Introduction
Lung cancer is a malignant tumor with extremely high
incidence (36.71 per 100,000) and mortality rates (28.49 per
100,000) in China in 2018 (1). Small
cell lung cancer (SCLC) is a common subtype of lung cancer.
Currently, 4–6 cycles of platinum-based chemotherapy (cisplatin or
carboplatin) remains the primary treatment option for patients with
extensive SCLC (2). Despite recent
advancements in novel chemotherapy regimens and improvements in the
use of immunotherapy, >75% of patients with SCLC relapse within
6 months (3), and the overall 5-year
survival rate of patients with SCLC remains <15% (4,5).
Immunotherapy has gained recent interest as a validated clinical
treatment for several types of cancer, including SCLC (6). However, given that immunotherapy is not
a first-line treatment for SCLC, the majority of patients who
receive immunotherapy have previously undergone chemotherapy
several times (7). Thus,
understanding the association between chemotherapy and immunity is
of great importance.
Natural killer (NK) cells serve a critical role in
the tumor cell immune response and their function is regulated via
signals initiated by a variety of activating and inhibitory
receptors on NK cells (8). NK
cell-activated receptor natural-killer group-2 member D (NKG2D) is
expressed on all NK cells (9). NKG2D
ligands (NKG2DLs), as homologues of major histocompatibility
complex (MHC) I molecules in human tissues, include two families:
MHC I chain-related proteins (MICs) (10,11) and
UL16-binding proteins (ULBPs) (12,13). MHC
class I polypeptide-related sequence A (MICA) is predominantly
expressed in epithelial-derived tumor cells, including lung,
breast, renal and ovarian cancer cells, as well as in certain
melanoma cells (14). The
combination of NKG2D with MICA on the tumor surface can directly
activate NK cells and enhance the killing activity of NK cells
against tumor cells (15).
Simultaneously, NKG2D, which recognizes non-MHC class I molecules,
serves a crucial role in mediating the immune response of NK cells
when recognizing and attacking tumor cells (16). Downregulation of NKG2D decreases the
sensitivity of NK cells to lung cancer cells, following MHC-I
deletion. Furthermore, a previous study have that the expression of
these ligands are associated with enhanced antibody-dependent
cell-mediated cytotoxic activity, which is one of the fundamental
molecular mechanisms underlying the antitumor effect of antibody
therapeutics (17). This topic is of
considerable interest as the potential to manipulate NKG2DL
expression may offer promise in the future treatment of different
types of tumor.
To the best of our knowledge, limited research has
focused on the association between chemotherapy and ligands. Thus,
the present study aimed to investigate the expression of NKG2DLs in
tissue samples of patients with SCLC to determine the association
between NKG2DLs and chemoresistance.
Materials and methods
Clinical characteristics and data
collection of patients
The present study was approved by the Ethics
Committee of Zhengzhou University (approval no. 2014079), and
performed in accordance with The Declaration of Helsinki. Written
informed consent was provided by all patients prior to study
enrolment. Biopsy tissue samples were collected from 115 patients
with advanced SCLC who received platinum-based chemotherapy at The
Affiliated Cancer Hospital of Zhengzhou University (Zhengzhou,
China), between January 2014 and March 2018. The median age of the
patients with advanced SCLC who were included in the study was
65.32 years (range, 56.25–77.14 years), of which 78 (67.83%) were
male and 37 (32.17%) were female. Histological diagnosis was based
on H&E and immunohistochemical staining, according to the 2015
World Health Organization criteria (18). The inclusion criteria were as
follows: i) Patients clinically diagnosed with SCLC; ii) patients
≥18 years old; iii) patients diagnosed by biopsy; and iv) patients
pathologically diagnosed with advanced SCLC. Patients who had a
history of other malignant tumors, or who had previously received
radiotherapy or chemotherapy were excluded from the present study.
The clinical characteristics of patients with SCLC are presented in
Table I.
 | Table I.Clinical characteristics of patients
with small cell lung cancer (n=115). |
Table I.
Clinical characteristics of patients
with small cell lung cancer (n=115).
| Characteristic | Patient, n | Cases, % | mPFS time, months ±
SD | P-value |
|---|
| Total, n | 115 |
| 6.29±2.24 |
|
| Sex |
|
|
| 0.063 |
|
Male | 78 | 67.83 | 6.02±1.78 |
|
|
Female | 37 | 32.17 | 6.75±1.90 |
|
| Age, years |
|
|
| 0.034 |
|
<57 | 77 | 66.96 | 6.34±2.03 |
|
|
≥57 | 38 | 33.04 | 5.97±1.35 |
|
| Smoker |
|
|
| 0.045 |
|
Never | 46 | 40.00 | 6.38±2.27 |
|
|
Previous/Current | 69 | 60.00 | 5.82±1.75 |
|
| Metastasis
site |
|
|
|
|
|
Brain | 21 | 18.26 | 5.73±1.68 |
<0.001b |
|
Liver | 18 | 15.65 | 4.44±1.93 | 0.026a |
| Distant
lymphoid | 35 | 30.43 | 6.60±2.11 | 0.045a |
|
Othersc | 41 | 35.65 | 7.20±1.96 |
|
Imaging examination (chest-abdomen CT and head MRI)
was performed regularly every two chemotherapy cycles until tumor
progression was observed. Routine blood examination, assessment of
liver-kidney function and ECG prior to chemotherapy were performed
for each patient. Chemotherapy response was evaluated according to
the Response Evaluation Criteria in Solid Tumors (version 1.1)
(19). Progression-free survival
(PFS) was measured following first-line chemotherapy. PFS was
defined from the beginning of chemotherapy until disease
progression or mortality from any cause. The method of follow-up
was via regular outpatient assessment every 3 weeks. The follow-up
period was between 1st February 2014 and 31st March 2018.
Reverse transcription-quantitative
(RT-q)PCR
Fresh lung puncture tissue samples were immediately
immersed in RNAlater Stabilization reagent (Qiagen AB) and stored
at −20°C until RNA extraction. Total RNA was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Total RNA (1 µg)
was reverse transcribed into cDNA using a OneStep RT-PCR kit,
according to the manufacturer's protocol (Tiangen Biotech Co.,
Ltd.). Temperature protocol were carried out at 25°C for 5 min and
42°C for 30 min and 85 for 5 min. qPCR was subsequently performed
to determine the mRNA expression of the six selected NKG2DLs [MICA,
MHC class I polypeptide-related sequence B (MICB), UL16-binding
proteins 1 (ULBP1), UL16-binding proteins 2 (ULBP2), UL16-binding
proteins 3 (ULBP3), UL16-binding proteins 4 (ULBP4)] (14) or 11 selected genes according to
previous research [ATP binding cassette subfamily B member 1
(ABCB1), ATP binding cassette subfamily C member 1 (ABCC1), ATP
binding cassette subfamily C member 2 (ABCC2), ATP binding cassette
subfamily G member 2 (ABCC2), ATP binding cassette subfamily C
member 9 (ABCC9), dihydropyrimidine dehydrogenase (DPYD),
glutathione S-transferase θ1 (GSTT1), glutathione S-transferase π 1
(GSTp1), glutathione S-transferase mu 1 (GSTM1), alcohol
dehydrogenase 1A class I α polypeptide (ADH1A), peroxisome
proliferator activated receptor α, (PPARA)] (20,21),
using FastStart Universal SYBR Green Master mix (Roche Diagnostics
GmbH) on a StepOne Plus real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). 115 patients' whole blood samples
were used as the control. The primer sequences used for qPCR are
listed in Table S1, and the final
concentration of each primer in a reaction was 0.5 µM. The
following thermocycling conditions were used for qPCR: Initial
denaturation at 95°C for 3 min; 35 cycles of amplification
(denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec,
elongation at 72°C for 20 sec) and a final elongation at 68°C for
10 min. Data were analyzed with the 2−ΔΔCq method
(22).
Cell culture
The NCI-H446 cell line, which was kindly provided by
Dr ZeLai He from Medical Oncology of The First Affiliated Hospital
of Bengbu Medical College (Bengbu, China), was cultured in
RPMI-1640 complete medium (HyClone; GE Healthcare Life Sciences)
supplemented with 10% FBS, 2 mM glutamine, 100 µg/ml streptomycin
and 100 U/ml penicillin (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C and 5% CO2.
Overexpression of MICA
The full-length MICA gene (Shanghai GeneChem Co.,
Ltd.) was subcloned into the pCDH-CMV-MCS lentivector (System
Biosciences, http://systembio.com/shop/pcdh-cmv-mcs-cdna-single-promoter-cloning-and-expression-lentivector/),
between the NheI and SalI sites. The infectious lentiviral
particles were generated and concentrated as previously described
(21). A total of 2×105
NCI-H446 cells were transduced with the recombinant lentivirus
carrying the human MICA gene or an empty vector to generate the
stable MICA-overexpressing cells (OE-MICA) or control cells
(VC-MICA), respectively.
MTT assay
To assess the chemosensitivity of tumor cells, cell
viability was measured via an MTT assay (Promega Corporation). Cell
suspensions were cultured in RPMI-1640 medium (Thermo Fisher
Scientific, Inc.) in 96-well flat-bottomed microtiter plates at a
density of 1×104 cells/well and incubated overnight at
37°C. For the nicardipine combination experiment, the concentration
of nicardipine (Sigma-Aldrich; Merck KGaG) in the culture medium
was adjusted to 10 µM and incubated at 37°C for 15 min in
advance.
The following concentrations of cisplatin
(Sigma-Aldrich; Merck KGaG) were used: 10−7 M,
10−6 M, 10−5 M, 10−4 M and
10−3 M. All experiments were performed in triplicate,
for each concentration. NCI-H446 cells cultured in RPMI-1640 medium
were used as the blank controls. MTT solution was added (1 mg/ml
per well) and incubated at 37°C for 48 h. Cell viability was
subsequently analyzed at a wavelength of 590 nm, using a
spectrophotometric microplate reader (Bio-Rad Laboratories, Inc.).
The response percentage of cell viability to different drug
concentrations was calculated as the inhibition rate (average
absorbance of treatment wells/control wells ×100%).
Western blotting
Cell or tumor lysates were obtained and equal
amounts of protein lysates from each sample were diluted with
loading buffer, denatured, and separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by
protein transfer to polyvinylidene fluoride membranes (PVDF). After
incubation in a blocking solution (5% nonfat milk powder) in TBST
buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% Tween-20) for
1 h at room temperature, the membranes were immunoblotted overnight
with primary monoclonal antibodies (Abs) against either ABCG2
(Santa Cruz Biotechnology Inc.) or Actin at a 1:1,000 dilution at
4°C. The membranes were then incubated for 2 h at room temperature
with the appropriate secondary antibody (1:1,000 dilution). The
protein antibody complex was detected by an enhanced
chemiluminescence detection system. The protein expression was
quantified by ImageJ software (http:/rsbweb.nih.gov/ij/).
GeneMANIA prediction
GeneMANIA datasets (http://genemania.org) were downloaded from to show the
interaction between 17 genes, including the six selected NKG2DLs
(MICA, MICB, ULBP1, ULBP2, ULBP3, ULBP4) (14) or 11 selected genes according to
previous research (ABCB1, ABCC1, ABCC2, ABCC2, ABCC9, DPYD, GSTT1,
GSTp1, GSTM1, ADH1A, PPARA) (20,21),
including: Co-expression data from Gene Expression Omnibus
(23); physical and genetic
interaction data from Biological General Repository for Interaction
Datasets (BioGRID) (24); predicted
protein interaction data, based on orthology, from I2D (25) and pathway and molecular interaction
data from Pathway Commons, which contains data from BioGRID
(24), Memorial Sloan-Kettering
Cancer Center, Human Protein Reference Database (26), HumanCyc (27), Systems Biology Center New York,
IntAct (28), MINT (29), NCI-Nature Pathway Interaction
Database (30) and Reactome
(31,32).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 19; IBM Corp.). Associations between NKG2DLs and
PFS were assessed using the coefficient of determination
(R2). The half maximal inhibitory concentration
(IC50) values were calculated using a non-linear
regression model within GraphPad Prism software (version 6.0;
GraphPad Software Inc.). Pearson's correlation analysis was
performed the correlation analysis of NKG2DLs and PFS. Unpaired
Student's t-tests were used to compare differences between two
groups, while one-way ANOVA followed by Bonferroni's correction was
used to compare cell viability between multiple groups, at 10
µmol/l of carboplatin. P<0.05 (two-tailed) was considered to
indicate a statistically significant difference.
Results
Clinical information of patients with
extensive SCLC
The age range of the 115 patients with extensive
SCLC was between 46.5 to 68.9 years and the total median PFS (mPFS)
associated with first-line chemotherapy was 6.29±2.24 months. There
were 78 men (67.83%) and 37 women (32.17%) in the study group.
There were no significant differences between the two groups
according to the median age (57 years; 77 patients <57 and 38
patients ≥57). There were 69 smokers (60.00%). As presented in
Table I, smoking and metastasis site
were significantly associated with the mPFS in relation to
chemotherapy.
The location of tumor metastasis was significantly
associated with the efficacy of chemotherapy and the rates of
metastasis to the brain, liver and lymph nodes were substantially
different compared with other sites, including bone, pleura and
adrenal gland. The mPFS of patients with liver metastasis
(4.44±1.93 months) was the least favorable, even shorter than that
of patients with brain metastasis, as presented in Table I.
MICA prolongs the PFS time associated
with chemotherapy
To detect the mRNAs of six NKG2DLs, the present
study used paracancerous tissues as a control. Pre-treatment NKG2DL
levels were determined in the tumor tissue samples of 115 patients
with SCLC. The results indicated that MICA was closely associated
with PFS time in patients treated with first-line chemotherapy
(Pearson r=0.4496; P<0.0001; Fig.
1A). However, no significant associations were observed between
PFS and the other five NKG2DLs. As presented in Fig. 1, the Pearson r values of MICB, ULBP1,
ULBP2, ULBP3 and ULBP4 were −0.0506, 0.0171, 0.0120, 0.0277 and
0.1156, respectively (all P>0.05). Clinically, a small number of
patients with drug-resistant SCLC relapse <3 months after the
first treatment, which is defined as rapid chemoresistance (QCR)
(33). In the present study, the 20
patients with the shortest PFS time were classified as having QCR,
while the 20 patients with the longest PFS time were classified as
having delayed chemotherapy resistance (DCR). The expression of the
NKG2DLs were subsequently analyzed in these two groups of patients
and the results demonstrated that MICA expression was significantly
increased in the DCR group, in which MICA expression was 1.6 times
higher compared with the QCR group (DCR, 1.5549±0.4506 vs. QCR,
1.0000±0.3557; P<0.0001; Fig.
1G). However, the other NKG2DLs failed to demonstrate any
significant differences between the two groups.
MICA significantly increases the
sensitivity of tumor cells to platinum chemotherapy
MICA may improve the PFS of patients with SCLC,
treated with chemotherapy; however, further investigations are
required to verify whether this is caused by increased
chemosensitivity. Thus, the present study constructed a NCI-H446
cell line which overexpressed MICA and verified the effect of MICA
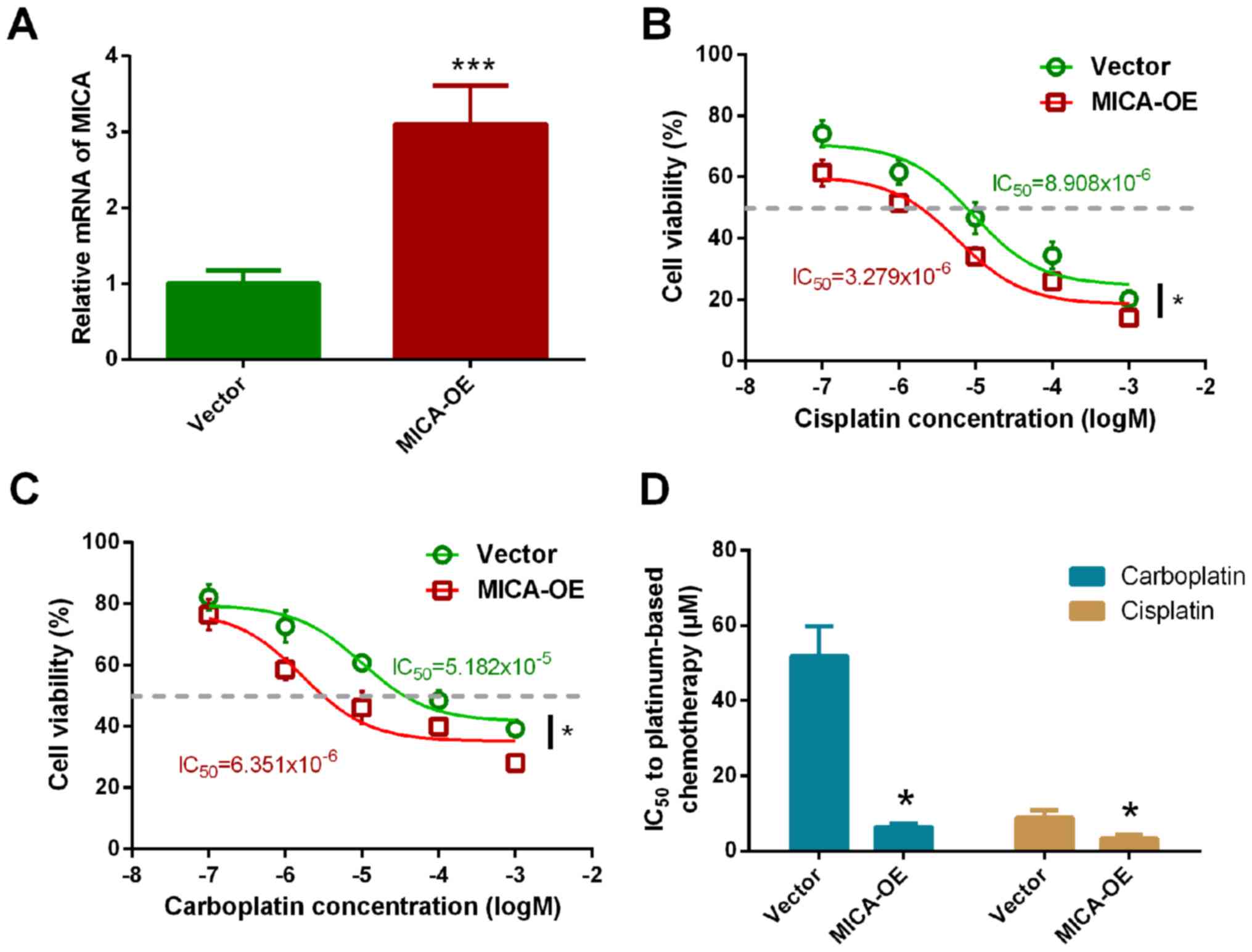
overexpression (Fig. S1). RT-qPCR
analysis demonstrated that MICA expression in the overexpressed
cell line was 3.103 times higher compared with the control group
(P<0.001; Fig. 2A).
Chemosensitivity of MICA-OE cells to carboplatin and
cisplatin was subsequently assessed, with vector-only cells as the
control. Overexpression of MICA significantly increased NCI-H446
chemosensitivity to cisplatin (Fig.
2B). The IC50 value of cisplatin in NCI-H446 cells
significantly decreased (vector cells, 9.279±3.931 µmol/l vs.
MICA-OE cells, 5.908±2.775 µmol/l; Fig.
2D). Furthermore, the chemosensitivity of MICA-OE cells to
carboplatin also significantly increased. The IC50 value
of carboplatin in the MICA-OE cells was 6.351±1.225 µmol/l, while
the IC50 value of carboplatin in the vector cells was
51.824±6.924 µmol/ (Fig. 2C and D).
The results demonstrated that the IC50 value
significantly decreased with overexpression of MICA, thus
suggesting that MICA may increase the sensitivity of tumor cells to
cisplatin and carboplatin.
ABCG2 serves a vital role in
regulating the chemotherapy sensitivity induced by MICA
To understand the molecular mechanism by which MICA
enhances chemosensitivity, the present study assessed the mRNA
expression of 11 genes associated with chemosensitivity according
to our previous research include: ABCB1, ABCC1, ABCC2, ABCG2,
ABCC2, ABCC9, DPYD, GSTT1, GSTp1, GSTM1, ADH1A, PPARA (20). The primer sequences of the associated
genes are listed in Table S1. The
results demonstrated that overexpressing MICA downregulated the
mRNA expression of ABCG2, ADH1 and GSTT1; however, no significant
differences were observed in the mRNA levels of the other genes
(ABCB1, ABCC1, ABCC2, ABCC9, DPYD, GSTT1, GSTp1 and GSTM1)
(Fig. 3A). According to the
GeneMANIA analysis, there is a complex interaction network between
the NKG2DLs and chemotherapeutic sensitivity-associated genes
(Fig. 3B). Western blot analysis was
performed to determine the protein expression levels of ABCG2, ADH1
and GSTT1 in MICA-OE cells compared with the vector group. The
results demonstrated that ABCG2 protein expression significantly
decreased in the MICA-OE group compared with the control (Fig. 3C); however, there was no significant
differences in ADH1 and GSTT1 protein expression between the two
groups (Fig. 3D and E). ImageJ
quantitative analysis of the western blots demonstrated that ABCG2
protein expression significantly decreased in the MICA-OE group
compared with the control (Fig.
3F).
 | Figure 3.ABCG2 serves a crucial role in
MICA-induced chemosensitivity. (A) mRNA expression levels of ABCG2,
ADH1A and GSTT1 were significantly downregulated compared with the
Vector control group. (B) According to the GeneMANIA analysis,
there is a complex interaction network between NKG2DLs and
chemotherapeutic sensitivity-associated genes. (C) ABCG2 protein
expression significantly decreased in the MICA overexpression
group, while (D) ADH1 and (E) GSTT1 protein expression levels did
not. (F) ImageJ western blot quantification suggested that ABCG2
expression was downregulated considerably compared with the Vector
group. (G) MICA-induced chemosensitivity significantly increased
following addition of nicardipine. Cell viability was compared
using ANOVA, at 10 µmol/l of carboplatin. *P<0.05, mRNA of GSTT1
and ADH1A in MICA-OE vs. vector. **P<0.01, mRNA of ABCG2 in
MICA-OE vs. vector (A) or cell viability of MICA-OE vs. vector in
response to 10−5 M carboplatin (G). ***P<0.001,
grayscale value of ABCG2 in MICA-OE vs. vector (F), or cell
viability of MICA-OE + nicardipine vs. vector in resopnse to
10−5 M carboplatin (G). ABCG2, ATP binding cassette
subfamily G member 2; MICA, major histocompatibility complex class
I polypeptide-related sequence A; ADH1A, alcohol dehydrogenase 1A
(class I), α polypeptide; GSTT1, glutathione S-transferase θ1. |
The results of western blotting were subsequently
validated via chemosensitivity testing. ABCG2 blockers
(nicardipine) was also added in order to clarify the role of ABCG2
in MICA chemotherapy sensitization (34). First, the effect on cell
proliferation was examined to determine whether nicardipine
improves the chemotherapeutic sensitivity of MICA-OE cells. A lower
concentration of nicardipine (10 µM) was selected as it has no
significant effect on the proliferation of NCI-H446 cells (35). The MTT assay demonstrated a
significantly lower IC50 value in the MICA-OE +
nicardipine group compared with the vector group (Fig. 3G; vector group, 2.663±1.235 µM;
vector + nicardipine group, 9.956±1.392 µM; MICA-OE + nicardipine
group, 4.475±1.178 µM; MICA-OE group, 9.279±1.931 µM).
Discussion
In recent years, the molecular mechanism of the
immune response against tumors has been recognized, along with the
synergetic effect of immunotherapy in combination with chemotherapy
(9). An in-depth understanding of
the role of NKG2D in tumor chemotherapy may contribute to the
establishment of a novel clinical therapy for SCLC. NKG2DLs serve
an essential role in the process of immune recognition and cell
killing mediated by NK cells, and they are also crucial to the
antitumor response (36).
Previous research on immune drugs has focused on the
effect on the tumor itself, without considering its interaction
with chemotherapeutic agents. The expression of NKG2DLs are lower
in tumor tissues compared with healthy tissues (10), which was confirmed in the present
study. Furthermore, this decreased expression is also considered to
be a molecular mechanism underlying tumor immune escape (37,38).
Zhang et al (39)
demonstrated that the expression of NKG2DLs by tumor cells may
result in the efficient development of antitumor immune responses
in vitro and in vivo (39). The present study assessed MICA
expression in patients with SCLC by determining the association
between MICA expression levels and PFS time and confirmed that it
may enhance the sensitivity of SCLC to platinum chemotherapy. MICA
serves a crucial role in the tumor chemotherapy response,
indicating that high MICA has a synergistic effect on chemotherapy.
Thus, MICA may represent a predictive factor for the efficacy of
chemotherapy and serve as a prognostic marker for patients
receiving chemotherapy.
The results of the present study demonstrated that
overexpressing MICA in vitro increased the sensitivity of
cells to cisplatin and carboplatin. This phenomenon may be due to
three factors: i) High MICA expression enhances the tumor killing
activity of NK and T cells (40);
ii) increased numbers of NK and T cells demonstrate a more specific
antitumor effect, particularly during chemotherapy and tumor
necrosis antigen exposure (41); and
iii) high NKG2D expression decreases the efflux pump activity of
the tumor cell membrane, while increasing the intracellular
concentration of drugs, which may enhance the chemosensitivity of
tumor cells.
To determine the molecular mechanism, the expression
of 11 genes in tumor specimens from patients with SCLC were
assessed. The expression changes associated with drug sensitivity
in tumor tissues with high MICA expression were predominantly
concentrated in the ATP binding cassette protein transport pathway
genes, while the expression changes associated with drug metabolism
were primarily concentrated in the glutathione S-transferase
pathway genes. RT-qPCR and western blot analyses were performed to
verify mRNA and protein expression in MICA-overexpressing tumor
cells, respectively. The results demonstrated that ABCG2 expression
decreased in tumor cells overexpressing MICA. This result supports
the hypothesis that high NKG2D expression decreases the activity of
the efflux pump, while increasing the intracellular concentration
of drugs. However, the present study has some limitations. Firstly,
a major limitation of the present study is that only one SCLC cell
line was used to verify the results. Secondly, the experimental
results were not verified using animal models and clinically in
patients. In subsequent studies, the growth rate of MICA-expressing
multi-tumor cells in mice and the response to chemotherapy need to
be studied. Subsequently, the key genes in the upstream and
downstream pathways of MICA will also be sequenced in whole
genes.
Previous studies have confirmed that downregulation
of ABCG2 serves a vital role in resistance to chemotherapy
(35,42,43).
Membrane transport proteins belong to the ATP-binding cassette
transport protein superfamily (42,43).
Members of this superfamily utilize energy from ATP hydrolysis to
transport a number of substrates, including peptides, lipids and
anticancer drugs, across the membrane. The human genome contains 48
ATP binding cassette subfamily genes, which can be further divided
into seven different subfamilies, named A-G, according to sequence
homology and domain similarity (42). These proteins mediate the development
of multidrug resistance in several types of cancer cells, including
breast and colon cancer (44–47).
ABCG2 transporter protein is a key ABC protein, which is
ubiquitously expressed in lung and liver, and contributes to the
disposition of a variety of endogenous substances, including
platinum-based regimens (48,49).
Previous studies have demonstrated an association between ABCG2
expression and prognosis and survival in patients with SCLC cancer
(45,46,50).
Peptides generated by the ubiquitin-proteasome pathway, including
MICA, are presented by MHC class I molecules (51). The predominant function of the MIC
protein family is to display peptides to cytotoxic CD8+
T cells in support of their crucial activity of immune surveillance
(36). Thus, the present study
hypothesized that MICA decreases ABCG2 expression on the cell
membrane via antigen presentation. ABCG2 functions as a xenobiotic
transporter and its involvement in multi-drug resistance has been
well-established and previously validated (35,52). For
exclude other pathway caused by MICA-mediated drug resistance,
nicardipine was selected to be the next phase of the experiment as
strong inhibitors of ABCG2. As expected, the IC50 of
MICA-OE was significant down-regulation by nicardipine in SCLC
cells to carboplatin. The results of the present study confirmed
that MICA is closely associated with the expression of membrane
transport proteins in tumors and that chemosensitivity can be
achieved by downregulating ABCG2. However, the molecular mechanisms
by which overexpression of MICA decreases ABCG2 expression remain
unclear, thus further investigation is required.
Currently, the focus is on programmed cell death-1
and programmed death-ligand 1, whereby several clinical trials have
been performed using immunotherapy combined with chemotherapy
(6). Most of these clinical studies
have achieved positive results; however, they have predominantly
prompted researchers to better investigate immune system-enhancing
drugs and assess the mechanism of synergy with chemotherapy.
To the best of our knowledge the present study was
the first to identify an association between immunotherapy and
chemotherapy in patients with SCLC. Taken together, the results of
the present study suggest that MICA may be used as a marker to
predict the response to chemotherapy and the prognosis of patients
with SCLC and it may improve sensitivity to cisplatin chemotherapy
via downregulation of ABCG2 expression.
Supplementary Material
Supporting Data
Acknowledgements
The authors of the present study would like to thank
Ms. Suxia Luo from the Affiliated Cancer Hospital of Zhengzhou
University, for valuable guidance in the initial stages of writing
the manuscript. The authors would also like to acknowledge and Ms.
Shujun Yang from the Affiliated Cancer Hospital of Zhengzhou
University for his help in the patient's investigation.
Funding
The present study was funded by the Science and
Technology Program of Henan Province (grant no. 162102310327), the
Natural Science Foundation of Henan Province (grant nos.
182300410297, 182300410297 and 162300410300), the Medical Science
and Technology Program of Henan Province (grant no. 201702249) and
the 51282 Project Leading Talent of Henan Provincial Health Science
and Technology Innovation Talents (grant no. [2016]32).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW wrote the manuscript. YW and HT performed data
analysis. RS, SX, QW and YW conceived the study. RS and SX designed
the study. RW and QW collected the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Zhengzhou University (approval no. 2014079) and
performed in accordance with the The Declaration of Helsinki.
Written informed consent was provided by all patients prior to the
study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NKG2D
|
natural killer cell-activated receptor
natural-killer group-2 member D
|
|
NKG2DLs
|
NKG2D ligands
|
|
ABCG2
|
ATP binding cassette subfamily G
member 2
|
|
PFS
|
progression-free survival
|
|
SCLC
|
small cell lung cancer
|
|
QCR
|
quick chemoresistance
|
|
DCR
|
delayed chemotherapy resistance
|
|
MHC
|
major histocompatibility complex
|
|
MIC
|
MHC class I chain-related protein
|
|
ULBP
|
UL16-binding protein
|
|
DPYD
|
dihydropyrimidine dehydrogenase
|
|
GSTT1
|
glutathione S-transferase θ1
|
|
GSTp1
|
glutathione S-transferase pi 1
|
|
GSTM1
|
glutathione S-transferase mu 1
|
|
ADH1A
|
alcohol dehydrogenase 1A (class I),
alpha polypeptide
|
|
PPARA
|
peroxisome proliferator activated
receptor alpha
|
References
|
1
|
Cao M and Chen W: Epidemiology of lung
cancer in China. Thorac Cancer. 10:3–7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kalemkerian GP: Small Cell Lung Cancer.
Semin Respir Crit Care Med. 37:783–796. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pelton K, Coticchia CM, Curatolo AS,
Schaffner CP, Zurakowski D, Solomon KR and Moses MA:
Hypercholesterolemia induces angiogenesis and accelerates growth of
breast tumors in vivo. Am J Pathol. 184:2099–2110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ott PA, Elez E, Hiret S, Kim DW, Morosky
A, Saraf S, Piperdi B and Mehnert JM: Pembrolizumab in patients
with extensive-stage small-cell lung cancer: Results from the phase
Ib KEYNOTE-028 Study. J Clin Oncol. 35:3823–3829. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fan Y and Mao W: Immune checkpoint
inhibitors in lung cancer: Current status and future directions.
Chin Clin Oncol. 6:172017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tanvetyanon T, Gray JE and Antonia SJ:
PD-1 checkpoint blockade alone or combined PD-1 and CTLA-4 blockade
as immunotherapy for lung cancer? Expert Opin Biol Ther.
17:305–312. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ready N, Farago AF, de Braud F, Atmaca A,
Hellmann MD, Schneider JG, Spigel DR, Moreno V, Chau I, Hann CL, et
al: Third-line nivolumab monotherapy in recurrent SCLC: CheckMate
032. J Thorac Oncol. 14:237–244. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Souza-Fonseca-Guimaraes F, Cursons J and
Huntington ND: The emergence of natural killer cells as a major
target in cancer immunotherapy. Trends Immunol. 40:142–158. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cai X, Caballero-Benitez A, Gewe MM,
Jenkins IC, Drescher CW, Strong RK, Spies T and Groh V: Control of
tumor initiation by NKG2D naturally expressed on ovarian cancer
cells. Neoplasia. 19:471–482. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bauer S, Groh V, Wu J, Steinle A, Phillips
JH, Lanier LL and Spies T: Activation of NK cells and T cells by
NKG2D, a receptor for stress-inducible MICA. Science. 285:727–729.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Diefenbach A, Jamieson AM, Liu SD, Shastri
N and Raulet DH: Ligands for the murine NKG2D receptor: Expression
by tumor cells and activation of NK cells and macrophages. Nat
Immunol. 1:119–126. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cosman D, Mullberg J, Sutherland CL, Chin
W, Armitage R, Fanslow W, Kubin M and Chalupny NJ: ULBPs, novel MHC
class I-related molecules, bind to CMV glycoprotein UL16 and
stimulate NK cytotoxicity through the NKG2D receptor. Immunity.
14:123–133. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eagle RA, Traherne JA, Hair JR, Jafferji I
and Trowsdale J: ULBP6/RAET1L is an additional human NKG2D ligand.
Eur J Immunol. 39:3207–3216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu H, Wang S, Xin J, Wang J, Yao C and
Zhang Z: Role of NKG2D and its ligands in cancer immunotherapy. Am
J Cancer Res. 9:2064–2078. 2019.PubMed/NCBI
|
|
15
|
Lopez-Soto A, Huergo-Zapico L,
Acebes-Huerta A, Villa-Alvarez M and Gonzalez S: NKG2D signaling in
cancer immunosurveillance. Int J Cancer. 136:1741–1750. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lanier LL: NKG2D Receptor and Its Ligands
in Host Defense. Cancer Immunol Res. 3:575–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Inagaki A, Ishida T, Yano H, Ishii T,
Kusumoto S, Ito A, Ri M, Mori F, Ding J, Komatsu H, et al:
Expression of the ULBP ligands for NKG2D by B-NHL cells plays an
important role in determining their susceptibility to
rituximab-induced ADCC. Int J Cancer. 125:212–221. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Travis WD, Brambilla E, Burke AP, Marx A
and Nicholson AG: Introduction to The 2015 World Health
Organization classification of tumors of the lung, pleura, thymus,
and heart. J Thorac Oncol. 10:1240–1242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fan Y, Gan Y, Shen Y, Cai X, Song Y, Zhao
F, Yao M, Gu J and Tu H: Leptin signaling enhances cell invasion
and promotes the metastasis of human pancreatic cancer via
increasing MMP-13 production. Oncotarget. 6:16120–16134. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu Y, Gan Y, Yuan H, Wang Q, Fan Y, Li G,
Zhang J, Yao M, Gu J and Tu H: Enriched environment housing
enhances the sensitivity of mouse pancreatic cancer to
chemotherapeutic agents. Biochem Biophys Res Commun. 473:593–599.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M,
Marshall KA, et al: NCBI GEO: Archive for high-throughput
functional genomic data. Nucleic Acids Res. 37:885–890. 2009.
View Article : Google Scholar
|
|
24
|
Breitkreutz BJ, Stark C, Reguly T, Boucher
L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bähler J,
Wood V, et al: The BioGRID Interaction Database: 2008 update.
Nucleic Acids Res. 36:D637–D640. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brown KR and Jurisica I: Online predicted
human interaction database. Bioinformatics. 21:2076–2082. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Keshava Prasad TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37:767–772. 2009.
View Article : Google Scholar
|
|
27
|
Romero P, Wagg J, Green ML, Kaiser D,
Krummenacker M and Karp PD: Computational prediction of human
metabolic pathways from the complete human genome. Genome Biol.
6:R22005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aranda B, Achuthan P, Alam-Faruque Y,
Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S,
Khadake J, et al: The IntAct molecular interaction database in
2010. Nucleic Acids Res. 38:D525–D531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ceol A, Chatr Arayamontri A, Licata L,
Peluso D, Briganti L, Perfetto L, Castagnoli L and Cesareni G:
MINT, the molecular interaction database: 2009 update. Nucleic
Acids Res. 38:D532–D539. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schaefer CF, Anthony K, Krupa S, Buchoff
J, Day M, Hannay T and Buetow KH: PID: The pathway interaction
database. Nucleic Acids Res. 37:D674–D679. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vastrik I, D'Eustachio P, Schmidt E,
Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S,
Matthews L, et al: Reactome: A knowledge base of biologic pathways
and processes. Genome Biol. 8:R392007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38((Web Server issue)): W214–W220. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu Y, Si R, Tang H, He Z, Zhu H, Wang L,
Fan Y, Xia S, He Z and Wang Q: Cholesterol reduces the sensitivity
to platinum-based chemotherapy via upregulating ABCG2 in lung
adenocarcinoma. Biochem Biophys Res Commun. 457:614–620. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shukla S, Robey RW, Bates SE and Ambudkar
SV: The calcium channel blockers, 1,4-dihydropyridines, are
substrates of the multidrug resistance-linked ABC drug transporter,
ABCG2. Biochemistry. 45:8940–8951. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu Y, Si R, Yang S, Xia S, He Z, Wang L,
He Z, Wang Q and Tang H: Depression induces poor prognosis
associates with the down-regulation brain derived neurotrophic
factor of serum in advanced small cell lung cancer. Oncotarget.
7:85975–85986. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu J, Zhu S, Xia X, Zhang L, Kleinerman ES
and Li S: CD8+T cell-specific induction of NKG2D receptor by
doxorubicin plus interleukin-12 and its contribution to CD8+T cell
accumulation in tumors. Mol Cancer. 13:342014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Raffaghello L, Prigione I, Airoldi I,
Camoriano M, Levreri I, Gambini C, Pende D, Steinle A, Ferrone S
and Pistoia V: Downregulation and/or release of NKG2D ligands as
immune evasion strategy of human neuroblastoma. Neoplasia.
6:558–568. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dhar P and Wu JD: NKG2D and its ligands in
cancer. Curr Opin Immunol. 51:55–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Basher F and Wu JD: NKG2D ligands
in tumor immunity: Two sides of a coin. Front Immunol. 6:972015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Deng W, Gowen BG, Zhang L, Wang L, Lau S,
Iannello A, Xu J, Rovis TL, Xiong N and Raulet DH: Antitumor
immunity. A shed NKG2D ligand that promotes natural killer cell
activation and tumor rejection. Science. 348:136–139. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zen K, Karsan A, Stempien-Otero A, Yee E,
Tupper J, Li X, Eunson T, Kay MA, Wilson CB, Winn RK and Harlan JM:
NF-kappaB activation is required for human endothelial survival
during exposure to tumor necrosis factor-alpha but not to
interleukin-1beta or lipopolysaccharide. J Biol Chem.
274:28808–28815. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Theodoulou FL and Kerr ID: ABC transporter
research: Going strong 40 years on. Biochem Soc Trans.
43:1033–1040. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boswell-Casteel RC, Fukuda Y and Schuetz
JD: ABCB6, an ABC transporter impacting drug response and disease.
AAPS J. 20:82017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gillet JP and Gottesman MM: Advances in
the molecular detection of ABC transporters involved in multidrug
resistance in cancer. Curr Pharm Biotechnol. 12:686–692. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang Y, Penchala S, Pham AN and Wang J:
Genetic variations and gene expression of transporters in drug
disposition and response. Expert Opin Drug Metab Toxicol.
4:237–254. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zinzi L, Capparelli E, Cantore M, Contino
M, Leopoldo M and Colabufo NA: Small and innovative molecules as
new strategy to revert MDR. Front Oncol. 4:22014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Colabufo NA, Berardi F, Contino M, Niso M
and Perrone R: ABC pumps and their role in active drug transport.
Curr Top Med Chem. 9:119–129. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xie N, Mou L, Yuan J, Liu W, Deng T, Li Z,
Jing Y and Hu Z: Modulating drug resistance by targeting BCRP/ABCG2
using retrovirus-mediated RNA interference. PLoS One.
9:e1034632014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
de Boussac H, Orban TI, Varady G, Tihanyi
B, Bacquet C, Brózik A, Váradi A, Sarkadi B and Arányi T:
Stimulus-induced expression of the ABCG2 multidrug transporter in
HepG2 hepatocarcinoma model cells involves the ERK1/2 cascade and
alternative promoters. Biochem Biophys Res Commun. 426:172–176.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Benderra Z, Faussat AM, Sayada L, Perrot
JY, Chaoui D, Marie JP and Legrand O: Breast cancer resistance
protein and P-glycoprotein in 149 adult acute myeloid leukemias.
Clin Cancer Res. 10:7896–7902. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yewdell JW: Not such a dismal science: The
economics of protein synthesis, folding, degradation and antigen
processing. Trends Cell Biol. 11:294–297. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Patel A, Li TW, Anreddy N, Wang DS, Sodani
K, Gadhia S, Kathawala R, Yang DH, Cheng C and Chen ZS: Suppression
of ABCG2 mediated MDR in vitro and in vivo by a novel inhibitor of
ABCG2 drug transport. Pharmacol Res. 121:184–193. 2017. View Article : Google Scholar : PubMed/NCBI
|