | 60 |
|
CADM1-unmet (R) |
CACCACAAACCCAACCCAAC |
|
|
| DAPK1 |
|
|
|
|
DAPK1-met (F) |
CGAGCGTCGCGTAGAATT | 114 | 53 |
|
DAPK1-met (R) |
CGAAAAACGACCGACAAACG |
|
|
|
DAPK1-unmet (F) |
TGAGTTTGGAGTGTGGAGTT | 96 | 53 |
|
DAPK1-unmet (R) |
AACACAACCCACCCACCT |
|
|
| CADM1
expression |
|
|
|
| CADM1
(F) |
TGACAGTGATCGAGGGAGAGGT | 236 | 60 |
| CADM1
(R) |
GGGATCGGTATAGAGCTGGCAA |
|
|
| DAPK1
expression |
|
|
|
| DAPK1
(F) |
CCACCACTCGGATCAAGATCATTG | 132 | 60 |
| DAPK1
(R) |
ATATCTGCCTCAAGACCAAGAGG |
|
|
| GAPDH
expression |
|
|
|
| GAPDH
(F) |
GTGGGCAAGGTATCCCTG | 90 | 60 |
| GAPDH
(R) |
GATTCAGTGTGGTGGGGGAC |
|
|
| CADM1
ChIP-PCR |
|
|
|
|
CADM1ChIP (F) |
ACTCCGCCTCCAGCGCATGT | 229 | 62 |
|
CADM1ChIP (R) |
CCCACACCTACCTGTGGGGAT |
|
|
Expression analysis
Quantitative PCR was performed to observe the
DAPK1 and CADM1 mRNA expression in SiHa C33a cells
and c33a transfected cells. Amplification was performed with 0.1 µM
DAPK1 and 0.1 µM CADM1 primers (Table I), including primers for 0.1 µM
GAPDH, which was used as a reference gene, and 19 Power SYBR
Green PCR master mix (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The cDNA was amplified using a 7500-fast real-time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with
initially denatured for 10 min at 95°C, followed by 35 cycles of 1
min at 95°C, 1 min at 60°C, and 1 min at 72°C, and 72°C for 7 min.
The fold changes in expression of DAPK1 and CADM1
between experimental and control cellswere then determined using
the ΔΔCt method (18).
The experiment was performed in triplicate.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was carried out in E7
recombinant plasmid- and empty plasmid-transfected C33a cells, as
previously described, with some modifications (19). To observe the binding of E7 protein
(with or without the CR3 region) at the CADM1 promoter, the
chromatin fragments were immunoprecipitated with 10 µg of the H3K4
(Abcam (Cambridge, UK), HPV16-E7 (Santa Cruz, CA, USA) and IgG
(Santa Cruz, CA, USA) antibodies. Next, the precipitated DNA was
further analyzed by PCR using 0.3 µM CADM1-ChIPF and 0.3 µM
CADM1-ChIPR (Table I) with
initially denatured for 10 min at 95°C, followed by 35 cycles of 1
min at 95°C, 1 min at 62°C, and 1 min at 72°C, and 72°C for 7 min.
Then, 10 µl PCR products were observed by gel electrophoresis using
a 8% acrylamide and 1% agarose gel stained with SYBR (Lonza Group,
Ltd.). The CADM1 bands were visualized using Storm840 and
ImageQuanNT Software (Amersham Biosciences; GE Healthcare). The
experiment was performed in triplicate.
Sequencing analysis
To confirm the accuracy of the E6 and
E7 recombinant plasmid sequences, E6 and E7
recombinant plasmids were transformed into competent Escherichia
coli XL-1 blue cells for cloning, and then the plasmids were
extracted using a GeneJET Plasmid Miniprep kit (Fermentas; Thermo
Fisher Scientific, Inc.) for sequencing analysis immediately after
transfection. After alignment between the sequence of recombinant
plasmid and GENBANK (www.ncbi.nlm.nig.gov), the accuracy of the E6
and E7 sequences were confirmed.
Statistical analysis
To test the significance of the E6- or E7-mediated
induction of CADM1 and DAPK1 promoter methylation and
the decrease in gene expression, two sample t-tests between two
groups of samples, E6 or E7 transfected cells and control cells,
were used. P<0.05 was considered to indicate a statistically
significant difference.
Results
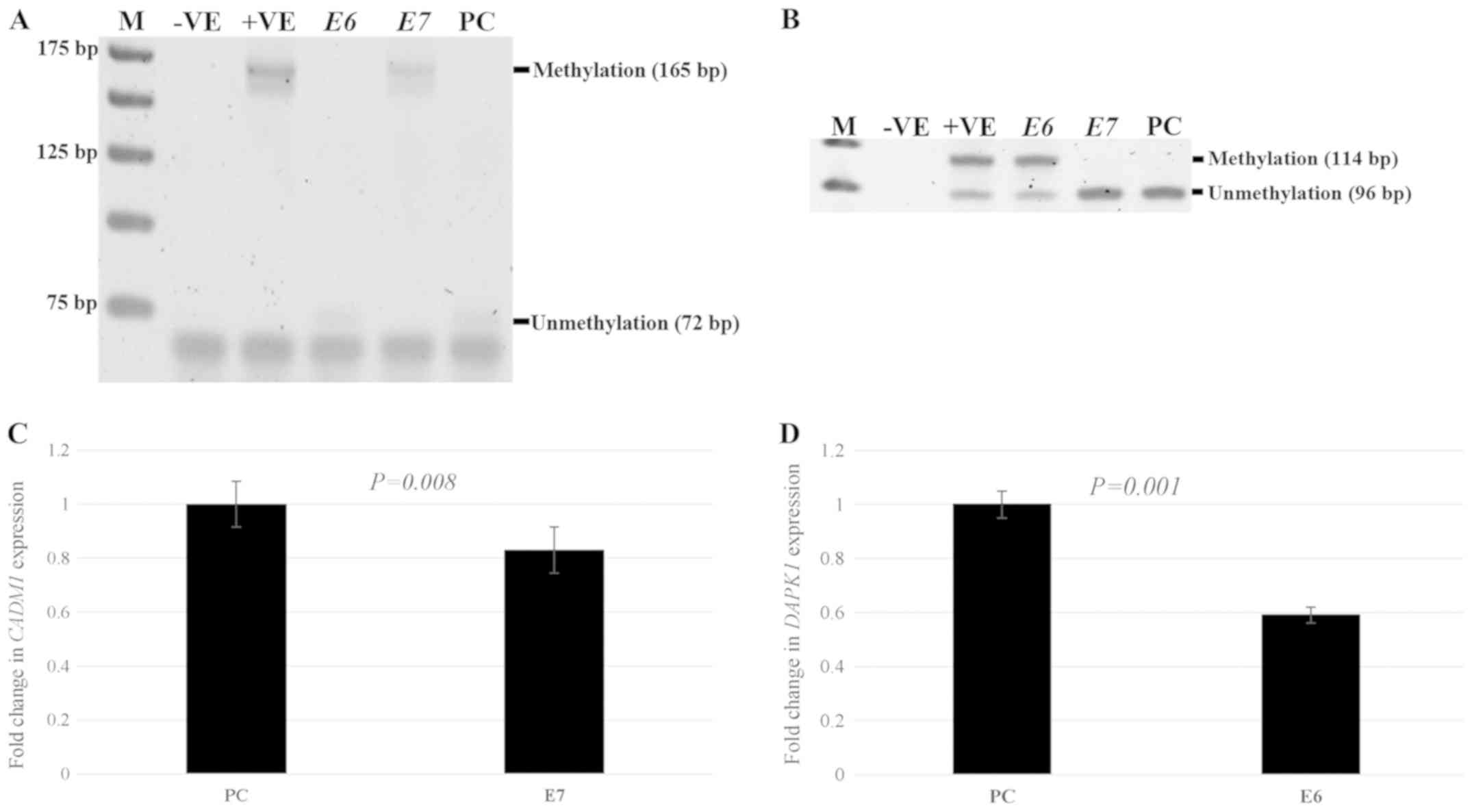
Methylation and expression status in
HPV+ and HPV-cervical cancer cell lines
To investigate the effect of HPV in mediating
promoter methylation and decreasing gene expression, the SiHa
cervical cancer cell line with HPV type 16 infection and the C33a
cervical cancer cell line without HPV infection were used for the
observation of methylation and the expression status of
CADM1 and DAPK1. There was methylation at the
CADM1 and DAPK1 promoters in SiHa cells, whereas
there was no methylation at the CADM1 and DAPK1
promoters in C33a cells (Fig. 1A and
B). There was no expression of CADM1 in SiHa cells,
while the expression of this gene in C33a cells was detected. For
DAPK1, significantly decreased expression was detected in
SiHa cells compared to that in C33a cells; P=0.005. (Fig. 1C and D).
HPV E6 and E7 induces DAPK1 and CADM1
promoter methylation and decreases their expression
To investigate whether E6 or E7 of HPV induce de
novo methylation of DAPK1 and CADM1, E6 or E7
recombinant plasmids and empty PCDNA3.1 (as a control) were
successfully transfected into C33a cell (Fig. 2A and B). CADM1 and
DAPK1 promoter methylation and their expression were
observed. For methylation status in E6- or
E7-transfected cells compared to control cells, significant
CADM1 promoter methylation was observed in
E7-transfected cells compared with control cells
(P<0.0001), while the promoter methylation of this gene could
not be detected in C33a cells transfected with E6. To
observe the methylation of DAPK1 in C33a-transfected cells,
the results showed that significantly increased DAPK1
promoter methylation was detected in E6-transfected cells
compared to cells with the empty vector (P=0.0003), whereas there
was no DAPK1 methylation in E7-transfected cells
(Fig. 3A and B). For the expression
study, after E7-transfected into C33a cells, there was a
significantly decreased CADM1 expression in E7-transfected
C33a cells (P=0.008) compared to cells with the empty vector. To
investigate the expression of DAPK1, after E6 and
empty vector transfection into C33a cells, there was significantly
decreased expression of DAPK1 (P=0.001) in E6-transfected
cells compared to cells with the empty vector (Fig. 3C and D).
E7 targets the CADM1 promoter
It was hypothesized that E7 promotes CADM1
methylation by forming a complex with Dnmt1 at the CADM1
promoter, which is the same mechanism as that for CCNA1. To
demonstrate that this hypothesis was viable, a chromatin
immunoprecipitation assay was carried out in C33a cells transfected
with E7 and the empty vector by precipitating with anti-HPV16 E7
and performing PCR to obtain a 229-bp CADM1 product. As
shown in Fig. 4, the product band of
the CADM1 promoter was detected in E7-overexpressing C33a
cells, but not in the cells with the empty vector. These data
suggested that E7 can bind at the CADM1 promoter.
Discussion
Epigenetics plays an important role in
tumorigenesis. DNA methylation at the promoter of tumor suppressor
genes is a process that decrease their expression, resulting in the
development of many cancer types, including cervical cancer. In
addition to promoter methylation, HPV is also a major cause of
cervical cancer. To date, there have been several studies showing
that HPV induces promoter methylation of tumor suppressor genes. It
has been found that E6 and E7 can promote the promoter methylation
of several genes. For example, a study by Li et al (20) reported that E6 and E7 gene silencing
led to a decrease in the methylation of six tumor suppressor genes,
MT1G, NMES1, RRAD, SFRP1, SPARC and TFP12, and
induced the phenotypic transformation of human cervical carcinoma
cell lines.
The mechanism by which E6 or E7 could induce the
promoter methylation of the genes has been explored. Our previous
study demonstrated that HPV 16 E7 had an interaction with Dnmt1
resulting in CCNA1 promoter methylation and a reduction in
expression (14). A study by Au
Yeung et al (21) indicated
that E6 increased the expression of Dnmt1 by degrading p53. Dnmt1
is a member of the Dnmt family, which can play role in de
novo methylation (22).
Therefore, the present study sought to discover genes whose
promoter methylation may be induced by E6 or E7. Here, CADM1
and DAPK1 were selected to be candidate genes because of
their functions as tumor suppressor genes, and the evidence showing
that the methylation of these two genes may be related to cervical
cancer progression. CADM1 is hypermethylated in CIN II–III
and cervical cancer, while DAPK1 is hypermethylated in CIN
III and cervical cancer (23).
Notably, it was found that the promoter methylation of both genes
was induced by HPV. Nevertheless, HPV16 E6 only induced methylation
of DAPK1, while HPV 16 E7 only induced methylation of
CADM1.
The present study demonstrated that HPV 16 E7 can
induce promoter methylation of CADM1, and not only of
CCNA1. Therefore, it was hypothesized that the mechanism by
which HPV16 E7 induced promoter methylation of CADM1 may be
the same as the mechanism for CCNA1. To elucidate this
mechanism, chromatin immunoprecipitation was carried out using an
E7 antibody to precipitate E7 protein binding at the CADM1
promoter. The results showed that E7 protein can bind at the
CADM1 promoter. Studies by Chalertpetch et al
(14) and Burgers et al
(24) found an interaction between
E7 and Dnmt1. This evidence indicated that that HPV16 E7 may bind
with Dnmt1 at the CADM1, promoter leading to aberrant
promoter methylation and decreasing the expression of the gene.
In the cells, it was hypothesized that E6 and E7 may
work together to induce promoter methylation. E6 was thought to
increase Dnmt1 expression (21), and
E7 to bind with Dnmt1 and target the promoters (14). However, it was found that
DAPK1 promoter methylation can be augmented by E6, but not
by E7. Therefore, it is possible that E6 might increase Dnmt1
activity and thus has another mechanism through which it targets
DAPK1 to induce its promoter methylation. The mechanism
underlying the increase in DAPK1 promoter methylation by E6
remains to be studied. However, in E7-transfected cells,
CADM1 promoter methylation was observed. This may be because
there is endogenous Dnmt1 in the cells, thus E7 protein can form
complexes with endogenous Dnmt1 and induce CADM1 promoter
methylation.
Understanding the mechanism by which HPV induces
promoter methylation of genes will be useful for drug discovery and
clinical diagnosis. At present, a Pap smear is still the gold
standard for cervical cancer diagnosis. Nevertheless, there is a
chance of receiving false negative results from a Pap smear test
(25,26). In addition, Pap smear tests
frequently detect atypical squamous cells of undetermined
significance (ASCUS) (27).
Therefore, molecular marker findings, together with HPV testing,
may be a good alternative for diagnosing the early stages of
cervical cancer, eventually leading to a decrease in the number of
cervical cancer patients worldwide.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by
Thailand Research Fund and Chulalongkorn University (grant no.
RSA5880065) and The National Science and Technology Development
Agency, Thailand (grant no P-15-50270).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
PY wrote the proposal for grants, designed the
experiments, analyzed data and wrote the manuscript. KC performed
the experiments and analyzed data. JS performed the experiments and
analyzed data. IN performed the experiments and analyzed data. PP
performed the experiments and analyzed data. AM designed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arbyn M, Castellsagué X, de Sanjosé S,
Bruni L, Saraiya M, Bray F and Ferlay J: Worldwide burden of
cervical cancer in 2008. Ann Oncol. 22:2675–2686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Virani S, Bilheem S, Chansaard W,
Chitapanarux I, Daoprasert K, Khuanchana S, Leklob A, Pongnikorn D,
Rozek LS, Siriarechakul S, et al: National and subnational
population-based incidence of cancer in thailand: Assessing cancers
with the Highest Burdens. Cancers (Basel). 9(pii): E1082017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bosch FX, Burchell AN, Schiffman M,
Giuliano AR, de Sanjose S, Bruni L, Tortolero-Luna G, Kjaer SK and
Muñoz N: Epidemiology and natural history of human papillomavirus
infections and type-specific implications in cervical neoplasia.
Vaccine. 26 (Suppl 10):K1–K16. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tulay P and Serakinci N: The role of human
papillomaviruses in cancer progression. J Cancer Metastasis Treat.
2:201–213. 2016. View Article : Google Scholar
|
|
5
|
Yeo-Teh NSL, Ito Y and Jha S: High-risk
human papillomaviral oncogenes E6 and E7 target key cellular
pathways to achieve oncogenesis. Int J Mol Sci. 19(pii): E17062018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sen P, Ganguly P and Ganguly N: Modulation
of DNA methylation by human papillomavirus E6 and E7 oncoproteins
in cervical cancer. Oncol Lett. 15:11–22. 2018.PubMed/NCBI
|
|
7
|
Yin F, Wang N, Wang S, Yu F, Sun X, Yu X,
Luo B, Zhao C and Wang Y: HPV16 oncogenes E6 or/and E7 may
influence the methylation status of RASSFIA gene promoter region in
cervical cancer cell line HT-3. Oncol Rep. 37:2324–2334. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yanatatsaneejit P, Mutirangura A and
Kitkumthorn N: Human papillomavirus's physical state and cyclin A1
promoter methylation in cervical cancer. Int J Gyneco. 21:902–906.
2011. View Article : Google Scholar
|
|
9
|
Chung CH and Gillson ML: Human
papillomavirus in head and neck cancer: Its role in pathogenesis
and clinical implications. Clin Cancer Res. 15:6758–6762. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sartor MA, Dolinoy DC, Jones TR, Colacino
JA, Prince ME, Carey TE and Rozek LS: Genome-wide methylation and
expression differences in HPV(+) and HPV(−) squamous cell carcinoma
cell lines are consistent with divergent mechanisms of
carcinogenesis. Epigenetics. 6:777–787. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lechner M, Fenton T, West J, Wilson G,
Feber A, Henderson S, Thirlwell C, Dibra HK, Jay A, Butcher L, et
al: Identification and functional validation of HPV-mediated
hypermethylation in head and neck squamous cell carcinoma. Genome
Med. 5:152013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kitkumthorn N, Yanatatsanajit P,
Kiatpongsan S, Phokaew C, Triratanachat S, Trivijitsilp P,
Termrungruanglert W, Tresukosol D, Niruthisard S and Mutirangura A:
Cyclin A1 promoter hypermethylation in human
papillomavirus-associated cervical cancer. BMC Cancer. 6:552006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chujan S, Kitkumthorn N, Siriangkul S and
Mutirangura A: CCNA1 promoter methylation: A potential marker for
grading Papanicolaou smear cervical squamous intraepithelial
lesions. Asian Pac J Cancer Prev. 15:7971–7975. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chalertpet K, Pakdeechaidan W, Patel V,
Mutirangura A and Yanatatsaneejit P: Human papillomavirus type 16
E7 oncoprotein mediates CCNA1 promoter methylation. Cancer Sci.
106:1333–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Steenbergen RD, Kramer D, Braakhuis BJ,
Stern PL, Verheijen RH, Meijer CJ and Snijders PJ: TSLC1 gene
silencing in cervical cancer cell lines and cervical neoplasia. J
Natl Cancer Inst. 96:294–305. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Banzai C, Nishino K, Quan J, Yoshihara K,
Sekine M, Yahata T and Tanaka K; Gynecological Cancer Registry of
Niigata, : Promoter methylation of DAPK1, FHIT, MGMT, and CDKN2A
genes in cervical carcinoma. Int J Clin Oncol. 19:127–132. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang HJ: Aberrant DNA methylation in
cervical carcinogenesis. Chin J Cancer. 32:42–48. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boyd KE and Farnham PJ: Coexamination of
site-specific transcription factor binding and promoter activity in
living cells. Mol Cell Biol. 19:8393–8399. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li L, Xu C, Long J, Shen D, Zhou W, Zhou
Q, Yang J and Jiang M: E6 and E7 gene silencing results in
decreased methylation of tumor suppressor genes and induces
phenotype transformation of human cervical carcinoma cell lines.
Oncotarget. 6:23930–23943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Au Yeung CL, Tsang WP, Tsang TY, Co NN,
Yau PL and Kwok TT: HPV-16 E6 upregulation of DNMT1 through
repression of tumor suppressor p53. Oncol Rep. 24:1599–1604.
2010.PubMed/NCBI
|
|
22
|
Fatemi M1, Hermann A, Gowher H and Jeltsch
A: Dnmt3a and Dnmt1 functionally cooperate during de novo
methylation of DNA. Eur J Biochem. 269:4981–4984. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng C, Dong J, Chang W, Cui M and Xu T:
The progress of methylation regulation in gene expression of
cervical cancer. Int J Genomics. 2018:82606522018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burgers WA, Blanchon L, Pradhan S, de
Launoit Y, Kouzarides T and Fuks F: Viral oncoproteins target the
DNA methyltransferases. Oncogene. 26:1650–1655. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van der Graaf Y and Vooijs GP: False
negative rate in cervical cytology. J Clin Pathol. 40:438–442.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sprenger E, Schwarzmann P, Kirkpatrick M,
Fox W, Heinzerling RH, Geyer JW and Knesel EA: The false negative
rate in cervical cytology. Comparison of monolayers to conventional
smears. Acta Cytol. 40:81–89. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jahic M and Jahic E: Diagnostic approach
to patients with atypical squamous cells of undetermined
significance cytologic findings on cervix. Med Arch. 70:296–298.
2016. View Article : Google Scholar : PubMed/NCBI
|