Introduction
Pancreatic cancer (PC) is one of the most vicious
malignancies with ~56,770 new cases and 45,750 deaths in the United
States in 2019 (1). It is generally
considered as an incurable disease, with an unsatisfactory
therapeutic outcome following general treatment methods, including
surgery, chemotherapy and radiotherapy (2). The 5-year survival rate of patients
with PC diagnosed between 2008 and 2014 was only 9% in the United
States, representing the lowest survival rate among all cancer
types according to the American Cancer Society (1). Therefore, it is urgent to investigate
the molecular mechanisms underlying PC and to identify more
effective therapeutic strategies and prognostic biomarkers.
The human transcriptome is composed of a
considerable number of protein-coding mRNAs as well as non-coding
transcripts. Long non-coding RNAs (lncRNAs) are transcripts >200
nucleotides in length without protein-coding ability, which possess
vital roles in gene expression regulation at the epigenetic,
transcriptional and post-transcriptional levels (3). MicroRNAs (miRNAs/miRs) are a series of
small non-coding RNAs <22 nucleotides in length originating from
hairpin precursors (4). miRNAs
couple with the 3′-untranslated region of their target genes and
decrease protein expression by inducing mRNA degradation, or by
inhibiting mRNA translation (5).
However, the biological and clinical roles of lncRNAs and miRNAs in
PC are not thoroughly understood, and require further
investigation. A common functional mechanism of lncRNAs in cancer
cells is to couple with their target miRNAs, leading to silencing
of downstream genes, which is generally known as the ‘competing
endogenous RNA (ceRNA)’ theory (6).
Construction of a ceRNA network using differentially expressed
transcripts in cancer could help to identify biomarkers or
therapeutic targets for PC.
LINC00460, located on chromosome 13:
106376564-106378595, has been reported to promote the progression
in numerous types of cancer, including nasopharyngeal carcinoma
(7), lung cancer (8), gastric cancer (9) and esophageal squamous cell carcinoma
(10). However, it remains unclear
whether LINC00460 is dysregulated and serves an important role in
PC tumorigenesis. In the present study, The Cancer Genome Atlas
(TCGA) database was used to construct a ceRNA network containing
differentially expressed lncRNAs, miRNAs and mRNAs. Furthermore,
tissue samples were collected and in vitro experiments were
performed to validate the pivotal lncRNA predicted in the ceRNA
network, LINC00460.
Materials and methods
Bioinformatic analysis
The expression profiles of mRNAs and miRNAs were
obtained from the TCGA-PDAC dataset (http://cancergenome.nih.gov/) (11), which contained 146 PC and 3 normal
samples. Based on the lncRNA information in the GENECODE database
(https://www.gencodegenes.org/), the
lncRNA expression profile was extracted from the mRNA dataset. The
package ‘edgeR’ v3.28.0 (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html)
of the R software v3.6.1 (https://www.r-project.org/) was utilized to screen out
differentially expressed transcripts using the threshold values of
|log2 fold change|>1 and P<0.05. The interactions
between lncRNAs and miRNAs were predicted using miRcode (http://www.mircode.org/index.php). Three
databases, including miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php),
miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/vert_72/), were used to
predict targeted mRNAs of miRNAs. All differentially expressed
mRNAs, miRNAs and lncRNAs were used to construct a ceRNA network
that was visualized and modified using the Cytoscape software
v3.7.0 (12). The prognostic value
of lncRNAs for the overall survival of patients with PC was
evaluated using a log-rank test through the Gene Expression
Profiling Interactive Analysis (GEPIA) website (http://gepia.cancer-pku.cn/index.html)
(13).
Tissue samples
The present study was approved by the Research
Ethics Committee of Anhui No. 2 Provincial People's Hospital
(Hefei, China). Written informed consent was obtained from all
subjects. A total of 59 patients with PC (35 males and 24 females,
aged between 52 and 75 years) receiving surgical excision therapy
at the aforementioned hospital, and admitted between March 2016 and
December 2017, were enrolled in the present study. All patients had
pathologically confirmed pancreatic ductal adenocarcinoma, and none
of the patients received preoperative adjuvant chemotherapy or
radiotherapy. All patients received standard gemcitabine-based
postoperative adjuvant chemotherapy. The last follow-up time point
was July 16, 2019. The cancer and adjacent normal tissues (>1 cm
from the margin of the cancer tissue) were collected from the
excised surgical specimens and stored in sterile enzyme-free tubes.
They were immediately frozen in liquid nitrogen and stored at −80°C
until subsequent experimentation. The general and
clinicopathological characteristics of the patients were recorded
using electronic health records.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from cancer and normal tissues was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). The RNA concentration and purity were
measured using the NanoDrop 2000 (Thermo Fisher Scientific, Inc.).
RT was performed using the PrimeScript™ RT reagent kit with gDNA
Eraser (Takara Biotechnology Co., Ltd.) at 37°C for 15 min and 85°C
for 5 sec to obtain cDNA. The quantitative measurement of LINC00460
expression was analyzed using the Applied Biosystems 7900 system
with the One-Step TB Green® PrimeScript™ PLUS RT-PCR kit
(Takara Biotechnology Co., Ltd.) with the following thermocycling
conditions: 42°C for 5 min and 95°C for 10 sec, followed by 40
cycles of 95°C for 3 sec and 60°C for 30 sec. The following primers
were used: GAPDH forward, 5′-TCGGAGTCAACGGATTTGGT-3′ and reverse,
5′-TTGGAGGGATCTCGCTCCT-3′; LINC00460 forward,
5′-ACAGCATGAGCCAGGACATC-3′ and reverse, 5′-GAAAGCTGCAACATGCTCCC-3′.
LINC00460 expression was normalized to that of GAPDH, and the
relative LINC00460 expression level was determined using the
2−ΔΔCq method (14).
Cell culture and small interfering RNA
(siRNA) transfection
The human PC PANC1 and SW1990 cell lines were
purchased from Wuhan Boster Biological Technology, Ltd., and were
cultured in DMEM supplemented with 10% FBS (both Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin at 37°C with 5%
CO2. The siRNA oligos (si-LINC00460;
5′-GUGUCAACAACCUGUUUAAUU-3′) and the negative control (si-NC;
5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Guangzhou RiboBio
Co., Ltd. PANC1 and SW1990 cells were transfected with 100 nM siRNA
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Subsequent experiments were conducted 48 h after siRNA
transfection.
Cell viability assay
A total of 1×103 cells/well were
incubated for 24, 48 and 72 h at 37°C. At the aforementioned time
points, MTT was added to the culture medium to form a 0.5 mg/ml MTT
solution. The spent culture medium was discarded 4 h later and
replaced with 100 µl DMSO to dissolve the purple formazan. After
mixing the plate gently for 10 min, the formazan crystals were
dissolved. Cell viability was analyzed at a wavelength of 570 nm
using the BioTek microplate reader (Agilent Technologies,
Inc.).
Colony formation assay
A total of 300 cells/well were uniformly seeded into
a 6-well plate. After 24 h, the medium was replaced and the cells
were incubated with 2 ml culture medium for a further 2 weeks. The
medium was then discarded, and the cells were washed twice with
PBS. The cells were stained using Giemsa stain for 30 min at room
temperature and the colony number in each well was counted manually
under a light microscope (magnification, ×4).
Statistical analysis
All statistical analyses were performed using SPSS
v19.0 (IBM Corp.). Data are presented as the mean ± SD (n=3).
Differences in the expression levels between the cancer tissues and
adjacent normal tissues were analyzed using a paired Student's
t-test. For ≥3 groups, one-way ANOVA followed by Fisher's least
significant difference post-hoc test were performed. The
χ2 test was used to assess the association between
LINC00460 expression and the clinicopathological features of
patients with PC. The Cox regression model was used for univariate
and multivariate analyses, in which clinicopathological features
(age, sex, differentiation, tumor size, lymph node metastasis and
vascular invasion) served as covariates. Kaplan-Meier analysis was
used to evaluate the prognostic value of LINC00460 expression.
LINC00460 expression higher than the average was defined as high
expression, while LINC00460 expression lower than the average was
defined as low expression. Pearson correlation was applied to
analyze the correlation between Ki-67 and LINC00460 expression.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Bioinformatic analysis identifies
LINC00460 as a key lncRNA in PC
To identify prognostic biomarkers for PC, an
integrated bioinformatics analysis was conducted using PC data from
the TCGA. This analysis identified differentially expressed mRNAs
(476 upregulated and 436 downregulated; Fig. 1A), miRNAs (12 upregulated and 13
downregulated; Fig. 1B) and lncRNAs
(142 upregulated and 218 downregulated; Fig. 1C) in cancer tissues, compared with
normal tissues. miRcode identified 18 differentially expressed
lncRNAs (AC005280.1, GRIK1-AS1, AC020907.1, WT1-AS, LINC00365,
COL4A2-AS2, LINC00092, SACS-AS1, MIR205HG, VIPR1-AS1, IDI2-AS1,
LINC00460, TM4SF19-AS1, NEXN-AS1, EGOT, ZNF295-AS1, SYNPR-AS1 and
PVT1) and seven differentially expressed lncRNA-targeted miRNAs
(hsa-mir-424, hsa-mir-206, hsa-mir-503, hsa-mir-192, hsa-mir-383,
hsa-mir-100 and hsa-mir-215). miRTarBase, TargetScan and the miRDB
database identified 13 differentially expressed miRNA-targeted
mRNAs: Chromobox 2, gap junction protein alpha 1, cell division
cycle 25A, checkpoint kinase 1, cyclin D1 (CCND1), osteoclast
associated Ig-like receptor, centrosomal protein 55, protein
tyrosine phosphatase receptor type D, kinesin family member 23,
homeobox A10, E2F transcription factor 7, FH2 domain containing 1
and cyclin E1. Using these significantly upregulated or
downregulated transcripts in PC, a ceRNA network was constructed
using the Cytoscape platform (Fig.
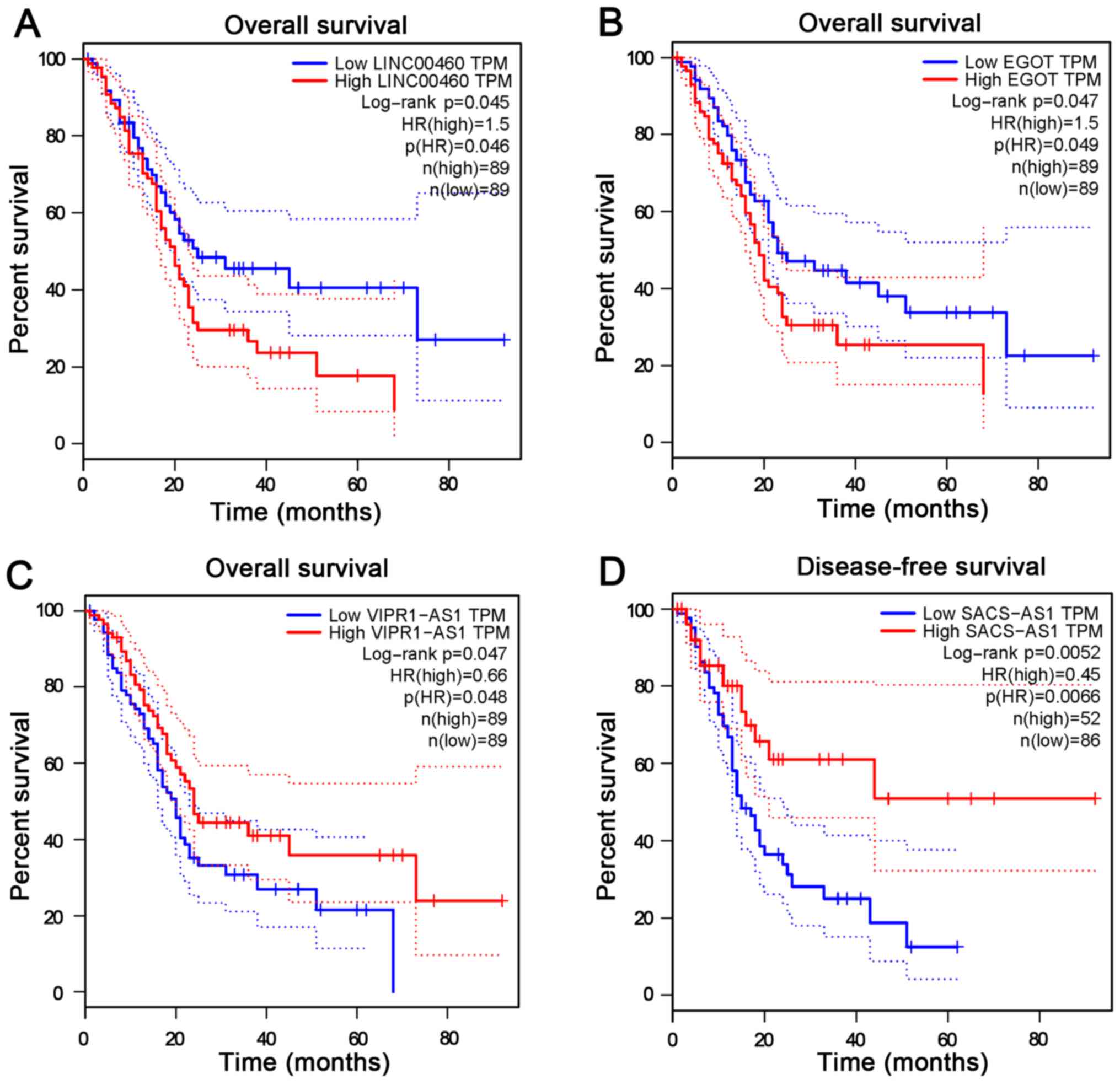
1D). The prognostic significance of lncRNAs involved in the
network was evaluated using the GEPIA platform. The results
revealed that expression levels of LINC00460 (Fig. 2A), EGOT (Fig. 2B), VIPR1-AS1 (Fig. 2C) and SACS-AS1 (Fig. 2D) were significantly associated with
the survival rate of patients with PC (P<0.05). Among these four
prognosis-associated lncRNAs, the fold change of LINC00460
expression was the highest (3.47 times higher than the normal
control; data not shown). Therefore, LINC00460 was selected as the
target to be validated in the PC cohort enrolled in the present
study.
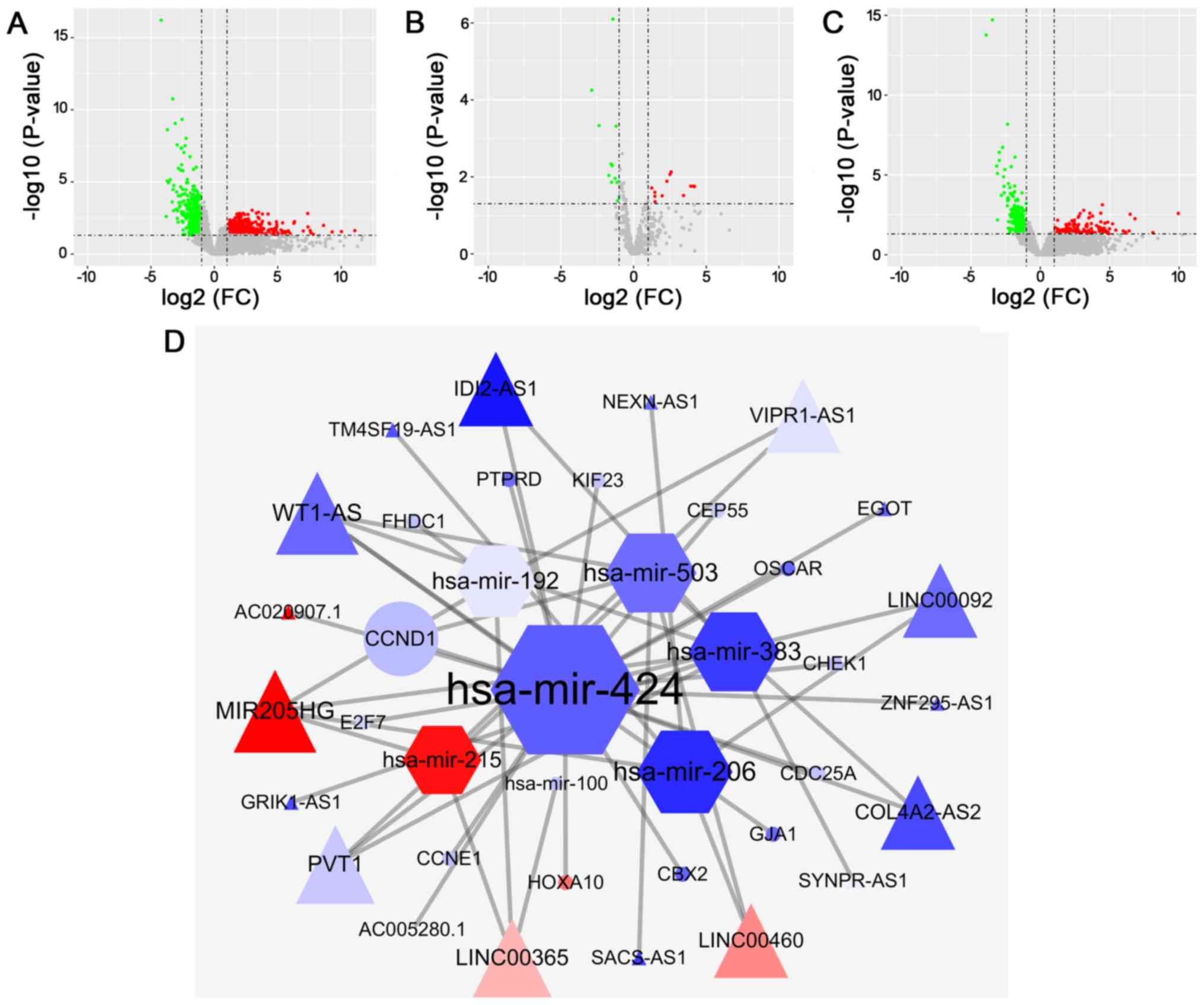 | Figure 1.LINC00460 is a key lncRNA in PC
according to bioinformatics analysis. Volcano plots of
differentially expressed (A) mRNAs, (B) miRNAs and (C) lncRNAs in
PC, compared with normal tissues. (D) Competing endogenous RNA
network constructed using differentially expressed mRNAs (circles),
miRNAs (hexagons) and lncRNAs (triangles) based on miRcode,
miRTarBase, miRDB and TargetScan. The red and blue in the network
represent upregulated and downregulated transcripts, respectively.
PC, pancreatic cancer; miRNA, microRNA; lncRNA, long non-coding
RNA; FC, fold change. |
LINC00460 is an independent prognostic
biomarker for PC
To test the results of the bioinformatic analysis,
LINC00460 expression was examined in 59 paired PC and normal
tissues. The expression levels of LINC00460 quantified by RT-qPCR
were significantly upregulated in cancer tissues compared with
those in normal tissues (P<0.001; Fig. 3A). Additionally, the Kaplan-Meier
plot revealed that high LINC00460 expression predicted worse
survival of patients with PC compared with low expression
(P<0.05; Fig. 3B), which was
consistent with the predicted result from GEPIA. LINC00460
expression and clinical parameters of patients are summarized in
Table I. The expression levels of
LINC00460 were statistically associated with tumor size
(P<0.05), but not with age, sex, differentiation, vascular
invasion, lymph node metastasis and tumor stage. The univariate
analysis revealed that lymph node metastasis [hazard ratio (HR),
0.721; 95% CI, 0.545–0.898; P<0.01], tumor size (HR, 0.640; 95%
CI, 0.442–0.925; P<0.01) and expression levels of LINC00460 (HR,
0.698; 95% CI, 0.433–0.916; P<0.05) were prognostic parameters
for PC. Furthermore, multivariate analysis confirmed that LINC00460
expression (HR, 0.751; 95% CI, 0.518–0.993; P<0.05), tumor size
(HR, 0.724; 95% CI, 0.479–0.962; P<0.05) and lymph node
metastasis (HR, 0.848; 95% CI, 0.663–0.975; P<0.05) were
independent prognostic biomarkers for PC (Table II).
 | Table I.Association between LINC00460
expression and clinical parameters of 59 patients with pancreatic
cancer. |
Table I.
Association between LINC00460
expression and clinical parameters of 59 patients with pancreatic
cancer.
|
| LINC00460, n |
|
|---|
|
|
|
|
|---|
| Parameters | Low expression | High expression | P-value |
|---|
| Age, years |
|
| 0.358 |
| ≤60 | 12 | 17 |
|
|
>60 | 16 | 14 |
|
| Sex |
|
| 0.205 |
| Male | 19 | 16 |
|
|
Female | 9 | 15 |
|
| Differentiation |
|
Well/moderate | 18 | 14 | 0.141 |
| Poor | 10 | 17 |
|
| Tumor size, cm |
|
| 0.028a |
| ≤4 | 17 | 10 |
|
|
>4 | 11 | 21 |
|
| Lymph node
metastasis |
|
| 0.500 |
| N0 | 11 | 14 |
|
| N1 | 19 | 17 |
|
| Vascular
invasion |
|
| 0.253 |
| No | 15 | 12 |
|
|
Yes | 13 | 19 |
|
| Stage |
|
I–IIA | 11 | 14 | 0.500 |
|
IIB-IV | 19 | 17 |
|
 | Table II.Univariate and multivariate analysis
identifying prognostic biomarkers for pancreatic cancer. |
Table II.
Univariate and multivariate analysis
identifying prognostic biomarkers for pancreatic cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years (≤60 vs.
>60) | 0.893 | 0.668–1.314 | 0.452 |
|
|
|
| Sex (male vs.
female) | 1.371 | 0.875–1.585 | 0.527 |
|
|
|
| Differentiation
(well/moderate vs. poor) | 1.065 | 0.881–1.242 | 0.468 |
|
|
|
| Tumor size, cm (≤4
vs. >4) | 0.640 | 0.442–0.925 | 0.007 | 0.724 | 0.479–0.962 | 0.012 |
| Lymph node
metastasis (N0 vs. N1) | 0.721 | 0.545–0.898 | 0.005 | 0.848 | 0.663–0.975 | 0.020 |
| Vascular invasion
(no vs. yes) | 0.855 | 0.710–1.245 | 0.496 |
|
|
|
| LINC00460
expression (low vs. high) | 0.698 | 0.433–0.916 | 0.022 | 0.751 | 0.518–0.993 | 0.039 |
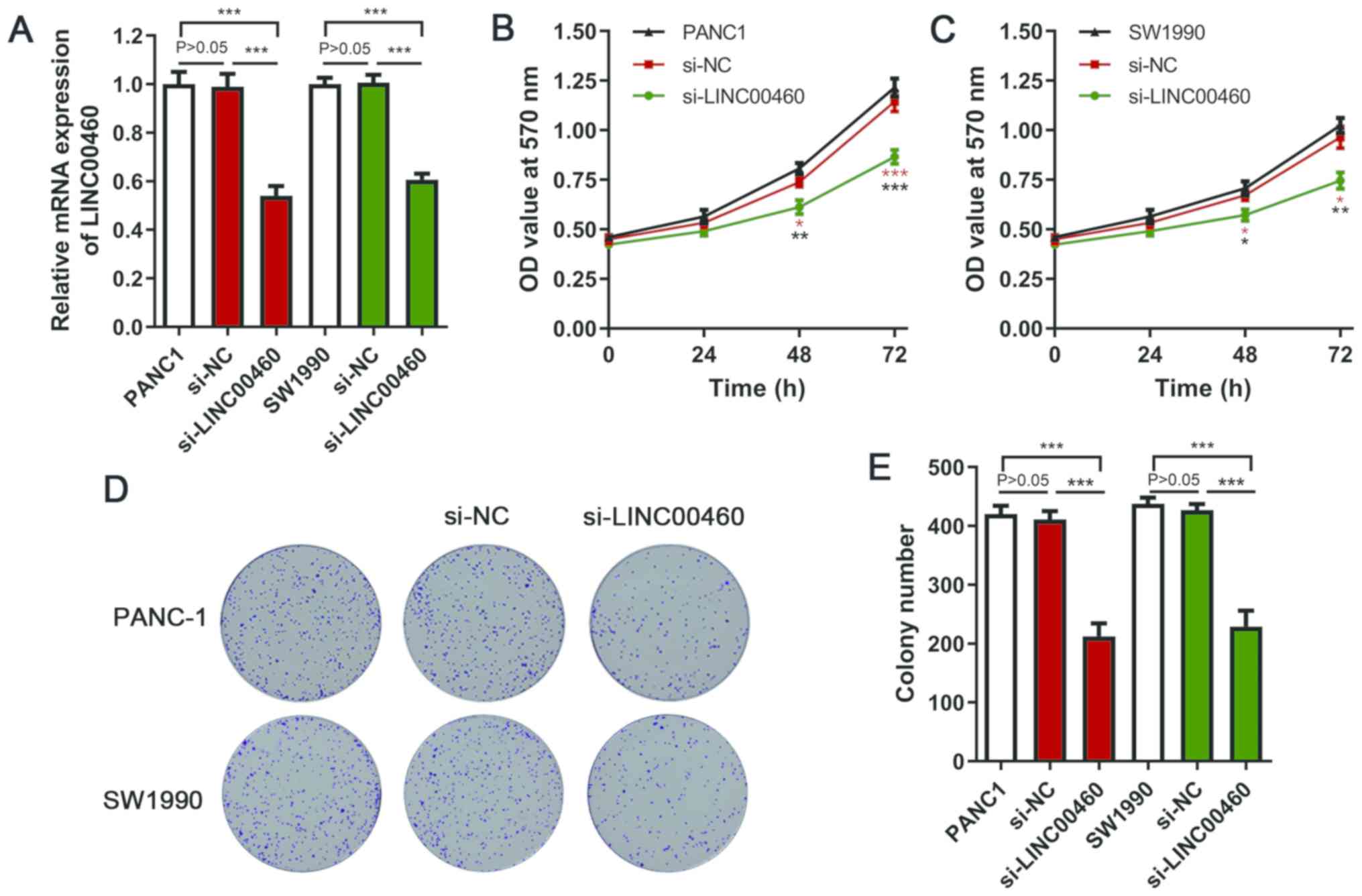
Silencing LINC00460 inhibits PC cell
proliferation
The ceRNA network demonstrated that LINC00460 may
act as a molecular sponge of miR-503 and further regulate CCND1,
which has been recognized as an oncogene in different types of
cancer (15,16). Therefore, it was hypothesized that
LINC00460 may regulate PC cell proliferation. In PC tissues, high
expression levels of LINC00460 were significantly associated with
high Ki67 expression (r=0.343; P<0.05; Fig. 3C), which reflected the proliferative
potential of cells. siRNA transfection significantly suppressed
LINC00460 expression in PANC-1 and SW1990 cells (Fig. 4A). The viability of PANC-1 and SW1990
cells in the si-LINC00460 groups was significantly decreased
compared with that in the si-NC and blank groups at 48 and 72 h
(Fig. 4B and C). As shown in
Fig. 4D, silencing LINC00460
expression notably reduced colony numbers, with statistical
significance identified compared with the si-NC and blank groups
(Fig. 4E). These in vitro
results suggest that LINC00460 may have oncogenic ability and may
regulate the proliferation of PC cells.
Discussion
Exploring specific biomarkers to improve survival
prediction and therapeutic strategies for PC is an urgent
requirement. Accumulating evidence has demonstrated that lncRNAs
regulate various critical biological functions, including
transcription (17), RNA processing
(18) and chromatin organization
(19). Additionally, lncRNAs exert
vital functions in the modulation of a large number of genes that
regulate cancer cell proliferation, invasion, apoptosis and
stemness (20,21). Exploring key lncRNAs associated with
pathological parameters and the survival of patients with PC may
help to identify prognostic biomarkers and vital molecules involved
in tumorigenesis. In the present study, a regulatory network of
differentially expressed miRNAs, mRNAs and lncRNAs was constructed,
which revealed potential molecular mechanisms, as well as helped to
identify pivotal biomarkers for PC.
The clinical and biological roles of LINC00460,
which is located on the human chromosome 13q33.2, have been
validated in several types of cancer. Zhang et al (22) demonstrated that LINC00460 expression
is notably increased in colorectal cancer and is significantly
associated with clinicopathological parameters and a shorter
patient survival time; additionally, they demonstrated that
LINC00460 targets the miR-939-5p/LIM domain kinase 2 axis to
facilitate colorectal cancer cell progression. In glioma, LINC00460
has been demonstrated to be overexpressed in cancer tissues and
cell lines, and to promote cancer cell progression by negatively
regulating miR-320a (23).
Additionally, a recent study reported that LINC00460 could promote
breast cancer cell migration, invasion and proliferation by
targeting the miR-489-5p/fibroblast growth factor 7/AKT axis
(24). Notably, nicotine has been
found to facilitate carcinogenesis in the bronchus by activating
LINC00460 and the PI3K/AKT signaling pathway (25). In the present study, the prognostic
value of LINC00460 in PC was first predicted using the TCGA
database and was subsequently examined in a cohort of patients with
PC. It was demonstrated that high expression levels of LINC00460
were associated with poor overall survival and that LINC00460 was
an independent prognostic biomarker for patients with PC.
It has been reported that the binding of miRNAs to
lncRNAs is able to decrease miRNA levels, resulting in the
overexpression of miRNA-targeted genes (26). The ceRNA PC network constructed in
the present study may help to comprehensively understand the
regulatory association between differentially expressed mRNAs and
non-coding RNAs. Bioinformatics prediction revealed that in PC,
LINC00460 was likely to couple with miR-503 to further regulate
CCND1, a well-recognized oncogene that regulates the cell cycle and
cellular proliferation (27,28). This prediction is in accordance with
the in vitro cell experiments of the present study, which
indicated that LINC00460 modulated cell viability in PANC-1
cells.
However, the current study presents several
limitations. Firstly, pathological confirmation of the
non-cancerous tissues was not performed, which may have led to
inaccurate expression levels of LINC00460. Secondly, the
bioinformatics prediction of the ceRNA network was not confirmed by
in vitro experiments. Thirdly, LINC00460 expression was only
evaluated in association with disease-free survival, and no
follow-up of the enrolled patients was conducted for overall
survival analysis.
In summary, the present study identified LINC00460
as an independent prognostic biomarker for PC and suggests that it
may serve as an oncogenic lncRNA that promotes PC cell
proliferation. Further in depth exploration is required to reveal
the specific mechanism of LINC00460 in PC cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JS performed the bioinformatics analysis and wrote
the manuscript. JY and KL helped to collect the data and performed
the in vitro experiments. JG designed the study and drafted
and revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Anhui No. 2 Provincial People's Hospital
(Hefei, China). Written informed consent was obtained from all
subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeSantis CE, Miller KD, Goding Sauer A,
Jemal A and Siegel RL: Cancer statistics for African Americans,
2019. CA Cancer J Clin. 69:211–233. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chiorean EG and Coveler AL: Pancreatic
cancer: Optimizing treatment options, new, and emerging targeted
therapies. Drug Des Devel Ther. 9:3529–3545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mansoori B, Mohammadi A, Shirjang S and
Baradaran B: MicroRNAs in the diagnosis and treatment of cancer.
Immunol Invest. 46:880–897. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kong YG, Cui M, Chen SM, Xu Y, Xu Y and
Tao ZZ: LncRNA-LINC00460 facilitates nasopharyngeal carcinoma
tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene.
639:77–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y,
Zhou JK, Guo C, Xia Z, Liu L, et al: Long non-coding RNA linc00460
promotes epithelial-mesenchymal transition and cell migration in
lung cancer cells. Cancer Lett. 420:80–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang F, Liang S, Liu X, Han L, Wang J and
Du Q: LINC00460 modulates KDM2A to promote cell proliferation and
migration by targeting miR-342-3p in gastric cancer. OncoTargets
Ther. 11:6383–6394. 2018. View Article : Google Scholar
|
|
10
|
Liang Y, Wu Y, Chen X, Zhang S, Wang K,
Guan X, Yang K, Li J and Bai Y: A novel long noncoding RNA
linc00460 up-regulated by CBP/P300 promotes carcinogenesis in
esophageal squamous cell carcinoma. Biosci Rep. 37:BSR201710192017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cancer Genome Atlas Research Network.
Electronic address, . andrew_aguirre@dfci.harvard.edu; Cancer
Genome Atlas Research Network: Integrated genomic characterization
of pancreatic ductal adenocarcinoma. Cancer Cell. 32:185–203.e13.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ai B, Kong X, Wang X, Zhang K, Yang X,
Zhai J, Gao R, Qi Y, Wang J, Wang Z, et al: LINC01355 suppresses
breast cancer growth through FOXO3-mediated transcriptional
repression of CCND1. Cell Death Dis. 10:5022019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qie S and Diehl JA: Cyclin D1, cancer
progression, and opportunities in cancer treatment. J Mol Med
(Berl). 94:1313–1326. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dykes IM and Emanueli C: Transcriptional
and post-transcriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han P and Chang CP: Long non-coding RNA
and chromatin remodeling. RNA Biol. 12:1094–1098. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schmitt AM and Chang HY: Long Noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Flynn RA and Chang HY: Long noncoding RNAs
in cell-fate programming and reprogramming. Cell Stem Cell.
14:752–761. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Liu X, Li Q and Zhang Y: lncRNA
LINC00460 promoted colorectal cancer cells metastasis via
miR-939-5p sponging. Cancer Manag Res. 11:1779–1789. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng L, Rao M, Zhou Y, Zhang Y and Zhu Y:
Long noncoding RNA 00460 (LINC00460) promotes glioma progression by
negatively regulating miR-320a. J Cell Biochem. 120:9556–9563.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu Y, Yang L, Chong QY, Yan H, Zhang W,
Qian W, Tan S, Wu Z, Lobie PE and Zhu T: Long noncoding RNA
Linc00460 promotes breast cancer progression by regulating the
miR-489-5p/FGF7/AKT axis. Cancer Manag Res. 11:5983–6001. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao H, Wang Y and Ren X: Nicotine
promotes the development of non-small cell lung cancer through
activating LINC00460 and PI3K/Akt signaling. Biosci Rep.
39:BSR201824432019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Awan HM, Shah A, Rashid F and Shan G:
Primate-specific Long Non-coding RNAs and MicroRNAs. Genomics
Proteomics Bioinformatics. 15:187–195. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang R, Lin JY and Chi YJ: MiR-519d
reduces the 5-fluorouracil resistance in colorectal cancer cells by
down-regulating the expression of CCND1. Eur Rev Med Pharmacol Sci.
22:2869–2875. 2018.PubMed/NCBI
|
|
28
|
Zhu Y, Wen X and Zhao P: MicroRNA-365
inhibits cell growth and promotes apoptosis in melanoma by
targeting BCL2 and Cyclin D1 (CCND1). Med Sci Monit. 24:3679–3692.
2018. View Article : Google Scholar : PubMed/NCBI
|