Introduction
Inflammatory bowel disease (IBD), including Crohn's
disease and ulcerative colitis, is a chronic, relapsing
inflammatory disorder of the digestive tract (1,2). Chronic
inflammation is considered to play an important role in the
development of cancer (3,4). Colorectal cancer (CRC) is the most
serious complication of IBD, and patients with IBD are at a
significantly increased risk of developing CRC compared with the
general population, worldwide (5,6). This
may be a representative example reflecting the association between
chronic inflammation and tumorigenesis. Lutgens et al
(7) demonstrated that the pooled
standardized incidence ratio of CRC in all patients with IBD in
worldwide population-based studies was 1.7, while the cumulative
risk for CRC in IBD patients was 1, 2 and 5% at 10, 20 and >20
years of disease duration, respectively.
CD73, also referred to as ecto-5′-nucleotidase, is a
membrane-bound glycoprotein, the primary function of which is to
hydrolyze extracellular nucleoside monophosphates into bioactive
nucleoside intermediates, leading to the generation of
extracellular adenosine (8).
Adenosine has multiple functions aimed at maintaining tissue
homeostasis, and mediates its immunosuppressive effects mainly via
A2A and A2B receptors (9). CD73 is
upregulated in several types of cancer and increasing evidence
suggested that CD73 plays a crucial role in the control of tumor
progression (10–12). It was demonstrated that inhibition of
CD73 activity or CD73 knockdown on tumor cells inhibited tumor
growth by enhancing the antitumor T-cell response (13,14). By
using CD73-deficient mice, it was demonstrated that CD73 on
hematopoietic cells (including Foxp3+ Treg cells)
impairs the antitumor T-cell-mediated immune response. These
effects are attributed to the regulation of extracellular adenosine
generated by CD73 within the tumor microenvironment (15,16).
Additionally, CD73 research on IBD revealed that transfer of
CD73+ B cells to CD73−/− mice decreased the
severity of colitis, suggesting that B-cell CD73/CD39/adenosine can
modulate dextran sulfate sodium (DSS)-induced colitis (17).
The understanding of the role of CD73 in tumor
initiation in patients with IBD remains limited (11). The aim of the present study was to
determine the role of CD73 in IBD-associated tumorigenesis in a
mouse model by using the CD73 inhibitor adenosine
5′-(α,β-methylene) diphosphate (APCP) and the non-selective
adenosine receptor agonist
1-(6-amino-9H-purin-9-yl)-1-deoxy-N-ethyl-β-D-ribofuranuronamide
(NECA).
Materials and methods
Mice
A total of 39 female C57BL/6 mice (age, 6–8 weeks;
weight, ~20 g) were obtained from the Laboratory Animal Center of
Sun Yat-sen University (Guangzhou, China). The mice were kept in a
special pathogen-free facility with free access to drinking water
and a pellet-based diet, and were quarantined for 7 days prior to
the experiment. They were maintained at acontrolled temperature
(22±1°C), humidity (50–70%) and a 12 h light/dark cycle. The
experimental protocol was approved by the Ethics Committee of Sun
Yat-sen University (approval no. SYSU-IACUC-2020-B0038). All animal
studies were conducted with the approval of the Institutional
Animal Care and Use Committee of Sun Yat-sen University.
Reagents
Azoxymethane (AOM), APCP and NECA were purchased
from Sigma-Aldrich; Merck-KGaA. DSS was purchased from MP
Biomedicals, LLC.
Animal model induction and
treatment
The C57BL/6 mice were divided into four groups (6
mice in the negative control group and 11 mice per experimental
group), including the negative control group (receiving no AOM/DSS
or other treatment), the model control group (receiving AOM/DSS and
PBS treatment), the CD73 inhibitor group (APCP group; receiving
AOM/DSS and APCP) and the adenosine receptor agonist group (NECA
group; receiving AOM/DSS and NECA).
AOM and DSS were used to induce colitis-associated
tumorigenesis (CAT) in the mice. Briefly, the mice were injected
intraperitoneally with a single dose (10 mg/kg) of AMO, followed by
three cycles of DSS, with each cycle consisting of 1 week of 2% DSS
in the drinking water and 2 weeks of normal drinking water. While
drinking 2% DSS water, mice in the three experimental groups were
injected intraperitoneally with APCP (80 µg, 0.2 ml) (18,19) or
NECA (24.66 µg, 0.2 ml) (20) or PBS
(0.2 ml) every other day. Mice in the control group were
intraperitoneally injected with PBS and received pure water for 9
weeks. No measurable toxicity was observed after using the
indicated doses of APCP and NECA.
The mice were monitored for body weight, stool
consistency and the presence of blood in the excreta on days 1, 3,
5 and 7 when drinking water with 2% DSS, and every 3 days when
drinking normal water in each cycle. At the end of week 9, the mice
were sacrificed by cervical dislocation. Colon length (from the
ileocecal junction to the anal verge) were measured. Subsequently,
the colon was incised longitudinally and macroscopically visible
tumor number per mouse, tumor diameter and tumor burden (determined
by summation of total tumor volume) were measured with a caliper.
Segments of the distal colon were fixed in 10% neutral buffered
formalin (pH7.4) at room temperature for at least 24 h for
subsequent paraffine embedding, or kept in RNAlater™ stabilization
solution (Ambion; Thermo Fisher Scientific, Inc.) as tissue samples
for further analysis. Details of the humane endpoints in our mouse
model are shown in Table SI. Humane
endpoints in the AOM/DSS-induced CAT mouse model experiment
suggested to have a total score ≥8 or with the highest point in at
least two individual items or with non-CAT-associated symptoms,
such as skin lesion and allergy, based on the humane endpoints
scoring system (21). After last
record, analgesic administration and euthanasia were carried out
for animals that reached the humane endpoints. Anticipated weight
loss of over 20% in the NECA group, which did not reach the humane
endpoints was considered and approved by the ethics committee.
Histopathological evaluation
Sections (4-µm) of formalin-fixed (10% neutral
buffered formalin at room temperature for at least 24 h),
paraffin-embedded tissues were stained with hematoxylin for 2–3 min
and eosin for 1–2 min (HE) at room temperature to evaluate the
severity of inflammation using a DMI400B inverted microscope
(magnification ×10; Leica Microsystems GmbH). Colitis was scored in
a blinded manner as previously described (22), with a combined score for tissue
injury (score, 0–3) and infiltration of inflammatory cells (score,
0–3). Briefly, for tissue injury, 0 = normal colonic mucosa; 1 =
discrete lymphoepithelial lesions; 2, surface mucosal erosion or
focal ulceration and 3, extensive mucosal damage and extension into
deeper layers. For infiltration of inflammatory cells: 0,
occasional presence of inflammatory cells in the lamina propria; 1,
increasing number of inflammatory cells in the lamina propria; 2,
inflammatory cells extending into the submucosa; and 3, transmural
extension of the infiltration. The histological score was defined
as the sum of the two aforementioned parameters (score, 0–6).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from colonic segments using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA quality and concentration were then assessed using a
NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific,
Inc.). RNA (1 µg) was then reverse-transcribed using the ReverTra
Ace qPCR RT kit (cat. no. FSQ-101; Toyobo Life Science), according
to the manufacturer's protocol. qPCR was performed using SYBR Green
Realtime PCR master mix (cat. no. QPK-201; Toyobo Life Science) on
an Applied Biosystems 7500 Real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used: Initial denaturation at 95°C
for 10 min; 40 cycles of denaturation at 95°C for 10 sec, annealing
at 60°C for 20 sec and elongation at 72°C for 30 sec; and a final
extension at 72°C for 7 min. Levels of tumor necrosis factor
(TNF)-α, TNF-β, interleukin (IL)-10 and IL-6 were measured. All
reactions were performed in triplicate, with 3 samples from
different groups. The quantification of target mRNA was normalized
against GAPDH. The relative expression levels of each gene were
calculated and normalized using the 2−∆∆Cq method
(23). All primer sequences are
listed in Table SII.
Cell culture, transfection and
infection
The human colorectal cancer cell lines HCT8 and RKO
were purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Cells were routinely grown in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and incubated at 37°C in 5%
CO2 and humidified atmosphere. CD73-overexpressed cell
lines were generated by lentiviral transduction of a CD73 human
cDNA open reading frame clone using a plasmid reformed from
pMSCV-EcoRI-BglII-Zeocin (15 µg, 1 µg/µl; cat. no. 75088; Addgene,
Inc.). The empty vector was used as a control. All plasmids were
verified by sequencing. Lipofectamine™ 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) was used for transfection, according to
the manufacturer's protocol. Cells were cultured for 48 h until
they reached >90% confluence, and subsequently digested using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
collected for RT-qPCR analysis.
RNA sequencing and analysis
RNA samples extracted from colonic segments were
collected from the three experimental groups (model, APCP and NECA
groups). Library construction and sequencing were performed using
the BGISEQ-500 platform of the Beijing Genomic Institution
(www.bgitechsolutions.com; BGI Group).
The first step in the workflow involved purifying the
poly-A-containing mRNA molecules using poly-T oligo-attached
magnetic beads. Following purification, the mRNA was fragmented
into small pieces using divalent cations under 70°C. The cleaved
RNA fragments were copied into first-strand cDNA using reverse
transcriptase and random primers from the SuperScript™ III
First-Strand Synthesis System (Invitrogen; Thermo Fisher
Scientific, Inc.). This was followed by second-strand cDNA
synthesis using DNA polymerase I and RNase H. These cDNA fragments
then underwent addition of a single ‘A’ base and subsequent
ligation of the adapter. The products were then purified and
enriched with PCR amplification (Initial denaturation at 94°C for 2
min; 30 cycles of denaturation at 98°C for 2 sec, annealing at 55°C
for 30 sec and elongation at 68°C for 30 sec; and a final extension
at 68°C for 7 min). The PCR yield was then quantified by Qubit
(Thermo Fisher Scientific, Inc.) and samples were pooled together
to create a single-strand DNA (ssDNA) circle, which provided the
final library. DNA nanoballs (DNBs) were generated with the ssDNA
circle by rolling circle replication to enlarge the fluorescence
signals at the sequencing process. The DNBs were loaded into the
patterned nanoarrays and single-end reads of 50 bp were read
through on the BGISEQ-500 platform for the following data analysis
study. For this step, the BGISEQ-500 platform combined DNB-based
nanoarrays and stepwise sequencing using the Combinational
Probe-Anchor Synthesis Sequencing Method. The significance of the
differential expression of genes was defined by the bioinformatics
service of BGI according to the combination of the absolute value
of log2-ratio ≥1 and false discovery rate ≤0.001.
EuKaryotic Orthologous Group functional classification, Gene
Ontology (GO) and pathway annotation and enrichment analyses were
based on NCBI COG (https://www.ncbi.nlm.nih.gov/COG/), Gene Ontology
Database (http://www.geneontology.org/) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway database (http://www.genome.jp/kegg/), respectively.
Statistical analysis
Statistical analysis was performed using SPSS
version 22.0 (IBM Corp.). Values are expressed as the mean ± SEM
and multiple comparisons were analyzed using one-way ANOVA and
Tukey's post hoc test. Discrete data were compared using
Kruskal-Wallis test followed by Dunn's multiple comparison test.
Two-tailed unpaired Student's t-test was used to compare the
relative mRNA expression levels between CD73-overexpressed cell
lines and empty vector controls. P<0.05 was considered to
indicate a statistically significant difference.
Results
AOM/DSS-induced CAT model in C57BL/6J
mice
To investigate the role of CD73 and its downstream
pathway in CAT, the mice in a CAT model induced by AOM/DSS received
CD73 inhibitor (APCP) or adenosine receptor agonist (NECA)
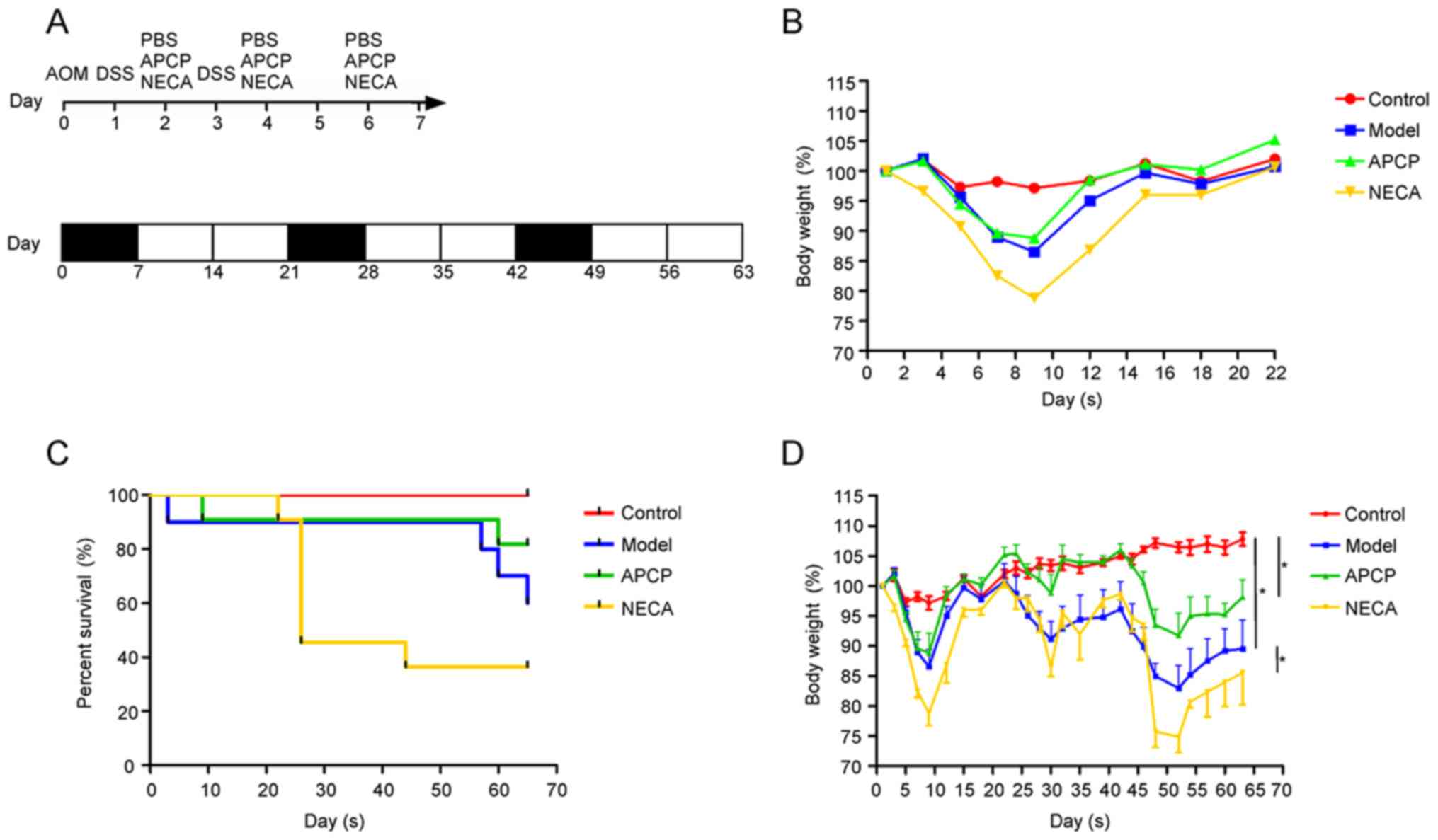
treatment. The experimental procedure and the effect on body weight
loss in the first cycle are shown in Fig. 1A and B.
By the end of the experiment, 6 mice in the control
group, 9 mice in the APCP group, 6 mice in the model group and 4
mice in the NECA group remained alive (Fig. 1C). Mice in the model group exhibited
a more significant body weight loss compared with the control
group, while mice receiving APCP exhibited a significantly lower
body weight loss and mice receiving NECA exhibited a higher body
weight loss compared with the mice in the model group (P<0.05;
Fig. 1D).
APCP attenuates, while NECA
aggravates, AOM/DSS-induced CAT
At the end of week 9, the mice were sacrificed and
the colon length (from the ileocecal junction to the anal verge)
was measured without tension. Subsequently, the colon was incised
longitudinally and macroscopically visible tumors were counted and
measured (Fig. 2A and C).
Shortening and swelling of the colon was observed in
both groups receiving AOM/DSS compared with the control group.
However, the colons in the APCP group were significantly longer
compared with the model group (68.29 vs. 62.67 mm, respectively;
P<0.05; Fig. 2B), while the
colons in the NECA group were shorter compared with the model group
(57.50 vs. 62.67 mm, respectively; P<0.05; Fig. 2B). After opening the colons, tumors
were observed between the mid colon and the distal rectum in the
experimental groups, while no tumors were found in the control
group. APCP treatment resulted in a significant reduction in the
number of tumors per mouse compared with mice in the model group
(5.1 vs. 7.7, respectively; P<0.05; Fig. 2D), while NECA treatment significantly
increased the number of tumors per mouse compared with mice in the
model group (10.3 vs. 7.7, respectively; P<0.05; Fig. 2D). The mean diameter of the existing
tumors was also significantly higher in the NECA group (2.96 mm in
the NECA group vs. 2.18 mm in the model group; P<0.05), while
the diameter of tumors of APCP-treated group were smaller compared
with the model group (1.85 mm in the APCP group vs. 2.18 mm in the
model group), although the difference was not statistically
significant (Fig. 2E).
APCP decreases, while NECA increases,
histological damage and levels of inflammatory cytokines in colonic
tissue
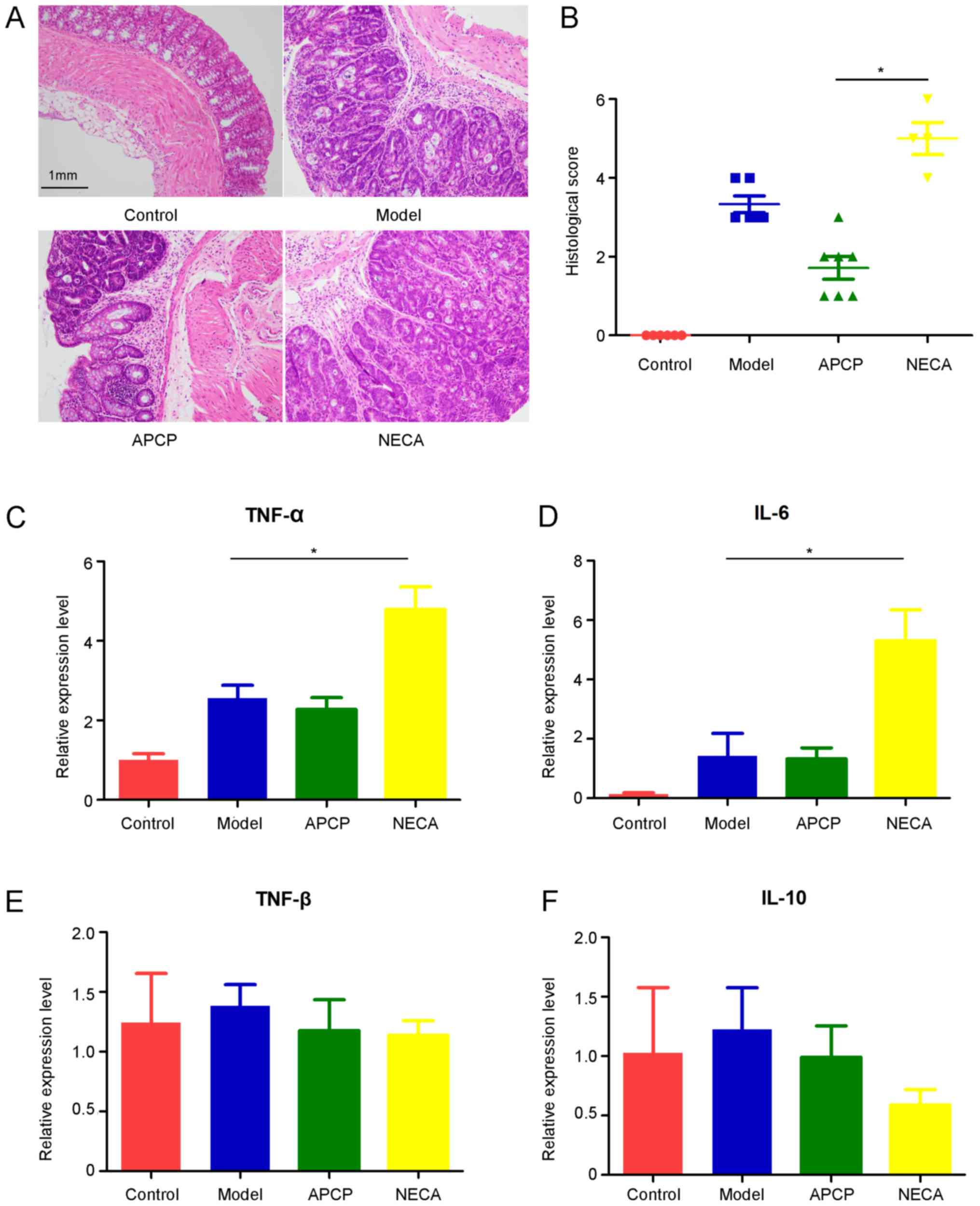
Following macroscopic inspection, segments of the
distal colon were stained with HE or kept in RNA stabilization
solution for further analysis.). All tumors from the experimental
groups were confirmed as adenomas with high-grade dysplasia
compared with the normal intestinal mucosa in the control group,
which was consistent with the histopathological score (Fig. 3A). The histopathological evaluation
score in the APCP group was significantly lower compared with the
NECA group (P<0.05; Fig. 3B).
Subsequently, qPCR was performed to detect the gene expression
levels of several inflammatory cytokines in colonic tissues,
including TNF-α, TNF-β, IL-10 and IL-6. The expression of TNF-α and
IL-6 significantly increased in the NECA group compared with the
model group, while the differences in the expression of TNF-α and
IL-6 between the APCP and model groups was not statistically
significant (Fig. 3C and D). There
was no significant difference in the expression of TNF-β and IL-10
among the four groups (Fig. 3E and
F).
Identification of differentially
expressed genes (DEGs) and genes associated with CAT by RNA
sequencing
To identify genes associated with CAT, RNA
sequencing of mice in the model, APCP and NECA groups was
conducted. A mean of 23.88 million clean reads were obtained that
mapped on the mouse genome with a reliable high mean mapping rate
of 93.69%, representing a mean of 20,677 genes that were expressed
in each sample. Subsequently, DEGs in the model vs. APCP groups,
model vs. NECA groups and NECA vs. APCP groups were analyzed.
Compared with the APCP group, a total of 180 upregulated and 222
downregulated genes were detected in the model group (Fig. S1A). Compared with the NECA group, a
total of 113 upregulated and 285 downregulated genes were detected
in the model group (Fig. S1B).
Compared with the APCP group, a total of 259 upregulated and 89
downregulated genes were detected in the NECA group (Fig. S1C). GO functional annotation of the
DEGs indicated that the upregulated and downregulated genes could
be classified into 6 major categories, including ‘cellular
processes’, ‘environmental information processing’, ‘genetic
information processing’, ‘human diseases’, ‘metabolism’ and
‘organismal systems’ (Fig. S1A-C).
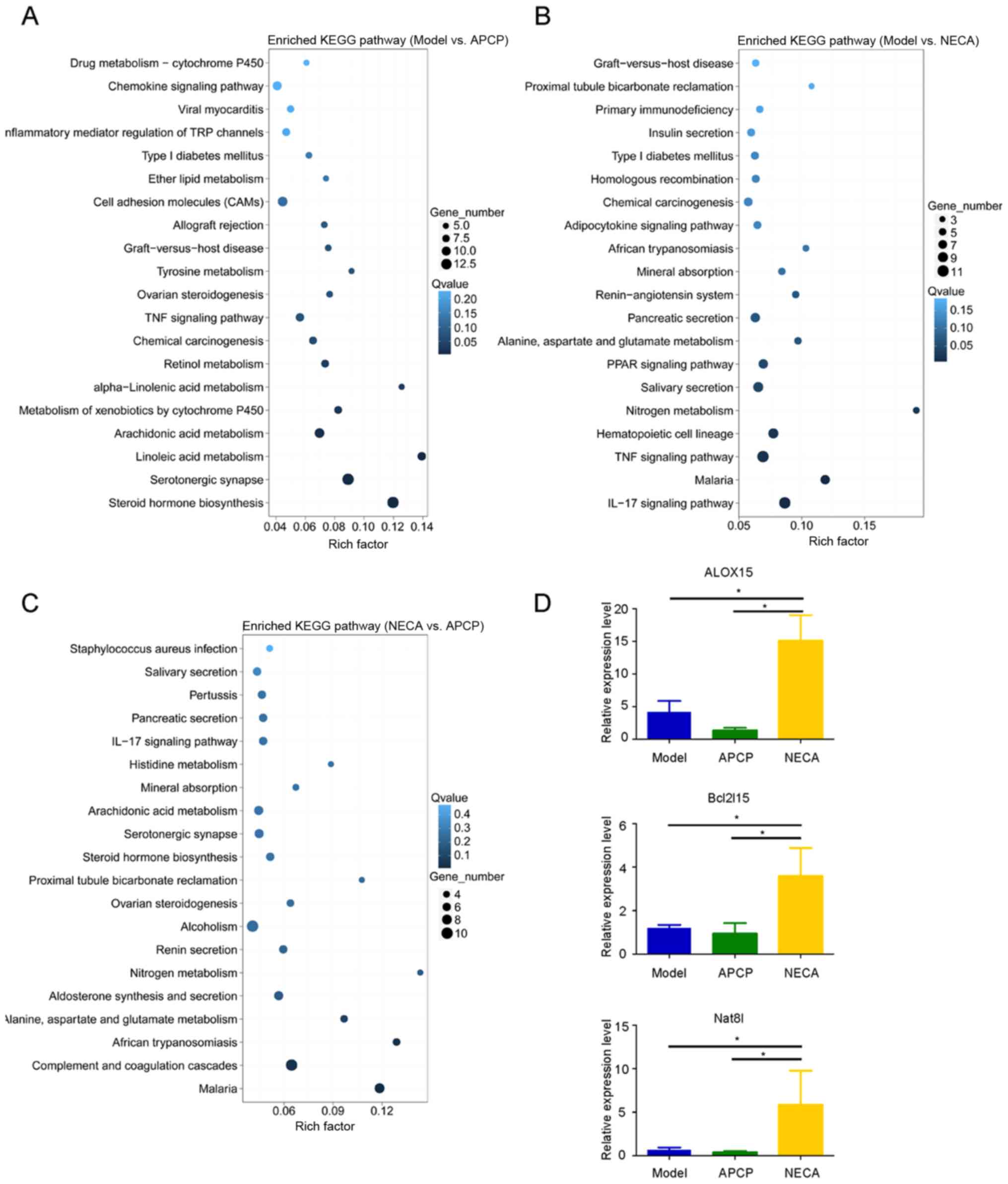
KEGG pathway analysis indicated the different pathway distributions
of DEGs in the model vs. APCP, model vs. NECA and NECA vs. APCP
groups (Fig. 4A-C). Among the
differentially upregulated and downregulated genes among the three
experimental groups, 8 genes were identified as having low
expression levels in the APCP group compared with the model group,
and high expression levels in the NECA group compared with the
model group (Fig. S1D). The 8 genes
were as follows: Arachidonate 15-lipoxygenase (ALOX15),
carcinoembryonic antigen-related cell adhesion molecule 2
(Ceacam2), mucosal pentraxin 1 (Mptx1), regulating synaptic
membrane exocytosis 2 (Rims2), Bcl-2-like protein 15 (Bcl2l15),
N-acetylaspartate synthetase (Nat8l), alpha-defensin 42 (Defa42)
and BC037156. Among these 8 genes, ALOX15, Bcl2l15 and Nat8l were
indicated as pathogenesis-associated genes, which were previously
reported to play an important role in tumorigenesis (24–26). The
expression levels of these three DEGs were then validated by
RT-qPCR, and the results were in accordance with the RNA sequencing
data. The expression levels of ALOX15, Bcl2l15 and Nat8l were
higher in the NECA group compared with the APCP and model groups
(P<0.05; Fig. 4D). In addition,
the expression of ALOX15 was significantly higher in
CD73-overexpressing cells compared with their vector-only
counterparts (Fig. S2).
Discussion
IBD is a multifactorial chronic relapsing disease
characterized by an abnormal systemic and local dysregulation of
the mucosal immune response (27,28).
Previous studies demonstrated that the risk factors for the
development of colorectal neoplasia in patients with IBD include
disease duration, anatomic extent of the disease, age, family
history of CRC and severity of inflammation (29–34).
Pro-inflammatory cytokines, such as TNF-α, IL-1β, IL-6 and IL-8,
can promote tumor cell proliferation, thereby promoting
carcinogenesis (35–37).
The primary function of CD73 is to hydrolyze
extracellular nucleoside monophosphates, leading to the generation
of extracellular adenosine, which is a well-known immunoregulatory
metabolite (38). Alam et al
(39) observed that
CD73−/− mice were more resistant to infection and
exhibited a greater inflammatory response and a significantly lower
bacterial load in the liver compared with wild-type mice. By
contrast, CD73 expression impaired murine immunity in
salmonellosis, leading to increased bacterial colonization and
prolonged infection (39). Doherty
et al (40) reported that the
CD73+/CD4+ T-cell population in patients with
active IBD are enriched with cells with a T-helper 17 (Th17)
phenotype, and may be used to monitor disease activity during
treatment. These peripheral CD73+/CD4+ T
cells expressed higher levels of pro-inflammatory markers (40). Forte et al (41) observed that inhibition of CD73 by
APCP improved B-cell-mediated antitumor immune response in a mouse
model of melanoma by affecting both the CD8+ T-cell- and
B-cell-mediated responses.
Given the crucial role of persistent inflammation in
the process of CAT and the immunoregulatory abilities of CD73, it
was hypothesized that CD73 may play an important role in
carcinogenesis in IBD by regulating pathways closely associated
with inflammation and immunity. Therefore, the CD73 inhibitor APCP
and the adenosine receptor agonist NECA were used in a mouse model
of CAT induced by AOM/DSS in immunocompetent C57BL/6J mice in the
present study.
The main cause of death in the three experiment
groups were due to the severe bloody diarrhea, followed by mucosa
inflammatory or intestinal obstruction caused by tumor formation
(42). These may reflect the
severity of AOM/DSS-induced CAT mouse model, as APCP attenuated
while NECA aggravated the survival rate compared with the model
group. CD73 was reported to be crucial for the regulation of murine
colonic inflammation due to the anti-inflammatory actions of
adenosine, mainly through A2 receptor stimulation (43,44).
However, other studies using mouse colitis models indicated that
inhibiting CD73 may reduce the histological damage of colonic
tissue (39,40). In line with these findings, the data
of the present study demonstrated that CD73 inhibition attenuated
AOM/DSS-induced CAT with reduced histological damage of the colon.
It was also observed that tumor number, diameter and tumor burden
were significantly increased in the NECA group and decreased in the
APCP group compared with the model group. Given the fact that
factors affecting tumor progression may lead to changes in the
tumor diameter, while those involved in tumor initiation may lead
to differences in tumor number, it may be inferred that inhibiting
the CD73 pathway may repress both AOM/DSS-induced tumor initiation
and progression. Compared with the model group, qPCR demonstrated
that the levels of inflammatory cytokines, including TNF-α and
IL-6, were significantly increased in the NECA group; however,
there were no significant differences in the decreased expression
levels of TNF-α and IL-6 between the APCP and model groups. TNF-α
is a pro-inflammatory cytokine secreted by Th17 cells in IBD and
IBD-associated cancer (45,46). In addition, IL-6 is one of the most
important pro-inflammatory cytokines mainly produced by myeloid
cells and was identified as a key promoter of carcinogenesis
(47). However, both NECA and APCP
act on all subtypes of the adenosine receptor, thus further studies
are required to determine which adenosine receptor subtype is
associated with the CAT process. The results of the present CAT
mouse model suggested that stimulating the adenosine receptors by
NECA might induce immunosuppression and finally lead to tumor
immune escape. However, the role of CD73 and adenosine receptors on
carcinogenesis induced by inflammation may be different compared
with other immunemodulatory effects, not only because it is a
complicated process, but because it also depends on the duration
that the agonist or inhibitor is administered.
Based on RNA sequencing results, DEGs, such as
ALOX15, Bcl2l15 and Nat8l, were found to be downregulated in the
APCP group and upregulated in the NECA group compared with the
model group. Namgaladze et al (48) reported that suppressing ALOX15
expression might promote an anti-inflammatory phenol type of
IL-4-stimulated human macrophages. Additionally, Kleinstein et
al (24) demonstrated that
genetic variability in 5-lipoxygenase-activating protein and ALOX15
might affect the risk of colorectal neoplasia, particularly for
rectal cancer. Bcl2l15 was reported to be involved in CRC cell
proliferation (25). Zand et
al (26) reported that Nat8l
plays a prominent role in promoting tumor growth and represents a
valuable target for anticancer therapy. Moreover, a higher Nat8l
expression level in colon cancer was associated with worse overall
survival (26). Taken together,
these studies strongly support the present finding that APCP
attenuated while NECA aggravated AOM/DSS-induced CAT in mice.
However, the mechanisms through which CD73 regulates ALOX15,
Bcl2l15 and Nat8l, and which of these three genes is the most
crucial as a CD73 downstream candidate involved in CAT, warrants
further investigation.
In conclusion, the results of the present study
suggested that CD73 and its downstream pathway might play an
important role in CAT. APCP, an inhibitor of CD73, can inhibit the
development of CAT through suppressing the expression of
pro-inflammatory cytokines, such as TNF-α, IL-6, and downregulating
the expression of colorectal tumorigenesis-associated genes, such
as ALOX15, Bcl2l15 and Nat8l.
Supplementary Material
Supporting Data
Acknowledgements
Part of this study has been presented in the 15th
congress of European Crohn's and Colitis Organisation (ECCO), 2020,
Vienna/Austria as a meeting text poster abstract.
Funding
The current study was supported by the National Key
R&D Program of China (grant no. 2017YFC1308800), National
Natural Science Foundation of China (grant nos. 81870383 and
81400603), Guangdong Natural Science Foundation (grant no.
2017A030313785), Science and Technology Planning Project of
Guangzhou City (grant no. 201804010014), Science and Technology
Planning Project of Guangdong Province (grant nos. 2015B020229001
and 20160916), and Clinical Innovation Research Program of
Guangzhou Regenerative Medicine and Health Guangdong Laboratory
(grant no. 2018GZR0201005).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XHL, XRW, PL and XJW contributed to study concept
and design. XHL, NL, XBZ, CZ and TH performed the animal model
experiments and acquired the data. XHL, XRW, YFC, ZRC, ZXC, PL, XJW
participated in the in vitro laboratory experiments and data
collection. XRW and XHL contributed to preliminary analysis and
interpretation of data and drafting of the manuscript. PL and XJW
reviewed the manuscript. All the authors took part in further
analysis and revising of the manuscript. XRW, PL and XJW supervised
the study. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol was approved by the Ethics
Committee of Sun Yat-sen University (approval no.
SYSU-IACUC-2020-B0038). All animal studies were conducted with the
approval of the Institutional Animal Care and Use Committee of Sun
Yat-sen University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
IBD
|
inflammatory bowel disease
|
|
CRC
|
colorectal cancer
|
|
CAT
|
colitis-associated tumorigenesis
|
|
APCP
|
adenosine 5′-(α,β-methylene)
diphosphate
|
|
NECA
|
1-(6-amino-9-H-purin-9-yl)-1-deoxy-N-ethyl-β-D-ribofuranuronamide
|
|
AOM
|
azoxymethane
|
|
DSS
|
dextran sulfate sodium
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
HE
|
hematoxylin and eosin
|
|
IHC
|
immunohistochemistry
|
|
SEM
|
standard error mean
|
References
|
1
|
Liu TC and Stappenbeck TS: Genetics and
pathogenesis of inflammatory bowel disease. Annu Rev Pathol.
11:127–148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khor B, Gardet A and Xavier RJ: Genetics
and pathogenesis of inflammatory bowel disease. Nature.
474:307–317. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Singh R, Mishra MK and Aggarwal H:
Inflammation, immunity, and cancer. Mediators Inflamm.
2017:60273052017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Navaneethan U, Zhu X, Lourdusamy D,
Lourdusamy V, Shen B and Kiran R: Colorectal cancer resection rates
in patients with inflammatory bowel disease: A population-based
study. Gastroenterol Rep (Oxf). 6:263–269. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bopanna S, Ananthakrishnan AN, Kedia S,
Yajnik V and Ahuja V: Risk of colorectal cancer in Asian patients
with ulcerative colitis: A systematic review and meta-analysis.
Lancet Gastroenterol Hepatol. 2:269–276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lutgens MW, van Oijen MG, van der Heijden
GJ, Vleggaar FP, Siersema PD and Oldenburg B: Declining risk of
colorectal cancer in inflammatory bowel disease: An updated
meta-analysis of population-based cohort studies. Inflamm Bowel
Dis. 19:789–799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zimmermann H: 5′-Nucleotidase: Molecular
structure and functional aspects. Biochem J. 285((Pt 2)): 345–365.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Haskó G and Cronstein BN: Adenosine: An
endogenous regulator of innate immunity. Trends Immunol. 25:33–39.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Antonioli L, Yegutkin GG, Pacher P,
Blandizzi C and Haskó G: Anti-CD73 in cancer immunotherapy:
Awakening new opportunities. Trends Cancer. 2:95–109. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang B: CD73: A novel target for cancer
immunotherapy. Cancer Res. 70:6407–6411. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ghalamfarsa G, Kazemi MH, Raoofi Mohseni
S, Masjedi A, Hojjat-Farsangi M, Azizi G, Yousefi M and
Jadidi-Niaragh F: CD73 as a potential opportunity for cancer
immunotherapy. Expert Opin Ther Targets. 23:127–142. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stagg J, Divisekera U, McLaughlin N,
Sharkey J, Pommey S, Denoyer D, Dwyer KM and Smyth MJ: Anti-CD73
antibody therapy inhibits breast tumor growth and metastasis. Proc
Natl Acad Sci USA. 107:1547–1552. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin D, Fan J, Wang L, Thompson LF, Liu A,
Daniel BJ, Shin T, Curiel TJ and Zhang B: CD73 on tumor cells
impairs antitumor T-cell responses: A novel mechanism of
tumor-induced immune suppression. Cancer Res. 70:2245–2255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stagg J, Divisekera U, Duret H, Sparwasser
T, Teng MW, Darcy PK and Smyth MJ: CD73-deficient mice have
increased antitumor immunity and are resistant to experimental
metastasis. Cancer Res. 71:2892–2900. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Fan J, Thompson LF, Zhang Y, Shin
T, Curiel TJ and Zhang B: CD73 has distinct roles in
nonhematopoietic and hematopoietic cells to promote tumor growth in
mice. J Clin Invest. 121:2371–2382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kaku H, Cheng KF, Al-Abed Y and Rothstein
TL: A novel mechanism of B cell-mediated immune suppression through
CD73 expression and adenosine production. J Immunol. 193:5904–5913.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Grenz A, Zhang H, Eckle T, Mittelbronn M,
Wehrmann M, Köhle C, Kloor D, Thompson LF, Osswald H and Eltzschig
HK: Protective role of ecto-5′-nucleotidase (CD73) in renal
ischemia. J Am Soc Nephrol. 18:833–845. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eckle T, Krahn T, Grenz A, Köhler D,
Mittelbronn M, Ledent C, Jacobson MA, Osswald H, Thompson LF,
Unertl K and Eltzschig HK: Cardioprotection by ecto-5′-nucleotidase
(CD73) and A2B adenosine receptors. Circulation. 115:1581–1590.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mahamed DA, Toussaint LE and Bynoe MS:
CD73-generated adenosine is critical for immune regulation during
Toxoplasma gondii infection. Infect Immun. 83:721–729. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ray MA, Johnston NA, Verhulst S, Trammell
RA and Toth LA: Identification of markers for imminent death in
mice used in longevity and aging research. J Am Assoc Lab Anim Sci.
49:282–288. 2010.PubMed/NCBI
|
|
22
|
Yang X, Zhang F, Wang Y, Cai M, Wang Q,
Guo Q, Li Z and Hu R: Oroxylin A inhibits colitis-associated
carcinogenesis through modulating the IL-6/STAT3 signaling pathway.
Inflamm Bowel Dis. 19:1990–2000. 2013.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kleinstein SE, Heath L, Makar KW, Poole
EM, Seufert BL, Slattery ML, Xiao L, Duggan DJ, Hsu L, Curtin K, et
al: Genetic variation in the lipoxygenase pathway and risk of
colorectal neoplasia. Genes Chromosomes Cancer. 52:437–449. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao R, Li C and Chai B: miRNA-144
suppresses proliferation and migration of colorectal cancer cells
through GSPT1. Biomed Pharmacother. 74:138–144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zand B, Previs RA, Zacharias NM,
Rupaimoole R, Mitamura T, Nagaraja AS, Guindani M, Dalton HJ, Yang
L, Baddour J, et al: Role of increased n-acetylaspartate levels in
cancer. J Natl Cancer Inst. 108:djv4262016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Torres J, Mehandru S, Colombel JF and
Peyrin-Biroulet L: Crohn's disease. Lancet. 389:1741–1755. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ungaro R, Mehandru S, Allen PB,
Peyrin-Biroulet L and Colombel JF: Ulcerative colitis. Lancet.
389:1756–1770. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Farraye FA, Odze RD, Eaden J and Itzkowitz
SH: AGA technical review on the diagnosis and management of
colorectal neoplasia in inflammatory bowel disease.
Gastroenterology. 138:746–774, 774.e1-4; quiz e12-13. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Askling J, Dickman PW, Karlén P, Broström
O, Lapidus A, Löfberg R and Ekbom A: Colorectal cancer rates among
first-degree relatives of patients with inflammatory bowel disease:
A population-based cohort study. Lancet. 357:262–266. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fumery M, Dulai PS, Gupta S, Prokop LJ,
Ramamoorthy S, Sandborn WJ and Singh S: Incidence, risk factors,
and outcomes of colorectal cancer in patients with ulcerative
colitis with low-grade dysplasia: A systematic review and
meta-analysis. Clin Gastroenterol Hepatol. 15:665–674.e5. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gyde SN, Prior P, Allan RN, Stevens A,
Jewell DP, Truelove SC, Lofberg R, Brostrom O and Hellers G:
Colorectal cancer in ulcerative colitis: A cohort study of primary
referrals from three centres. Gut. 29:206–217. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Munkholm P, Langholz E, Davidsen M and
Binder V: Intestinal cancer risk and mortality in patients with
Crohn's disease. Gastroenterology. 105:1716–1723. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rogler G: Chronic ulcerative colitis and
colorectal cancer. Cancer Lett. 345:235–241. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Popivanova BK, Kitamura K, Wu Y, Kondo T,
Kagaya T, Kaneko S, Oshima M, Fujii C and Mukaida N: Blocking
TNF-alpha in mice reduces colorectal carcinogenesis associated with
chronic colitis. J Clin Invest. 118:560–570. 2008.PubMed/NCBI
|
|
36
|
Azer SA: Overview of molecular pathways in
inflammatory bowel disease associated with colorectal cancer
development. Eur J Gastroenterol Hepatol. 25:271–281. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Taniguchi K and Karin M: IL-6 and related
cytokines as the critical lynchpins between inflammation and
cancer. Semin Immunol. 26:54–74. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Regateiro FS, Cobbold SP and Waldmann H:
CD73 and adenosine generation in the creation of regulatory
microenvironments. Clin Exp Immunol. 171:1–7. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Alam MS, Kuo JL, Ernst PB, Derr-Castillo
V, Pereira M, Gaines D, Costales M, Bigley E and Williams K:
Ecto-5′-nucleotidase (CD73) regulates host inflammatory responses
and exacerbates murine salmonellosis. Sci Rep. 4:44862014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Doherty GA, Bai A, Hanidziar D, Longhi MS,
Lawlor GO, Putheti P, Csizmadia E, Nowak M, Cheifetz AS, Moss AC
and Robson SC: CD73 is a phenotypic marker of effector memory Th17
cells in inflammatory bowel disease. Eur J Immunol. 42:3062–3072.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Forte G, Sorrentino R, Montinaro A,
Luciano A, Adcock IM, Maiolino P, Arra C, Cicala C, Pinto A and
Morello S: Inhibition of CD73 improves B cell-mediated anti-tumor
immunity in a mouse model of melanoma. J Immunol. 189:2226–2233.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhu Y, Gu L, Li Y, Lin X, Shen H, Cui K,
Chen L, Zhou F, Zhao Q, Zhang J, et al: miR-148a inhibits colitis
and colitis-associated tumorigenesis in mice. Cell Death Differ.
24:2199–2209. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Odashima M, Bamias G, Rivera-Nieves J,
Linden J, Nast CC, Moskaluk CA, Marini M, Sugawara K, Kozaiwa K,
Otaka M, et al: Activation of A2A adenosine receptor attenuates
intestinal inflammation in animal models of inflammatory bowel
disease. Gastroenterology. 129:26–33. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sotnikov I and Louis NA: CD73-dependent
regulation of interferon alphaA and interleukin-10 in the inflamed
mucosa. ScientificWorldJournal. 10:2167–2180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mantzaris GJ: Previous cancer and/or
lymphoma in patients with refractory IBD-con: Anti-TNF or
conventional immunosuppressive treatment. Dig Dis. 32 (Suppl
1):S122–S127. 2014. View Article : Google Scholar
|
|
46
|
Bandzar S, Gupta S and Platt MO: Crohn's
disease: A review of treatment options and current research. Cell
Immunol. 286:45–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Namgaladze D, Snodgrass RG, Angioni C,
Grossmann N, Dehne N, Geisslinger G and Brüne B: AMP-activated
protein kinase suppresses arachidonate 15-lipoxygenase expression
in interleukin 4-polarized human macrophages. J Biol Chem.
290:24484–24494. 2015. View Article : Google Scholar : PubMed/NCBI
|