Introduction
Ovarian cancer is one of the most malignant types of
gynecological cancer, and is the 11th most common type of cancer
among women, as well as the 5th leading cause of cancer-associated
mortality in the USA (1). In
addition, ovarian cancer is the leading cause of gynecological
malignancy-associated mortality (1,2). The
American Cancer Society estimated there were 22,530 new cases of
ovarian cancer and 13,980 mortalities from ovarian cancer in the
USA in 2019 (1). Furthermore, the
incidence of new ovarian cancer cases has been decreasing on
average by 2.5% each year in the past decade; however, the overall
survival rate has not improved in recent years (2). The current 5-year survival rate for all
the stages of ovarian cancer cases in the US is approximately 47%
(1). However, approximately 60% of
the new cases are diagnosed at advanced stages, and in those cases,
the 5-year survival rate is only 29% (1). Moreover, there is a high rate of
recurrence even after aggressive multimodal treatment, which
further worsens the prognosis (3).
Ovarian cancer belongs to a group of heterogeneous
tumors that arise spontaneously largely from the ovaries, but may
evolve from various other potential sources (4–6). In
addition, ovarian cancer can be morphologically classified into
epithelial and non-epithelial types, of which 80–90% of all ovarian
cancer cases are epithelial type (5). Based on their aggressiveness,
epithelial ovarian cancers (EOCs) are further subdivided into high-
and low-grade categories; or morphologically, they are subdivided
into serous, endometrioid, mucinous and clear cell varieties
(7,8). High-grade serous ovarian cancer (HGSOC)
accounts for 50–60% of all ovarian neoplasms (8). Furthermore, advanced HGSOC accounts for
approximately 50% of all EOCs (7–9), but the
precise etiological factors underlying ovarian cancer have not been
fully elucidated. However, hereditary susceptibility is considered
an important risk factor, as approximately 35% of HGSOC cases
harbor a germline mutation of the tumor suppressor genes Breast
cancer type 1 susceptibility protein (BRCA1) or BRCA2
(10).
The current therapeutic measure used to treat
ovarian cancer is a multimodal regimen, and a combination of
platinum and paclitaxel is used as the primary chemotherapeutic
regimen (11). However, the relapse
rate remains high due to chemoresistance (12). Poly ADP-ribose polymerase inhibitor
(PARPi) has been introduced as a promising therapeutic agent to
improve the prognosis of HGSOC (13). Olaparib is the most commonly used
PARPi, and exhibits favorable outcomes in lowering disease
progression and mortality rates (14). Moreover, olaparib has been approved
by the Food and Drug Administration (FDA) as the first monotherapy
to combat advanced epithelial ovarian cancer cases harboring
germline BRCA mutations (15).
IQ motif containing GTPase Activating Proteins
(IQGAPs) are a family of GTPase activating proteins, which
have been evolutionarily conserved from yeast to mammals (16,17). In
a review by Hedman et al (18), the varied functions of IQGAPs,
in addition to serving as scaffolding proteins are discussed. In
total, three members of the IQGAP family have been described
in humans (18). Furthermore, all
three members are equipped with four IQ motifs and a Ras
GTPase-activating protein (GAP)-related domain (18); the GAP-related domain of
IQGAPs mediates its binding to the Rho family of GTPases
(19). A member of the Rho family of
GTPases, Cell Division Cycle 42 (CDC42) has been revealed to
serve critical roles in cell proliferation, survival, adhesion and
migration, and is correlated with a less favorable prognosis in
various types of cancer (20–23). Of
the three IQGAP family members, IQGAP1 has been
reported to play a synergistic role in cancer progression and aid
in cellular motility (24–26). However, IQGAP2 exhibits a
tumor suppressive function (26).
Moreover, IQGAP3 is hypothesized to be involved in the
proliferation of epithelial cells (27), and is a novel member of the
IQGAP family, which was discovered in 2007 (28). IQGAP3 is located on chromosome
1 at 1q21.3 loci and has been reported to act as an oncogene in
several types of cancer (29–35).
Furthermore, IQGAP3 is a transmembrane protein, and has been
speculated to be a potential therapeutic target (35).
The present study aimed to analyze the differential
expression of IQGAP3 in HGSOC and healthy tissues, and the
effect of IQGAP3 knockdown on various functional processes,
such as cell proliferation, migration, invasion and apoptosis, to
determine whether IQGAP3 could serve as a potential
oncogenic prognostic and therapeutic target for patients with
HGSOC.
Materials and methods
Tissue samples
A total of 149 ovarian cancer tissue samples
(patient age range, 34–79 years; median age, 56 years) and 64
healthy fallopian tube epithelial tissues (patient age range, 26–74
years; median age, 47 years) with detailed clinical information
were collected from the Pathology Department at Qilu Hospital of
Shandong University (Ji'nan, China) between January 2005 and
January 2015. All the malignant samples were diagnosed in
accordance with the International Federation of Gynecology and
Obstetrics criteria (36). The
healthy samples were collected from patients who underwent surgery
for benign conditions. Signed consents were collected from all the
patients and the study was approved by the Ethics Committee of Qilu
Hospital of Shandong University.
Survival analysis was performed on datasets from the
Gene Expression Omnibus (GEO) database, including 523 patients for
overall survival analysis using datasets GSE18520 (37), GSE26193 (38), GSE30161 (39), GSE63885 (40) and GSE9891 (41), and 483 patients for the
progression-free survival analysis using datasets GSE26193,
GSE30161, GSE63885, GSE9891, GSE65986 (42) on Kaplan-Meier Plotter (43).
Cell lines and cell culture
Human ovarian cancer cells A2780 (cat. no. CL-0013;
Procell Life Science & Technology Co., Ltd.) were cultured in
RPMI-1640 medium supplemented with 10% FBS and penicillin (100
IU/ml) and streptomycin (100 µg/ml) (all Gibco; Thermo Fisher
Scientific, Inc.). HEY cells (gifted from Dr Jianjun Wei;
Laboratory at Northwestern University) were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
All the cells were maintained in a humidified incubator at 37°C
with 5% CO2.
Immunohistochemistry (IHC)
IHC staining of the tissue microarray (TMA) was
performed on 4-µm sections sliced from each TMA receiver block
fixed with 4% paraformaldehyde at room temperature for 48 h and
embedded in paraffin. Tissue slides were deparaffinized in xylene
and rehydrated in a graded series of ethanol (10 min each in 100,
95, 80 and 70% ethanol). Antigen retrieval was performed using a
heat-induced epitope retrieval method with 10 mmol/l EDTA buffer
(pH 8.0) at 98°C for 15 min. Endogenous peroxidase activity was
quenched with 3% hydrogen peroxide in methanol for 15 min at 37°C,
and non-specific binding was blocked by incubation with donkey
serum as part of the SP9000 IHC kit (OriGene Technologies, Inc.;
cat. no. SP9000) for 30 min at 37°C. The slides were subsequently
incubated overnight at 4°C in a humid chamber with
anti-IQGAP3 (Abcam; cat. no. ab219354) antibody at a
dilution of 3 µg/ml. Staining was visualized using I–View
3,3′-diaminobenzidine staining detection system (OriGene
Technologies, Inc.; cat. no. ZLI-9018). The IHC score was
determined using a semi-quantitative method based on the extent and
intensity of positively stained cells. The percentage of positive
cells within each sample was scored independently from 0 to 100%
upon observation under a light microscope (magnification, ×10). The
intensity of immunostaining was graded as follows: 0, Negative; 1,
weak; 2, moderate; and 3, strong. The final IHC score was generated
by multiplying the percentage extent with the staining intensity
score. Then, two gynecological pathologists independently reviewed
the IHC staining. High IQGAP3 expression grade was defined
as a final IHC score ≥100.
Stable and transient transfection
For stable transfection, lentiviral vector GV493
(hU6-MCS-CBh-gcGFP-IRES-puromycin) was packaged with IQGAP3
short hairpin (sh)RNA along with the respective negative control
(NC), which were purchased from Shanghai GeneChem Co., Ltd. A total
of 1×105 cells were plated into 6-well plates 24 h prior
to stable transfection. Multiplicity of infection (MOI) was
determined and the lentivirus was added to the culture medium
complemented with the transfection reagent HiTransGA (Shanghai
GeneChem Co., Ltd.) with a MOI value of 20–50. After 24 h
incubation, the medium was replaced with fresh culture medium
containing 2 µg/ml puromycin (Sigma-Aldrich; Merck KGaA) for
selection of the stably transfected colonies.
Transient transfection was performed using small
interfering (si)RNAs purchased from Shanghai GenePharma Co., Ltd.
at a concentration of 20 µM. RNAi-mediated knockdown was performed
with the following siRNAs: si-IQGAP3−1,
5′-GGCAGAAACUAGAAGCAUA-3′; si-IQGAP3−2,
5′-GAGCCAACCAGGACACUAA-3′; si-CDC42,
5′-GGACGGAUUGAUUCCACAU-3′; and si-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′.
Cells were transfected with Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The subsequent experiments were performed
24–48 h after transfection.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from tissue samples and
cultured cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
mRNA was then reverse-transcribed into cDNA using PrimeScript cDNA
Synthesis kit (Takara Bio, Inc.) at 37°C for 1 h and then at 85°C
for 5 min according to the manufacturer's protocol. qPCR was
performed using SYBR-Green Premix Ex Taq II (Takara Bio, Inc.) with
a StepOne Plus RT PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The reaction conditions were as follows: Initial
denaturation at 95°C for 5 sec, followed by 40 cycles of annealing
at 60°C for 10 sec and an extension at 72°C for 30 sec. β-actin was
used as the endogenous control. The primers were designed based on
the GeneBank sequences. The primer sequences used were:
IQGAP3 forward, 5′-GTGCAGCGGATCAACAAAGC-3′ and reverse,
5′-ACGATGCAACAGGGTACACTG-3′; and β-actin forward,
5′-GAGGCACTCTTCCAGCCTTC-3′ and reverse, 5′-GGATGTCCACGTCACATTC-3′.
The comparative threshold cycle method, 2−ΔΔCq, was used
to calculate the relative gene expression level (44).
Western blotting
Cells were harvested and lysed in RIPA lysis buffer
(Beyotime Institute of Biotechnology) with PMSF (1%) and NaF (1%).
Protein samples were incubated for 30 min on ice and cell debris
were removed by centrifugation at 12,000 × g at 4°C for 15 min. The
protein concentration was determined using a bicinchoninic acid
assay kit (Thermo Fisher Scientific, Inc.). Protein samples (30 µg)
were separated by SDS-PAGE (5% stacking gel and 10% separating gel)
and transferred to a PVDF membrane (EMD Millipore) using a Bio-Rad
Trans-blot system (Bio-Rad Laboratories, Inc.). After blocking with
5% skimmed milk for 1 h at room temperature, the membrane was
incubated overnight at 4°C with the primary antibodies. The
membranes were then rinsed with TBST (0.1% Tween-20) followed by
incubation with a horseradish peroxidase-conjugated secondary
antibody for 1 h at room temperature. Signals were detected using
enhanced chemiluminescence (PerkinElmer, Inc.) with ImageQuant LAS
4000 (GE Healthcare Life Sciences). β-actin was used as the
endogenous control. Densitometry analysis was performed using
ImageJ version 1.52 g (National Institutes of Health).
The antibodies used were: Rabbit anti-human
IQGAP3 (1:1,000; Abcam; cat. no. ab219354), rabbit
anti-human CDC42 (1:1,000; Affinity Biosciences; cat. no.
DF6322), rabbit anti-human Zinc Finger E-Box Binding Homeobox 1
(ZEB-1; 1:1,000; CST Biological Reagents Co., Ltd.; cat. no.
3396), rabbit anti-human N-cadherin (N-CAD; 1:1,000; CST Biological
Reagents Co., Ltd.; cat. no. 13116), rabbit anti-human E-cadherin
(E-CAD; 1:1,000; CST Biological Reagents Co., Ltd.; cat. no. 3195),
rabbit anti-human Vimentin (1:1,000; CST Biological Reagents
Co., Ltd.; cat. no. 5741), rabbit anti-human Snail (1:1,000;
CST Biological Reagents Co., Ltd.; cat. no. 3879), rabbit anti
phospho-(p-)AKT (1:1,000; Abcam; cat. no. ab66138), rabbit
anti-human AKT (1:1,000; Abcam; cat. no. ab179463), rabbit
anti-human PI3K (1:1,000; Abcam; ab182651), rabbit
anti-human phosphorylated (p-)mTOR (1:1,000; CST Biological
Reagents Co., Ltd.; cat. no. 2971), rabbit anti-human mTOR
(1:1,000; CST Biological Reagents Co., Ltd.; cat. no. 2983), rabbit
anti-human Bcl2 (1:1,000; CST Biological Reagents Co., Ltd.;
cat. no. 2876), rabbit anti-human caspase3 (1:1,000; CST Biological
Reagents Co., Ltd.; cat. no. 8G10), rabbit anti-human p-ATM
Serine/Threonine Kinase (1:1,000; CST Biological Reagents Co.,
Ltd.; cat. no. 5883), rabbit anti-human ATM (1:1,000; CST
Biological Reagents Co., Ltd.; cat. no. 2873), rabbit anti-human
Checkpoint Kinase 2 (CHK2; 1:1,000; CST Biological Reagents
Co., Ltd.; cat. no. 2662), rabbit anti-human RAD51
(1:10,000; Abcam; cat. no. ab133534) mouse anti-human Bax
(1:1,000; CST Biological Reagents Co., Ltd.; cat. no. 2772), mouse
anti-human caspase9 (1:1,000; CST Biological Reagents Co., Ltd.;
cat. no. 9508), rabbit anti-human Pgp (1:1,000; Abcam; cat.
no. ab103477) and mouse anti-human β-actin (1:1,000; CST
Biological Reagents Co., Ltd.; cat. no. 3700).
The secondary antibodies used were: Horseradish
peroxidase-conjugated anti-rabbit (1:5,000; Sigma-Aldrich; Merck
KGaA; cat. no. A0545) or anti-mouse secondary antibody (1:5,000;
Sigma-Aldrich; Merck KGaA; cat. no. A9044).
Cell proliferation assay
The proliferative ability of cells was measured
using an MTT assay. Each cell line was seeded in quintuplicate into
96-well plates (0.8–1×103 cells/well) for 0–4 days. At
specified time points, 20 µl MTT reagent (Sigma-Aldrich; Merck
KGaA) at 5 mg/ml concentration was added to each well, and the
cells were incubated for an additional 3.5 h at 37°C. Subsequently,
the supernatants were discarded and 100 µl DMSO (Sigma-Aldrich;
Merck KGaA) was added to each well. The absorbance at 490 nm was
measured using a Varioskan Flash microplate reader (Thermo Fisher
Scientific, Inc.).
Cell migration and invasion assay
Cell migration and invasion were analyzed using
Boyden chamber-style cell culture inserts, with and without
Matrigel (BD Biosciences), respectively. Matrigel was thawed at 4°C
and then coated onto the Transwell inserts, after which the gel was
allowed to set at 37°C for 1 h. Ovarian cancer cells
(2×105 cells) were seeded in the upper chamber of the
Transwell inserts (24-well plate; 8-µm pore size; BD Biosciences)
with 200 µl serum-free media. The lower chambers were filled with
700 µl culture media containing 10% FBS as the chemoattractant.
After 6–48 h of incubation, the cells on the lower surface of the
membrane were washed with PBS and fixed in 100% methanol for 15 min
at room temperature. Then, cells were stained with 0.1% crystal
violet for 20 min at room temperature to quantify migration and
invasion. Transwell inserts were observed under a light microscope
(magnification, ×10) and cells in 10 random fields were
counted.
Clonogenic assay
For the colony formation assay, 500 cells were
seeded into each well of a 6-well plate and maintained in media
containing 10% FBS at optimum conditions of 37°C with 5%
CO2 for 10–14 days, until the colonies became visible to
the naked eye. Colonies were then fixed with 100% methanol at room
temperature for 15 min and stained with 0.1% crystal violet at room
temperature. Colonies with >50 cells were counted manually under
a light microscope (magnification, ×10) for quantification.
Apoptosis assay
Apoptosis was detected using an Annexin V-FITC and
propidium iodide (PI) kit (BD Biosciences), according to the
manufacturer's protocol. A2780 and HEY cells were transfected with
20 µM si-IQGAP3 or si-NC, and were harvested 48 h after
transfection with EDTA-free trypsin, centrifuged at 800 × g for 5
min at room temperature, washed twice with cold PBS, resuspended at
a concentration of 1×106 cells/ml and mixed with 100 µl
1X binding buffer. Subsequently, cells were stained with 5 µl
Annexin V-FITC and 5 µl PI at room temperature for 15 min in the
dark, after which 300 µl 1X binding buffer was added and the cells
were analyzed by flow cytometry (FACSCalibur; BD Biosciences)
within 1 h. The results were analyzed using FlowJo software version
X.0.7 (FlowJo, LLC).
Cell viability assay
A total of 2×103 cells/well were seeded
in 96-well plates. The A2780 and HEY cells were exposed to olaparib
(Selleck Chemicals; cat. no. AZD2281) at various final
concentrations (0, 5, 10, 20, 40 and 80 µmol/ml) at 37°C for 36–72
h. Each concentration was repeated in quintuplicate wells.
Subsequently, 20 µl 5 mg/ml MTT was added to each well. After
incubation for 3.5 h, the medium was replaced with 100 µl DMSO, and
cell viability was determined by analyzing the absorbance values at
490 nm on a Varioskan Flash microplate reader (Thermo Fisher
Scientific, Inc.).
Mouse xenograft models
HEY cells that were stably transfected with
IQGAP3-shRNA and the corresponding NC were used for the
in vivo experiments. For in vivo experiments, eight
female athymic BALB-c nude mice (age, 5 weeks; weight, 20–30 g)
were purchased from Nanjing Biochemical Research Institute and
housed in a standard pathogen-free condition in individually
ventilated cages with HEPA filters at the ambient temperature of
30–31°C and humidity of 50–60% with 12 h light/dark cycle, and
adequate access to food and water. For tumor formation assays,
1×107 cells (knockdown or control), resuspended in 200
µl PBS were subcutaneously injected into either side of the
axilla.
For metastasis assays, 1×107 cells were
intraperitoneally injected individually in the experimental and
control groups. After 2–3 weeks, bioluminescence images were
captured on an In-vivo imaging system (Kodak 2000 Imager).
The mice were euthanized via intraperitoneal injection of 200 mg/kg
sodium phenobarbital and the tumors were excised, fixed with 4%
paraformaldehyde at room temperature for 48 h, paraffin-embedded
and sectioned into 5-µm slices for hematoxylin and eosin staining.
The tissue slides were stained with hematoxylin for 5 min and eosin
for 10 min at room temperature and observed under a light
microscope (magnification, ×4).
Statistical analysis
GraphPad Prism version 7 (GraphPad Software, Inc.)
was used to analyze data. A χ2 test was used to analyze
the differences in clinical characteristics. Survival analysis was
performed using Kaplan-Meier analysis and a log-rank test. An
unpaired Student's t-test and a one-way ANOVA were used to
determine the statistically significant differences between
different groups. Fisher's least significant difference was used
for the post-hoc test following ANOVA. Data are presented as the
mean ± standard deviation of ≥3 independent experiments. P<0.05
was considered to indicate a statistically significant
difference.
Results
IQGAP3 expression is upregulated in
HGSOC
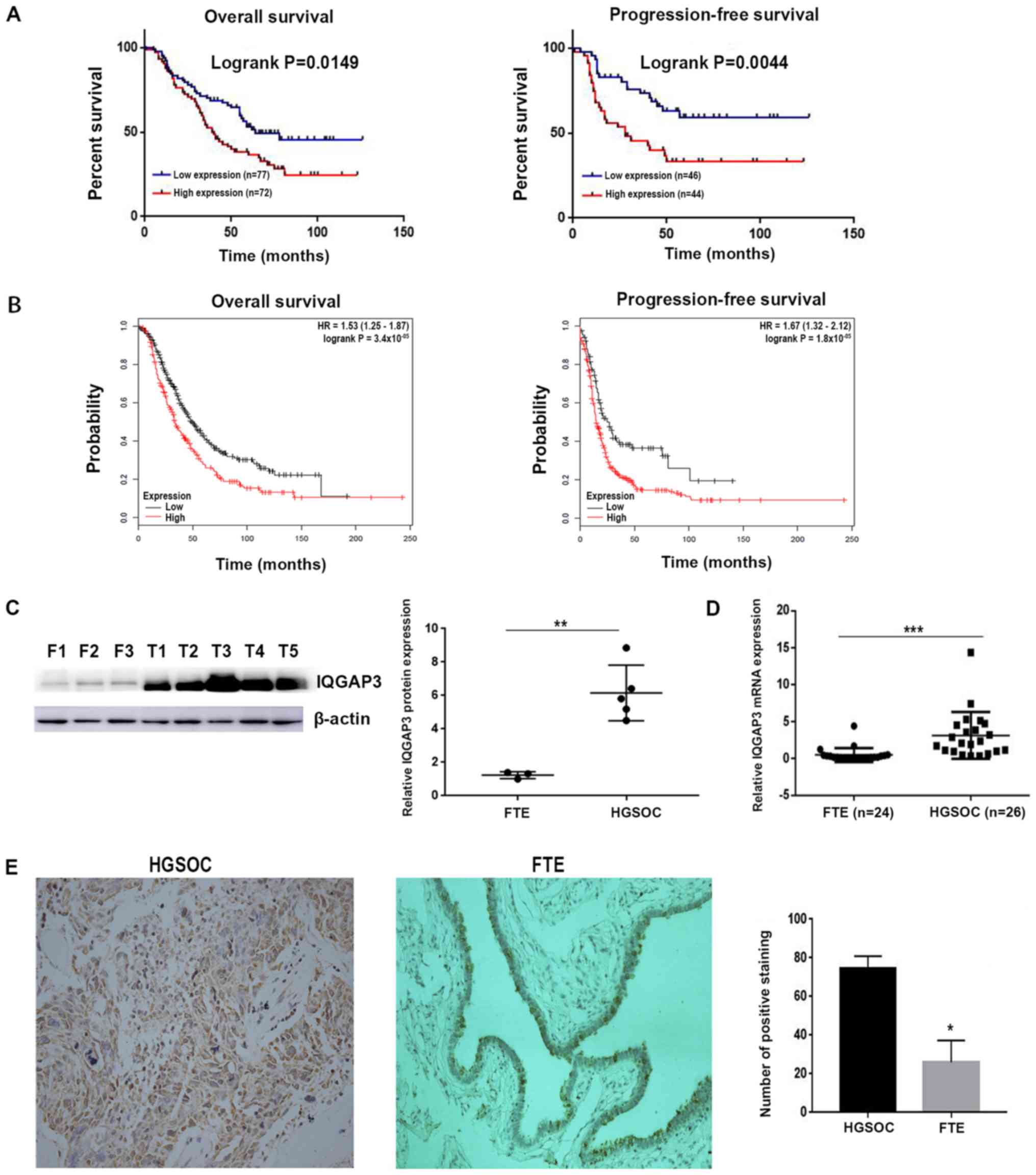
The mRNA and protein expression levels of
IQGAP3 in healthy fallopian tube and HGSOC tissues were
determined using RT-qPCR and western blotting, respectively. The
mRNA expression level of IQGAP3 was significantly higher in
HGSOC tissues compared with the control samples (Fig. 1D). Furthermore, IQGAP3 protein
expression was significantly upregulated in HGSOC tissues compared
with the fallopian tubal samples (Fig.
1C).
Upregulated expression of IQGAP3 is
associated with a less favorable prognosis
To examine whether upregulated expression of
IQGAP3 was associated with clinical prognosis, IHC staining
was performed on 149 HGSOC samples (Fig.
1E). Most positive staining was observed in the cytoplasm and
at the cell membrane. Moreover, a high expression of IQGAP3
was observed in 53.02% (79/149) of tissues. Subsequently, the
relationship between IQGAP3 and clinicopathological
characteristics were assessed (Table
I). The patients with a lower expression of IQGAP3 had
longer survival times compared with those with higher IQGAP3
expression levels. A log-rank test demonstrated that the
upregulated expression of IQGAP3 was significantly
associated with overall survival (P=0.0149), as well as
progression-free survival (P=0.0044; Fig. 1A).
 | Table I.Association between IQGAP3 expression
and clinicopathologic characteristics. |
Table I.
Association between IQGAP3 expression
and clinicopathologic characteristics.
| Clinicopathologic
characteristics | High
expression | Low expression | P-value |
|---|
| Age, years |
|
| 0.2089 |
|
<55 | 42 | 30 |
|
|
≥55 | 37 | 40 |
|
| FIGO stage |
|
| 0.4367 |
| I +
II | 21 | 15 |
|
| III +
IV | 57 | 55 |
|
| CA125, U/ml |
|
| 0.0147a |
|
<600 | 20 | 38 |
|
|
≥600 | 50 | 41 |
|
| Ascites, ml |
|
| 0.3255 |
|
Yes | 59 | 58 |
|
| No | 13 | 19 |
|
| Peritoneal
metastasis |
|
| 0.0007b |
|
Yes | 33 | 20 |
|
| No | 32 | 64 |
|
| Lymph node
metastasis |
|
| 0.1703 |
|
Positive | 12 | 10 |
|
|
Negative | 23 | 38 |
|
| Recurrence |
|
| 0.0065b |
|
Yes | 56 | 41 |
|
| No | 8 | 20 |
|
Survival analysis performed on GEO cohorts using
Kaplan-Meier Plotter, showed a significant association between
IQGAP3 expression and both overall and progression-free
survival (Fig. 1B). In addition,
further analysis indicated that the expression of IQGAP3 was
associated with several other clinicopathological parameters,
including recurrence of the disease (P=0.0065), CA125 levels
(P=0.0147) and peritoneal metastasis (P=0.0007; Table I).
Downregulation of IQGAP3 reduces
proliferation and colony formation of HGSOC ovarian cancer cells,
and attenuates tumorigenicity in a xenograft model
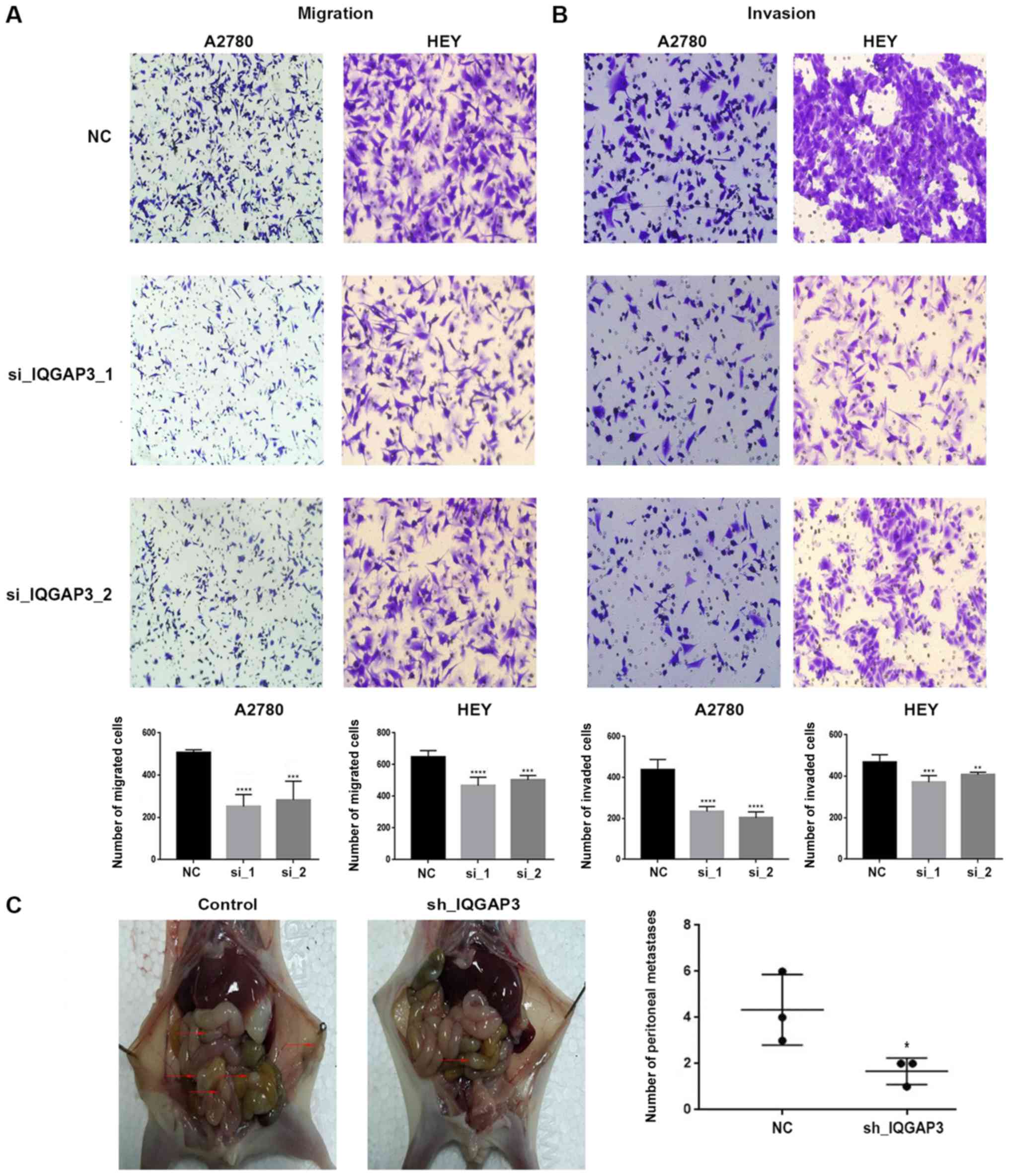
Downregulation of IQGAP3 resulted in the
reduced proliferation of ovarian cancer cells in vitro.
Moreover, two siRNAs, si-IQGAP3−1 and si-IQGAP3−2,
were used to silence IQGAP3 in A2780 and HEY cells. MTT
assays results identified a significant suppression of the
proliferative capacity in the two cell lines following transfection
with the siRNAs compared with the NCs (Fig. 2A).
These findings were further assessed in the in
vivo experiments, where xenografts of BALB-c nude mice were
established with injection of HEY cells stably transfected with
sh-IQGAP3 or NC (Fig. 2C).
After 3 weeks, the mice were euthanized, imaged on a
bioluminescence imaging system, and the tumors were excised and
weighed. It was found that there was a significant decrease in
tumor size and tumor weight in the sh-IQGAP3 group compared
with the NC group (Fig. 2D and E),
supporting the in vitro results. Therefore, the results
demonstrated the contribution of IQGAP3 to tumor
proliferation.
Knockdown of IQGAP3 also significantly
reduced colony formation in both A2780 and HEY cell lines (Fig. 2B).
IQGAP3 increases migration and
invasion of ovarian cancer cells via epithelial-to-mesenchymal
transition (EMT)
Transwell assays were used to examine the role of
IQGAP3 on migration and invasion in vitro. A2780 and
HEY cells both had significantly decreased migratory and invasive
capacities when IQGAP3 was knocked down compared with the
respective NC (Fig. 3A and B).
Furthermore, the underlying mechanism contributing
to this increase in tumorigenic features was determined by
analyzing EMT-related factors. Knockdown of IQGAP3 had an
effect on the expression of several EMT markers (Fig. 4). Silencing of IQGAP3 resulted
in downregulation of mesenchymal markers, including ZEB-1,
Vimentin, N-CAD and Snail, while the expression of the
epithelial marker E-CAD was upregulated. Thus, it was suggested
that IQGAP3 induced the migration and invasion of ovarian
cancer cells via induction of EMT.
 | Figure 4.Western blot analysis revealed the
changes in protein expression of several proteins following
IQGAP3 knockdown using two siRNAs. Both A2780 and HEY cells
had an altered protein expression following the knockdown. The
alteration in the protein expression included EMT-related proteins
(E-CAD, N-CAD, ZEB-1, Vimentin and Snail),
apoptosis-related proteins (Caspase-3, Caspase-9, Bcl2 and
Bax), proteins associated with DNA damage and
chemoresistance (Rad51, p-ATM, ATM, CHK2 and
Pgp), and proteins involved in the regulation and mechanism
of the effect of IQGAP3 (CDC42, PI3K, p-AKT,
AKT, p-mTOR and mTOR). β-actin was used as
the internal control. IQGAP3, IQ motif containing GTPase
Activating Protein 3; EMT, epithelial-to-mesenchymal transition;
p-, phosphorylated; E-CAD, E-cadherin; N-CAD, N-cadherin;
Pgp, phosphoglycolate phosphatase; Rad51, RAD51
recombinase; CHK2, checkpoint kinase 2; NC, negative
control; si, small interfering RNA; ZEB-1, zinc finger E-Box
binding homeobox 1. |
IQGAP3 reduces tumor metastasis in
vivo
To evaluate the role of IQGAP3 on metastasis
of ovarian cancer in vivo, female nude mice were injected
intraperitoneally with sh-IQGAP3-HEY cells or their
corresponding NCs. Then, 3 weeks after injection, the mice were
euthanized and the peritoneal cavities were examined for
metastases. Consistent with the in vitro experimental
results, mice injected with IQGAP3-silenced cells exhibited
significantly lower numbers of metastatic nodules compared with the
respective NC group (P<0.05; Fig.
3C). Bioluminescence imaging also identified larger metastatic
foci in the control group compared with the knockdown group
(Fig. 2E).
The excised metastatic nodules were fixed with
formalin and paraffin embedded and 4-µm thick slices were
sectioned. Subsequently, the slides were stained using hematoxylin
and eosin staining (Fig. S1).
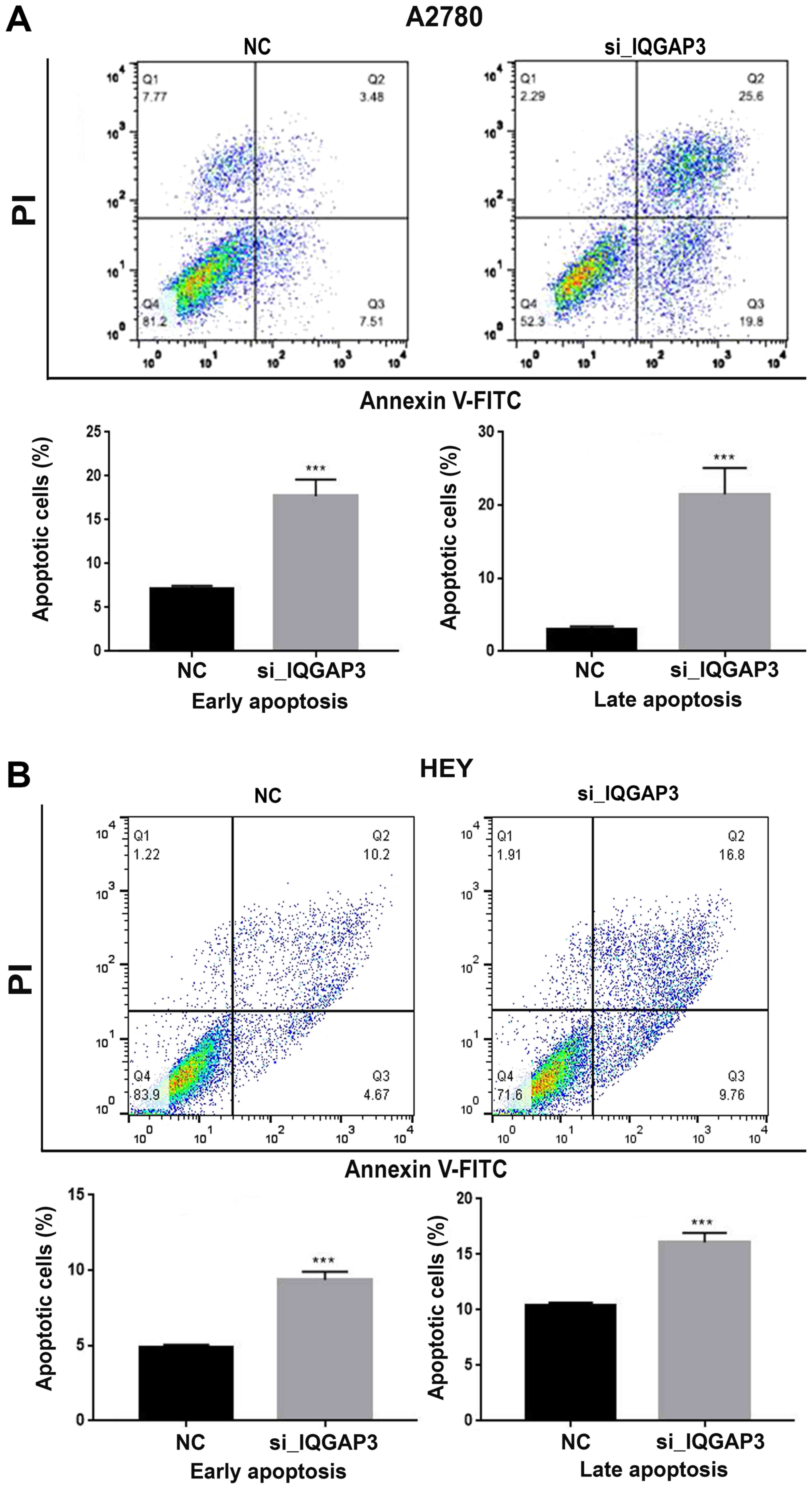
IQGAP3 knockdown promotes apoptosis in
ovarian cancer cells
To assess the effects of IQGAP3 knockdown on
apoptosis of ovarian cancer cells, Annexin V-FITC/PI dual staining
was performed following transfection with si-IQGAP3 or NC.
Both A2780 and HEY cells exhibited significantly increased
apoptosis following knockdown of IQGAP3 compared with the
respective NC group (Fig. 5A and B).
These results were further validated by the increased expression of
the pro-apoptotic proteins Bax, Caspase 3 and Caspase 9, and
decreased expression of Bcl-2 following IQGAP3
knockdown (Fig. 4).
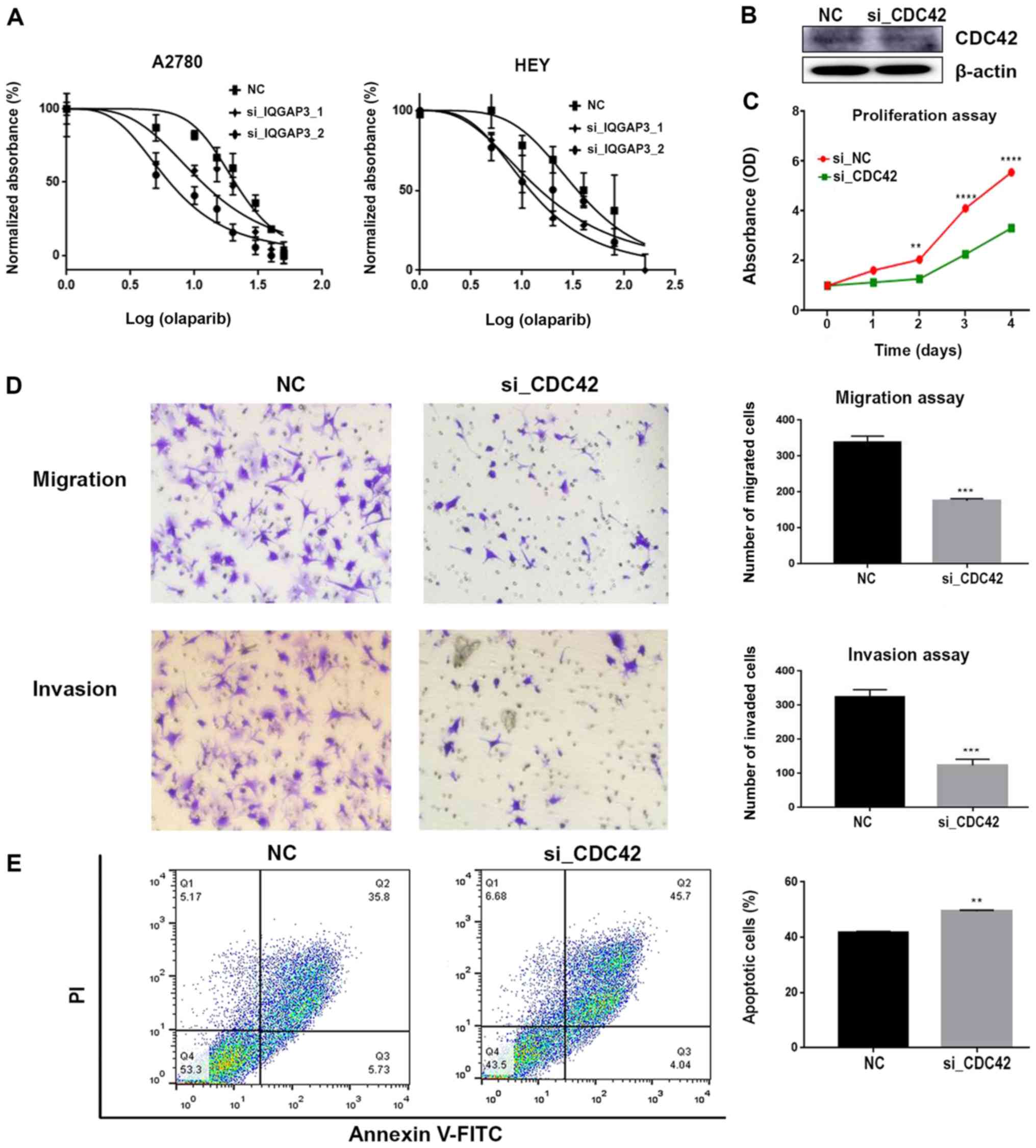
IQGAP3 knockdown increases sensitivity
to chemotherapy with PARPi
The si-IQGAP3 transfected A2780 and HEY cells
were exposed to various concentrations of olaparib (5, 10, 20, 40
or 80 µmol/ml) for 36–72 h, after which, the cell viability was
assessed using an MTT assay. Cells transfected with
si-IQGAP3 exhibited increased sensitivity to olaparib
compared with the respective control group (Fig. 6A). Western blotting results
identified the downregulation of the expression levels of
Rad51, p-ATM (normalized to total ATM) and
CHK2 when IQGAP3 expression was knocked down
(Fig. 4). Thus, knockdown of
IQGAP3 may have sensitized cells to olaparib by
downregulating key factors involved in the DNA damage response.
Phosphoglycolate Phosphatase (Pgp) is a
multidrug resistance protein that is localized in the cell membrane
and is responsible for extruding several xenobiotics (including
chemotherapeutic agents) outside the cells, rendering the cells
chemoresistant (45). IQGAP3
knockdown reduced the expression of Pgp, which in turn
attenuated chemoresistance (Fig. 4).
Therefore, it was speculated that this effect may underlie the
enhanced sensitivity of cells towards olaparib following
IQGAP3 knockdown.
IQGAP3 exerts its function via the
regulation of CDC42
It has been reported that IQGAP3 is an
effector of CDC42 (28). Xu
et al (35) also revealed
that IQGAP3 may exert its oncogenic function in pancreatic
cancer via the regulation of CDC42. To determine whether
IQGAP3 was associated with CDC42 in ovarian cancer,
the protein expression levels of CDC42 in ovarian cancer
cells were assessed by knocking down IQGAP3 expression. It
was identified that knockdown of IQGAP3 decreased the
expression of CDC42 (Fig.
4).
Therefore, the effects of CDC42 on the cancer
cells were assessed. Knockdown of CDC42 expression (Fig. 6B) resulted in a significant decrease
in the proliferative potential of HEY cells (Fig. 6C). Furthermore, migration and
invasion were inhibited, while apoptosis was enhanced following
CDC42 knockdown (Fig. 6D and
E). Collectively, these results suggest that IQGAP3 may
exert its effects via the regulation of CDC42.
Discussion
The principle dilemma when dealing with ovarian
cancer is the rate of distant metastasis at the time of diagnosis
and its resistance to chemotherapy, which frequently results in
negative consequences (1,46–48).
Thus, there is an unmet need for an improved understanding of the
molecular mechanisms involved in the proliferation, metastasis and
chemoresistance of ovarian cancer.
Out of the three primary members of the
IQGAP family, IQGAP1 has been reported to be an
oncogene, and IQGAP2 a tumor suppressor (24–26,49).
Furthermore, IQGAP3 is a scaffolding protein, which
interacts with various structural proteins that influence the
cytoskeletal dynamics and intracellular signaling (28). IQGAP3 has also previously been
implicated in the proliferation of epithelial cells (27). Moreover, previous studies have
revealed the role of IQGAP3 in the proliferation and
metastasis of lung, gastric, breast, pancreatic cancer, and
colorectal cancer as well as hepatocellular carcinoma (29–35).
Therefore, the role of IQGAP3 is crucial in the malignant
transformation of several types of cancer. Yang et al
(29) also reported that
IQGAP3 promotes the metastasis of lung cancer cells by
activating the epidermal growth factor receptor/ERK signaling
pathway. In addition, Yang et al (29) used bioinformatics analysis to show
that IQGAP3 is upregulated in several malignancies,
including ovarian cancer. Furthermore, Wu et al (50) reported there were alterations in the
genes regulating cytoskeleton remodeling in metastatic lung
adenocarcinoma and that IQGAP3 was a marker of a less
favorable prognosis.
The present results indicated that IQGAP3
was upregulated in ovarian cancer, and this enhanced expression
resulted in increased proliferative and metastatic capacities in
vitro and in vivo. Upon silencing IQGAP3, the
aggressive nature of ovarian cancer cells was significantly
abrogated. Thus, IQGAP3 may be a putative oncogene in HGSOC.
Moreover, the upregulated expression of IQGAP3 was
associated with a shorter overall and progression-free survival,
cancer recurrence and CA125 expression. Kaplan-Meier survival
analysis on data obtained from the online GEO database also
demonstrated that patients with an upregulated expression of
IQGAP3 exhibited reduced survival rates, further validating
the in vitro and in vivo results. However, whether
IQGAP3 is an independent poor prognostic factor of HGSOC is
yet to be determined.
A number of in vitro and in vivo
experiments were designed to establish the oncogenic potential of
IQGAP3 in HGSOC. IQGAP3 expression was significantly
upregulated in HGSOC compared with the healthy control. Cell
proliferation and tumorigenesis assays in nude mice demonstrated
the decreased proliferative capacity of ovarian cancer cells when
IQGAP3 was knocked down in vitro and in
vivo.
Metastasis is a culmination of cancer cells gaining
migratory and invasive abilities (51). Furthermore, distant metastasis at the
time of diagnosis is one of the major obstacles negatively
impacting the prognosis of ovarian cancer (52). The molecular mechanisms of metastasis
in ovarian cancer are yet to be fully elucidated; however, EMT has
been considered to be a potential contributing factor (52–54).
The present results suggested that IQGAP3
serves a substantial role in migration and invasion of ovarian
cancer, and knocking down IQGAP3 reduced the metastatic
potential. These findings were also observed in vivo, where
fewer metastatic foci formed in the mice injected with
IQGAP3-silenced cells compared with the control group.
EMT is initiated by several EMT-inducing
transcription factors (53–58). In the present study, it was found
that several of the EMT-inducing factors were affected by
alterations in the expression of IQGAP3, which suggests a
pivotal role of IQGAP3 in the induction of EMT in ovarian
cancer.
Several studies have reported that the
PI3K/AKT/mTOR signaling pathway is a crucial pathway by which
cancer cells exhibit increased proliferative and metastatic
potential (59–64). This signaling pathway is involved in
several fundamental processes in ovarian cancer, such as cell
proliferation, survival, autophagy, transcription regulation and
angiogenesis (63,64). Therefore, to determine the mechanism
underlying the effects of IQGAP3 in ovarian cancer, the
effects of altering IQGAP3 gene expression of the
PI3K/AKT/mTOR pathway were determined. Western blot analysis
revealed a significant downregulation in the expression levels of
PI3K, p-AKT and p-mTOR when IQGAP3
expression was knocked down. Thus, these results suggested that
IQGAP3 may promote tumor progression and metastasis via the
PI3K/AKT/mTOR signaling pathway.
Previous studies have shown that increased
apoptosis may underlie decreased tumor growth, chemoresistance and
metastasis in several types of cancer (65,66). It
has also been reported that IQGAP3 in certain types of
cancer is closely associated with apoptosis (35). In the present study, it was
demonstrated that the apoptotic potential of cells was increased
when IQGAP3 expression was knocked down, and this may
underlie the effects of IQGAP3 on tumor growth.
CDC42 is a member of the Rho family of
GTPases, and is ubiquitously expressed (23). Moreover, CDC42 participates in
the regulation of cytoskeletal dynamics, cellular proliferation,
motility, polarity and cytokinesis (67). Wang et al (28) also identified a direct interaction
between IQGAP3 with CDC42, and revealed IQGAP3
is an indispensable effector of CDC42-mediated cell
proliferation. Furthermore, Xu et al (35) hypothesized that IQGAP3 may
serve as an oncogene in pancreatic cancer by regulating the
CDC42 signaling pathway. Morgan et al (68) reported there was an interaction
between IQGAP3 and CDC42 using immunoprecipitation
assays. The results of the present study also demonstrated that
knocking down IQGAP3 expression resulted in the
downregulation of CDC42 expression. Therefore, the role of
CDC42 in ovarian cancer cells was further investigated. It
was found that knockdown of CDC42 resulted in a significant
decrease in proliferation, migration and invasion, and increased
apoptosis in ovarian cancer cells. Thus, it was speculated that
IQGAP3 may exert its function via modulation of
CDC42, but further studies are required to verify this
hypothesis.
PARP serves a key role in the DNA damage
response of the cell (10,11,13). The
PARP inhibitor olaparib has been recently approved by the
FDA for the treatment of patients with ovarian cancer who harbor
BRCA1/2 mutations (15).
Moreover, BRCA1/2 mutations are responsible for 18–40% of
lifetime risk ovarian cancer cases in women, and 5–15% of all
diagnosed cases harbor one of these mutations (69). Thus, the introduction of PARPi, such
as olaparib, may improve the prognostic prospects of patients.
However, this drug is not effective for all HGSOC cases (70). IQGAP3 is associated with
olaparib drug sensitivity, and knockdown of IQGAP3 in the
present study resulted in increased efficacy of olaparib,
suggesting that the effectiveness of the treatment may be dependent
upon specific clinical aspects. Additionally, the expression
profiles of proteins involved in DNA damage response of cell,
including ATM and CHK, were assessed. In the present study, there
was a significant decrease in the expression levels of these
proteins following IQGAP3 knockdown. Furthermore, similar
effects were observed in Rad51, which possesses a crucial role in
the homologous recombination repair of DNA (10). Therefore, it was hypothesized that
downregulation of DNA repair factors may result in defective DNA
repair in cells, thus increasing the sensitivity to PARPi.
However, further investigations focusing on the
mechanistic role of IQGAP3 in proliferation and metastasis
of ovarian cancer are required before IQGAP3 may be
considered a diagnostic and prognostic marker, and as a potential
therapeutic target for ovarian cancer. To the best of our
knowledge, the present study was the first to report the role of
IQGAP3 in the progression of HGSOC.
In conclusion, IQGAP3 exhibited oncogenic
features in HGSOC. In addition, the expression of IQGAP3 was
upregulated in HGSOC, and its expression was associated with a poor
outcome in patients. However, more studies are required to further
validate IQGAP3 as a prognostic marker and a therapeutic
target for ovarian cancer.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to acknowledge the
contribution of Dr Kun Song from the Department of OB/GYN and Dr
Ning Yang from the Department of Pathology at Qilu Hospital of
Shandong University (Ji'nan, China), and Dr Shi Yan, Dr Rongrong Li
and Dr Cunzong Yuan from the Key Laboratory of Gynecologic Oncology
of Shandong (Ji'nan, China), who contributed greatly to the
completion of this manuscript.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81874107 and
81572554).
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
SD, QZ and BK contributed to the conceptualization
of the study. SD drafted the manuscript. SD, CQ, CS, ZZ and HW
contributed to data acquisition, carried out the data analysis and
revised the manuscript. QZ and BK were involved in analyzing the
critical intellectual content and gave the final approval of the
version to be published. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Signed consents were collected from all the
patients and the study was approved by Ethics Committee of Qilu
hospital of Shandong University. Approval of Shandong University
Animal Care and Use Committee was acquired for all the animal
experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Stat Facts: Ovarian Cancer. 2019.
PubMed/NCBI
|
|
3
|
Ozols RF: Systemic therapy for ovarian
cancer: Current status and new treatments. Semin Oncol. 33 (Suppl
6):S3–S11. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Karnezis AN, Cho KR, Gilks CB, Pearce CL
and Huntsman DG: The disparate origins of ovarian cancers:
Pathogenesis and prevention strategies. Nat Rev Cancer. 17:65–74.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cho KR and Shih IeM: Ovarian cancer. Annu
Rev Pathol. 4:287–313. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kurman RJ and Shih IeM: The origin and
pathogenesis of epithelial ovarian cancer: A proposed unifying
theory. Am J Surg Pathol. 34:433–443. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bast RC Jr, Hennessy B and Mills GB: The
biology of ovarian cancer: New opportunities for translation. Nat
Rev Cancer. 9:415–428. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Seidman JD, Horkayne-Szakaly I, Haiba M,
Boice CR, Kurman RJ and Ronnett BM: The histologic type and stage
distribution of ovarian carcinomas of surface epithelial origin.
Int J Gynecol Pathol. 23:41–44. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peres LC, Cushing-Haugen KL, Köbel M,
Harris HR, Berchuck A, Rossing MA and Doherty JA: Invasive
epithelial ovarian cancer survival by histotype and disease stage.
J Natl Cancer Inst. 111:60–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pilié PG, Tang C, Mills GB and Yap TA:
State-of-the-art strategies for targeting the DNA damage response
in cancer. Nat Rev Clin Oncol. 16:81–104. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
AlHilli MM, Becker MA, Weroha SJ, Flatten
KS, Hurley RM, Harrell MI, Oberg AL, Maurer MJ, Hawthorne KM, Hou
X, et al: In vivo anti-tumor activity of the PARP inhibitor
niraparib in homologous recombination deficient and proficient
ovarian carcinoma. Gynecol Oncol. 143:379–388. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McGuire WP III and Markman M: Primary
ovarian cancer chemotherapy: Current standards of care. Br J
Cancer. 89 (Suppl 3):S3–S8. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pokhriyal R, Hariprasad R, Kumar L and
Hariprasad G: Chemotherapy resistance in advanced ovarian cancer
patients. Biomark Cancer. 11:1179299X198608152019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moore K, Colombo N, Scambia G, Kim BG,
Oaknin A, Friedlander M, Lisyanskaya A, Floquet A, Leary A, Sonke
GS, et al: Maintenance olaparib in patients with newly diagnosed
advanced ovarian cancer. N Engl J Med. 379:2495–2505. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim G, Ison G, McKee AE, Zhang H, Tang S,
Gwise T, Sridhara R, Lee E, Tzou A, Philip R, et al: FDA approval
summary: Olaparib monotherapy in patients with deleterious germline
BRCA-mutated advanced ovarian cancer treated with three or more
lines of chemotherapy. Clin Cancer Res. 21:4257–4261. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Briggs MW and Sacks DB: IQGAP proteins are
integral components of cytoskeletal regulation. EMBO Rep.
4:571–574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weissbach L, Settleman J, Kalady MF,
Snijders AJ, Murthy AE, Yan YX and Bernards A: Identification of a
human rasGAP-related protein containing calmodulin-binding motifs.
J Biol Chem. 269:20517–20521. 1994.PubMed/NCBI
|
|
18
|
Hedman AC, Smith JM and Sacks DB: The
biology of IQGAP proteins: Beyond the cytoskeleton. EMBO Rep.
16:427–446. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Swart-Mataraza JM, Li Z and Sacks DB:
IQGAP1 is a component of Cdc42 signaling to the cytoskeleton. J
Biol Chem. 277:24753–24763. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sahai E and Marshall CJ: RHO-GTPases and
cancer. Nat Rev Cancer. 2:133–142. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Y, Olufemi L, Wang MT and Nie D: Role
of Rho GTPases in breast cancer. Front Biosci. 13:759–776. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Erickson JW and Cerione RA: Multiple roles
for Cdc42 in cell regulation. Curr Opin Cell Biol. 13:153–157.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arias-Romero LE and Chernoff J: Targeting
Cdc42 in cancer. Expert Opin Ther Targets. 17:1263–1273. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Johnson M, Sharma M and Henderson BR:
IQGAP1 regulation and roles in cancer. Cell Signal. 21:1471–1478.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mataraza JM, Briggs MW, Li Z, Entwistle A,
Ridley AJ and Sacks DB: IQGAP1 promotes cell motility and invasion.
J Biol Chem. 278:41237–41245. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schmidt VA, Chiariello CS, Capilla E,
Miller F and Bahou WF: Development of hepatocellular carcinoma in
Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol.
28:1489–1502. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nojima H, Adachi M, Matsui T, Okawa K and
Tsukita S and Tsukita S: IQGAP3 regulates cell proliferation
through the Ras/ERK signalling cascade. Nat Cell Biol. 10:971–978.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Watanabe T, Noritake J, Fukata M,
Yoshimura T, Itoh N, Harada T, Nakagawa M, Matsuura Y, Arimura N
and Kaibuchi K: IQGAP3, a novel effector of Rac1 and Cdc42,
regulates neurite outgrowth. J Cell Sci. 120:567–577. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Y, Zhao W, Xu QW, Wang XS, Zhang Y
and Zhang J: IQGAP3 promotes EGFR-ERK signaling and the growth and
metastasis of lung cancer cells. PLoS One. 9:e975782014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu G, Xu Y, Chen W, Wang J, Zhao C and
Wang M: RNA Interference of IQ motif containing GTPase-activating
protein 3 (IQGAP3) inhibits cell proliferation and invasion in
breast carcinoma cells. Oncol Res. 24:455–461. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi Y, Qin N, Zhou Q, Chen Y, Huang S,
Chen B, Shen G and Jia H: Role of IQGAP3 in metastasis and
epithelial-mesenchymal transition in human hepatocellular
carcinoma. J Transl Med. 15:1762017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao H, Wang Q, Gao Z, Xu X, Lu Q and Wu Y:
Clinical value of detecting IQGAP3, B7-H4 and cyclooxygenase-2 in
the diagnosis and prognostic evaluation of colorectal cancer.
Cancer Cell Int. 19:1632019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu J, Chen Z, Cao H, Yu Z, Feng J, Wang K,
Lu Q and Wu Y: High expression of IQGAP3 indicates poor prognosis
in colorectal cancer patients. Int J Biol Markers. 34:348–355.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yasui W, Sentani K, Sakamoto N, Anami K,
Naito Y and Oue N: Molecular pathology of gastric cancer: Research
and practice. Pathol Res Pract. 207:608–612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu W, Xu B, Yao Y, Yu X, Cao H, Zhang J,
Liu J and Sheng H: Overexpression and biological function of IQGAP3
in human pancreatic cancer. Am J Transl Res. 8:5421–5432.
2016.PubMed/NCBI
|
|
36
|
Javadi S, Ganeshan DM, Qayyum A, Iyer RB
and Bhosale P: Ovarian cancer, the revised FIGO staging system, and
the role of imaging. AJR Am J Roentgenol. 206:1351–1360. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Birrer MJ, Johnson ME, Hao K, Wong KK,
Park DC, Bell A, Welch WR, Berkowitz RS and Mok SC: Whole genome
oligonucleotide-based array comparative genomic hybridization
analysis identified fibroblast growth factor 1 as a prognostic
marker for advanced-stage serous ovarian adenocarcinomas. J Clin
Oncol. 25:2281–2287. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mateescu B, Batista L, Cardon M, Gruosso
T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P,
Sastre-Garau X and Mechta-Grigoriou F: miR-141 and miR-200a act on
ovarian tumorigenesis by controlling oxidative stress response. Nat
Med. 17:1627–1635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ferriss JS, Kim Y, Duska L, Birrer M,
Levine DA, Moskaluk C, Theodorescu D and Lee JK: Multi-gene
expression predictors of single drug responses to adjuvant
chemotherapy in ovarian carcinoma: Predicting platinum resistance.
PLoS One. 7:e305502012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lisowska KM, Olbryt M, Dudaladava V,
Pamuła-Piłat J, Kujawa K, Grzybowska E, Jarzab M, Student S,
Rzepecka IK, Jarzab B and Kupryjańczyk J: Gene expression analysis
in ovarian cancer-faults and hints from DNA microarray study. Front
Oncol. 4:62014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tothill RW, Tinker AV, George J, Brown R,
Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro
B, et al: Novel molecular subtypes of serous and endometrioid
ovarian cancer linked to clinical outcome. Clin Cancer Res.
14:5198–5208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Uehara Y, Oda K, Ikeda Y, Koso T, Tsuji S,
Yamamoto S, Asada K, Sone K, Kurikawa R, Makii C, et al: Integrated
copy number and expression analysis identifies profiles of
Whole-arm chromosomal alterations and subgroups with favorable
outcome in ovarian clear cell carcinomas. PLoS One.
10:e01280662015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gyorffy B, Lánczky A and Szállási Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pokharel D, Roseblade A, Oenarto V, Lu JF
and Bebawy M: Proteins regulating the intercellular transfer and
function of P-glycoprotein in multidrug-resistant cancer.
Ecancermedicalscience. 11:7682017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Naora H and Montell DJ: Ovarian cancer
metastasis: Integrating insights from disparate model organisms.
Nat Rev Cancer. 5:355–366. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Campos SM and Ghosh S: A current review of
targeted therapeutics for ovarian cancer. J Oncol. 149362:2010.
|
|
48
|
Hogberg T: Chemotherapy: Current drugs
still have potential in advanced ovarian cancer. Nat Rev Clin
Oncol. 7:191–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
White CD, Khurana H, Gnatenko DV, Li Z,
Odze RD, Sacks DB and Schmidt VA: IQGAP1 and IQGAP2 are
reciprocally altered in hepatocellular carcinoma. BMC
Gastroenterol. 10:1252010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wu K, Zhang X, Li F, Xiao D, Hou Y, Zhu S,
Liu D, Ye X, Ye M, Yang J, et al: Frequent alterations in
cytoskeleton remodelling genes in primary and metastatic lung
adenocarcinomas. Nat Commun. 6:101312015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang RY, Chung VY and Thiery JP:
Targeting pathways contributing to epithelial-mesenchymal
transition (EMT) in epithelial ovarian cancer. Curr Drug Targets.
13:1649–1653. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Berx G, Raspe E, Christofori G, Thiery JP
and Sleeman JP: Pre-EMTing metastasis? Recapitulation of
morphogenetic processes in cancer. Clin Exp Metastasis. 24:587–597.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Barrallo-Gimeno A and Nieto MA: The Snail
genes as inducers of cell movement and survival: Implications in
development and cancer. Development. 132:3151–3161. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hugo H, Ackland ML, Blick T, Lawrence MG,
Clements JA, Williams ED and Thompson EW: Epithelial-mesenchymal
and mesenchymal-epithelial transitions in carcinoma progression. J
Cell Physiol. 213:374–383. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cantley LC: The phosphoinositide 3-kinase
pathway. Science. 296:1655–1657. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and Gonzalez-Baron M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Willems L, Tamburini J, Chapuis N, Lacombe
C, Mayeux P and Bouscary D: PI3K and mTOR signaling pathways in
cancer: New data on targeted therapies. Curr Oncol Rep. 14:129–138.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
McAuliffe P, Meric-Bernstam F, Mills GB
and Gonzalez-Angulo A: Deciphering the role of PI3K/Akt/mTOR
pathway in breast cancer biology and pathogenesis. Clin Breast
Cancer. 10 (Suppl 3):S59–S65. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gasparri ML, Bardhi E, Ruscito I, Papadia
A, Farooqi AA, Marchetti C, Bogani G, Ceccacci I, Mueller MD and
Benedetti Panici P: PI3K/AKT/mTOR pathway in ovarian cancer
treatment: Are we on the right track? Geburtshilfe Frauenheilkd.
77:1095–1103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mabuchi S, Kuroda H, Takahashi R and
Sasano T: The PI3K/AKT/mTOR pathway as a therapeutic target in
ovarian cancer. Gynecol Oncol. 137:173–179. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wong RS: Apoptosis in cancer: From
pathogenesis to treatment. J Exp Clin Cancer Res. 30:872011.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol
Cancer. 14:482015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Melendez J, Grogg M and Zheng Y: Signaling
role of Cdc42 in regulating mammalian physiology. J Biol Chem.
286:2375–2381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Morgan CJ, Hedman AC, Li Z and Sacks DB:
Endogenous IQGAP1 and IQGAP3 do not functionally interact with Ras.
Sci Rep. 9:110572019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ramus SJ and Gayther SA: The contribution
of BRCA1 and BRCA2 to ovarian cancer. Mol Oncol. 3:138–150. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Jiang X, Li X, Li W, Bai H and Zhang Z:
PARP inhibitors in ovarian cancer: Sensitivity prediction and
resistance mechanisms. J Cell Mol Med. 23:2303–2313. 2019.
View Article : Google Scholar : PubMed/NCBI
|