Introduction
The H2.0-like homeobox gene (HLX) encodes
transcription factors that play roles in promoting normal
hematopoietic cell proliferation and tumor immunity, and the HLX
protein plays a key role in the development of cancer (1). HLX is expressed at high levels in
various tumor cells but at low levels in normal cells; however, the
complete deletion of HLX results in cell death (2,3). A
previous study reported that the expression of HLX decreased during
CD34+ hematopoietic stem or progenitor cell
differentiation, suggesting that HLX may play a role in maintaining
the differentiation potential of hematopoietic stem or progenitor
cells (4).
Acute myelogenous leukemia (AML) is a class of
malignant tumors derived from hematopoietic stem or progenitor
cells. The main treatment methods for AML include chemotherapy,
radiotherapy, hematopoietic stem cell transplantation, cell
immunotherapy, targeted therapy and traditional Chinese medicine
treatment (5). Currently, it is
difficult to improve the complete remission rate and long-term
survival rate with existing treatment regimens (6). However, research and examination of
immunotarget therapy have revealed that tumor immunotherapy and
molecular targeted therapy may be potential therapeutic
breakthroughs. As previously demonstrated, the aim of Feiji Recipe
(Components of this prescription: Astragalus 30 g, Atractylodes
macrocephala 15 g, Poria 20 g, American ginseng 10 g, Trichosanthes
30 g, pinellia 12 g, Fritillaria Zhejiang 20 g, yam 15 g, job's
tears 30 g, Hedyotis Baihua 30 g, Chonglou 30 g, zhibaibu 15 g,
bayzha 15 g, zaojiaoci 30 g, Chenpi 12 g, liquorice 6 g, each
patient's dosage and content of the recipe are not exactly the
same) in the treatment of lung cancer was to restore the function
of T-cells in the cancer microenvironment by interfering with the
indoleamine 2,3-dioxygenase pathway (7). In recent years, fms related receptor
tyrosine kinase 3 (FLT3), Nucleophosmin 1 (NPM1), DNA
(cytosine-5)-methyltransferase 3A (DNMT3a) and isocitrate
dehydrogenase (NADP+) 2 (IDH2) have been the top four targeted
molecules in AML (7). Drug
development with NPM1 is difficult, and thus is not considered a
candidate (8). Moreover, the
mutation of DNMT3AR882H can inhibit the function of the wild-type
protein, and the development of inhibitors worsens the disease
(7). Thus, FLT3 and IDH2 have become
the main focus of drug development. IDH1 and IDH2 have similar
molecular structures, and related drugs are also in development
(9). Furthermore, molecular targeted
therapy has resulted in positive outcomes in the treatment of
hematologic malignancies (10).
Previous studies have identified HLX, which is highly expressed in
AML cells, as a new target for this malignancy and reported that
high HLX expression is associated with poor prognosis in patients
with AML, but the specific mechanism of HLX gene function in AML
remains unknown (10,11).
Previous studies have shown the effect of inhibiting
HLX on suppressing the proliferation of leukemia cells and have
revealed the relationship between HLX with p21-activated kinase 1
(PAK1) and B-cell translocation gene 1 (BTG1) (10–14);
however, to the best of our knowledge, these studies have not
examined the Janus kinase (JAK)/STAT signaling pathway. Thus, the
present study analyzed HLX expression in every subtype of AML
cells, and then focused on the NB4 cell line (AML/M3 subtype) to
elucidate the function and possible mechanism of action of HLX in
AML.
Materials and methods
Media and reagents
The human AML KG1a, NB4 and THP-1 cell lines and the
human acute lymphoblastic leukemia Jurkat cell line were obtained
from the Center Laboratory of Enze Medical Group (Zhejiang, China).
RPMI-1640 medium and FBS were obtained from Cytiva. DEPC water,
TRIzol®, RNAlater Stabilization Solution and Blockit
Alexa Fluor Red Fluorescent Oligo were obtained from Thermo Fisher
Scientific, Inc. SYBR-Green was obtained from Roche Diagnostics,
and PCR reagents were obtained from Axygen (Corning, Inc.).
Lipofectamine® RNAiMAX Reagent (www.lifetechnologies.com), STAT5 antibody (cat. no.
QC215910), small interfering siRNA-HLX1 and siRNA-HLX2 were
obtained from Thermo Fisher Scientific, Inc. The HLX1 antibody
(cat. no. GTX87590) was obtained from GeneTex International
Corporation. Lipofectamine® 2000 was obtained from
Invitrogen (Thermo Fisher Scientific, Inc.), and the Cell Cycle
Staining kit was obtained from Hangzhou Multi Sciences (Lianke)
Biotech Co., Ltd. The FACSCalibur flow cytometer and PCR machine
were obtained from BD Biosciences.
Cell culture
The human leukemia cell lines were cultured in RPMI
1640 medium with 10% FBS in an incubator at 37°C with 5%
CO2 and 95% humidity.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using the Qiagen
RNeasy® Mini kit (Qiagen, Inc.) and a RevertAid RT
Reverse Transcription kit (Thermo Fisher Scientific, Inc.) was used
to reverse transcribe RNA into cDNA at 65°C for 5 min. qPCR was
performed using the SYBR Premix Ex Tag kit (Takara Bio, Inc.) at
42°C for 60 min, 70°C for 5 min and 40°C for 10 min, for 40 cycles,
and an ABI 7500 Sequencing Detection system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocols. GAPDH was used as a quantitative control gene, and all
reactions were performed in triplicate.
The housekeeping gene GAPDH served as the reference
gene, and RT-qPCR was performed using an ABI 7500 Sequencing
Detection system. The results were calculated using the
2−ΔΔCq method (15), and
the median ΔCq value of GAPDH was used to calculate expression
levels in the control group. Primer sequences of genes HLX, PAK1,
neuropilin 1 (NRP1) and BTG1 for RT-qPCR were as follows
(www.generay.com): HLX: Forward,
5′-ATCTCACTTCCCTGCTAACCG-3′ and reverse,
5′-AGAAGCCTCGTTAATGGGATCT-3′; PAK1: Forward,
5′-CAGCCCCTCCGATGAGAAATA-3′ and reverse,
5′-CAAAACCGACATGAATTGTGTGT-3′; BTG1: Forward,
5′-AGCGGATTGGACTGAGCAG-3′ and reverse, 5′-GGTGCTGTTTTGAGTGCTACC-3′;
NRP1: Forward, 5′-ACGTGGAAGTCTTCGATGGAG-3′ and reverse,
5′-CACCATGTGTTTCGTAGTCAGA-3′; GAPDH: Forward,
5′-CTGGGCTACACTGAGACC-3′ and reverse,
5′-AAGTGGTCGTTGAGGGCAATG-3′.
Western blotting
Proteins were extracted from cells in the
logarithmic growth phase using RIPA buffer supplied by Enze Medical
Group Laboratory. For blocking, 5% skimmed milk was used at 4°C
overnight. Protein was determined using a BCA Protein Assay kit
(Sangon Biotech Co. Ltd). In total, 50 kDA protein was loaded per
lane onto a 5% gel. The proteins were then transferred to
polyvinylidene difluoride (PVDF) membranes (0.45 µm). The PVDF
membrane was incubated with the following primary antibodies
overnight at 4°C: HLX1 (cat. no. GTX87590, 1:1,000) and STAT5 (cat.
no. QC215910, 1:2,000), obtained from GeneTex International
Corporation. Then, the membranes were incubated with HLX secondary
antibody [goat anti-rabbit IgG (H+L) secondary antibody, cat. no.
31460, 1:1,000, Invitrogen; Thermo Fisher Scientific, Inc.], STAT
secondary antibody [goat anti-mouse IgG, IgM (H + L) secondary
antibody, cat. no. A-10677, 1:3,000, Invitrogen; Thermo Fisher
Scientific, Inc.] for 1 h at room temperature. Band intensity was
semi-quantitatively analyzed using ImageJ software version 1.8.0
(National Institutes of Health).
Construction of HLX-knockdown
cells
HLX-specific sequences were designed according to
the GenBank database (Invitrogen; Thermo Fisher Scientific, Inc.),
and siRNA-HLX1 and siRNA-HLX2 were generated (Table I). AML cells were transfected with
siRNA-HLX1, -HLX2 or non-targeting control siRNA-highGC using
Lipofectamine® RNAiMAX (www.lifetechnologies.com). The following solutions
were used: i) Liquid A, 25 µl Opti-MEM Medium + 0.5 µl siRNA; and
ii) Liquid B, 25 µl Opti-MEM + 1.5 µl Lipofectamine. Then, liquid B
was added to liquid A for 5 min at room temperature and mixed to
obtain Liquid C. Cells were added at a density of
(1-4×104) to liquid C after 20 min at room temperature,
followed by incubation for 1–3 days at 37°C. The transfected cells
were observed and counted manually under the fluorescence
microscope at 200× magnification. The cells were divided into the
blank control group (no added reagent), the negative control group
(transfected with non-specific high-GC siRNA) and experimental
group (transfected with siRNA-HLX1 or siRNA-HLX2). In total, three
wells were used for each group. The transfection efficiency was
determined at the time points of 12, 24, 48 or 72 h after
transfection, and the experiment was repeated three times.
 | Table I.Sequences of siRNA-HLX1 and
siRNA-HLX2. |
Table I.
Sequences of siRNA-HLX1 and
siRNA-HLX2.
| Gene | Cat. no. | Type | Sequence
(5′→3′) |
|---|
| siRNA-HLX1 | 10620318-296424
E04 | RNA |
CCCUUAAACUCGAACCCAAGAAAUU |
|
| 10620319-296424
E05 | RNA |
AAUUUCUUGGGUUCGAGUUUAAGGG |
| siRNA-HLX2 | 10620318-296424
E06 | RNA |
GCUGAGAGAUCUCACUUCCCUGCUA |
|
| 10620319-296611
A12 | RNA |
UAGCAGGGAAGUGAGAUCUCUCAGC |
MTS/PMS assay for cell
proliferation
NB4 cells were seeded into 96-well plates at a
density of 1×105 cells/ml. The following groups were
analyzed: Negative control group (transfected with non-specific
high-GC siRNA) and experimental group (siRNA-HLX1 or siRNA-HLX2).
Proliferation was determined using the MTS/PMS Cell Proliferation
Assay kit (www.liankebio.com) according to the
manufacturer's instructions. The absorbance was measured at 490 nm
on a multi-well plate reader at 12, 24, 48 and 72 h after
transfection. The survival rate was calculated [Survival inhibition
rate=(1-odvalue of experimental group/odvalue of control group)
×100%], and each assay was performed in triplicate. The results are
presented as the mean ± standard deviation (SD).
Cell cycle assay
NB4 cells were fixed with 70% ethanol at 4°C after
24 h of exposure to siRNA-HLX2. RNase was used (Thermo Fisher
Scientific, Inc.). Then, add the cell to −20 anhydrous ethanol,
stir it at high speed while adding, discard the ethanol, add PBS at
room temperature, place it for 15 min, added 1 ml Cell cycle
staining kit (www.liankebio.com) and shake it for 5–10 sec, and
incubate it at room temperature in dark for 30 min, and the cell
cycle was analyzed by flow cytometry (Flowjo®Flow data
analysis software version 10.5.2, BD Biosciences).
Changes in associated proteins and
genes after HLX gene knockdown
STAT5 protein expression in NB4 cells was analyzed
by western blotting after transfection with siRNA-HLX2, and the
expression levels of related genes, including PAK1, NRPl and BTG1,
were assessed by RT-qPCR.
Statistical analysis
The data were analyzed using SPSS 22.0 software
(SPSS, Inc.). A Kolmogorov-Smirnov test was used to analyze data
normality. Data are presented as the mean ± SD of ≥3 independent
experiments. A t-test was used to compare the means of two groups.
ANOVA (parametric) and Kruskal-Wallis (non-parametric) were used to
compare the means of multiple groups, and variations of statistical
significance were further subjected to post hoc pairwise analysis
by applying the Tukey's test and the Dunn's test, respectively.
P<0.05 was considered to indicate a statistically significant
difference.
Results
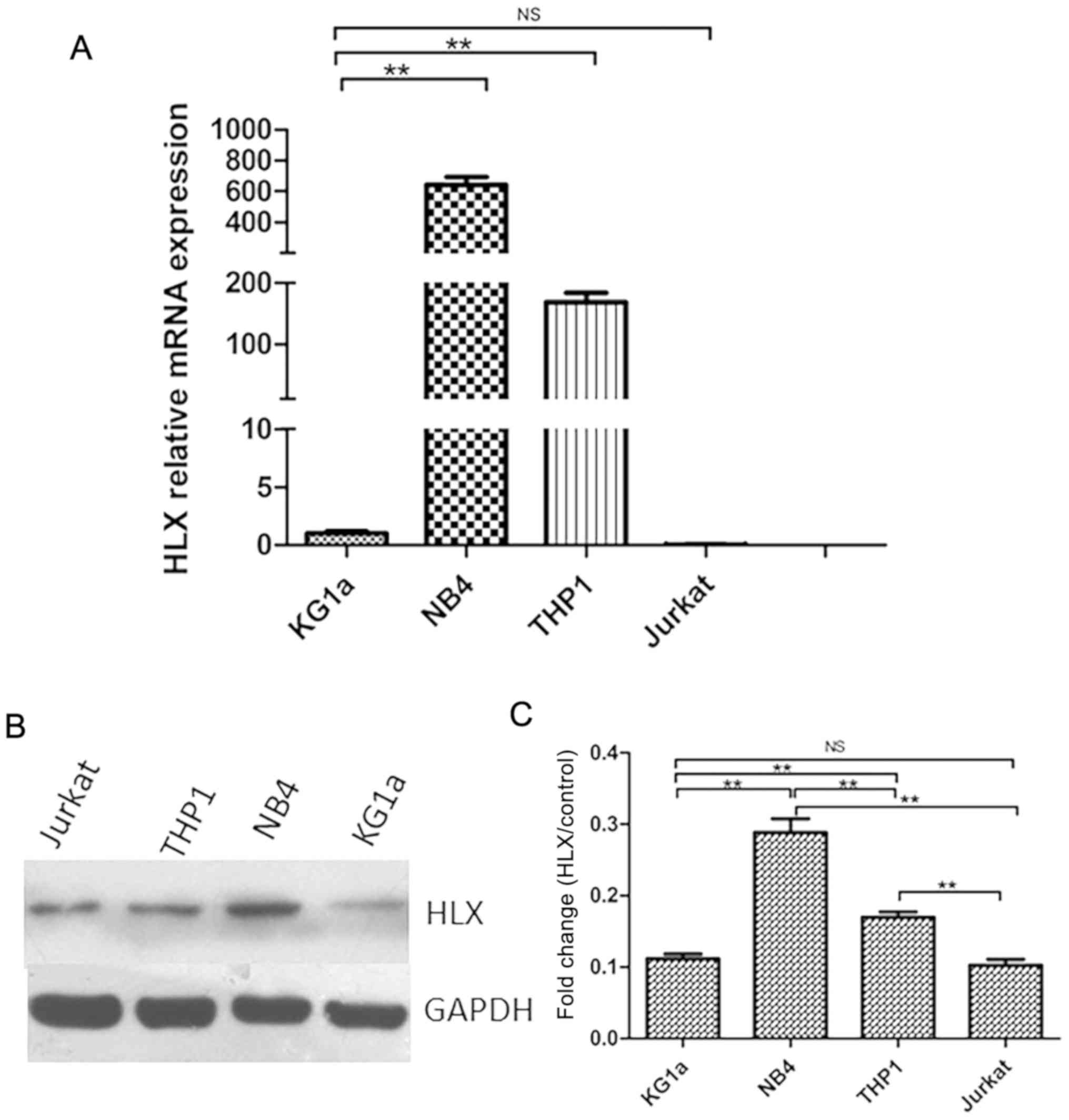
HLX expression is highest in the NB4
AML cell line
RT-qPCR was used to detect HLX gene expression in
KG1a, NB4, THP-1 and Jurkat cells. It was found that the HLX gene
was differentially expressed in KG1a (AML/M0 subtype), NB4 (AML/M3
subtype), and THP-1 (AML/M5 subtype) cells. Furthermore, HLX
expression was higher in NB4 and THP-1 cells compared with the
control group, but was highest in the NB4 cell line (Fig. 1A). This result was also demonstrated
by western blotting (Fig. 1B and
C).
HLX expression is lowest at 24 h in
NB4 cells after siRNA transfection
Cells in the blank control group (non-transfected),
negative control group (neg siRNA) and experimental group
(siRNA-HLX1 or siRNA-HLX2) were transfected as indicated, and HLX
gene expression was detected using RT-qPCR after 12, 24, 48 and 72
h. HLX mRNA expression was lowest at 24 h. Of the two
gene-targeting sequences, siRNA-HLX2 was more effective at
silencing HLX expression than the siRNA-HLX1 group compared with
the control group (Fig. 2A). HLX
protein expression in the aforementioned groups was detected by
western blotting after 24 h, and the results indicated that HLX
protein expression was decreased in the siRNA-HLX2 group compared
with the untreated control group (Fig.
2B and C).
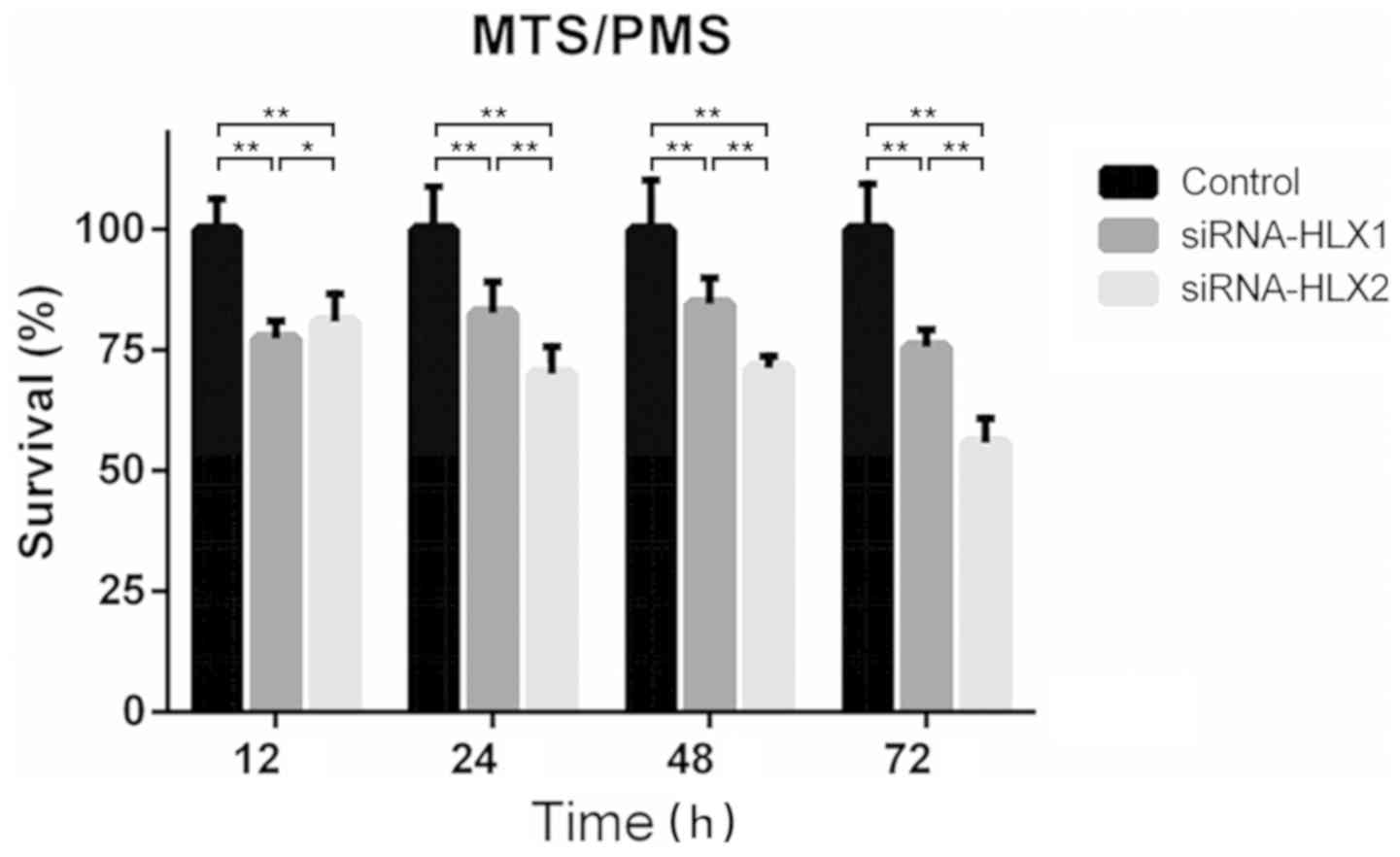
Knockdown of HLX using siRNA-HLX2
inhibits NB4 cell survival to a higher level compared with
siRNA-HLX1
The absorbance at 490 nm was measured using the
MTS/PMS method before transfection and at 12, 24, 48 and 72 h after
transfection with siRNA-HLX1, siRNA-HLX2 and high-GC siRNA. The
survival rate was calculated, and the results suggested that
siRNA-HLX2 had a stronger inhibitory effect on proliferation
compared with siRNA-HLX1. The survival rates of siRNA-HLX2 at 12,
24, 48 and 72 h were 80.87±4.99, 70.08±4.87, 71.29±2.15 and
55.73±4.46%, respectively, compared with the control (Fig. 3).
Knockdown of HLX using siRNA-HLX2
arrests NB4 cells in the G0/G1 phase
As HLX gene expression was lowest at 24 h after
siRNA-HLX2 transfection, the cell cycle distribution of NB4 cells
transfected with siRNA-HLX2 was analyzed at 24 h. At this time
point, the S phase population was reduced and the cell cycle was
blocked in the G0/G1 phase. After
transfection of NB4 cells with siRNA-HLX2, cell survival decreased
with the reduction in the S phase population from 12.32±0.76 to
4.29±2.85%, G2/M phase population from 23.92±1.22 to
20.13±2.56%, and the number of cells in the
G0/G1 phase increased from 63.57±1.87 to
72.24±2.64% (Fig. 4).
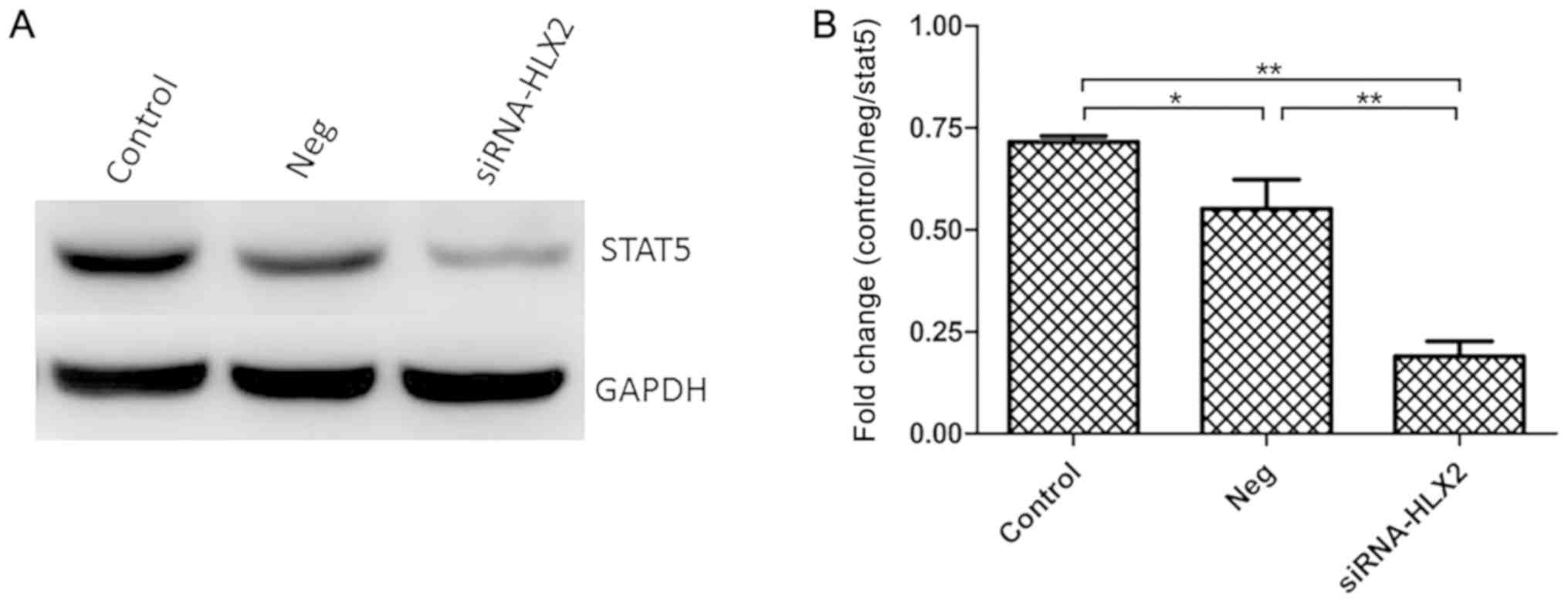
STAT5 protein expression is
downregulated in NB4 cells after siRNA-HLX2 transfection
After transfection of NB4 cells with siRNA-HLX2,
STAT5 protein expression was detected by western blotting. In
total, three groups were analyzed untreated control group, negative
control group and siRNA-HLX2 group), and the results indicated that
STAT5 protein expression decreased significantly in the siRNA-HLX2
group (Fig. 5). Compared with the
untreated control and the non-targeting control, the STAT5
expression levels in the siRNA-HLX2 group decreased
significantly.
Genes related to the JAK/STAT
signaling pathway show altered expression in NB4 cells after siRNA
transfection on the differences seen with time
The expression levels of JAK/STAT signaling pathway
related genes (PAK1, NRP1 and BTG1) were detected by RT-qPCR after
transfection of NB4 cells with siRNA-HLX1 or siRNA-HLX2. It was
demonstrated that knockdown of the HLX gene decreased the
expression levels of PAK1 (Fig. 6A and
B) and NRP1 (Fig. 6C and D), but
increased BTG1 expression (Fig. 6E and
F).
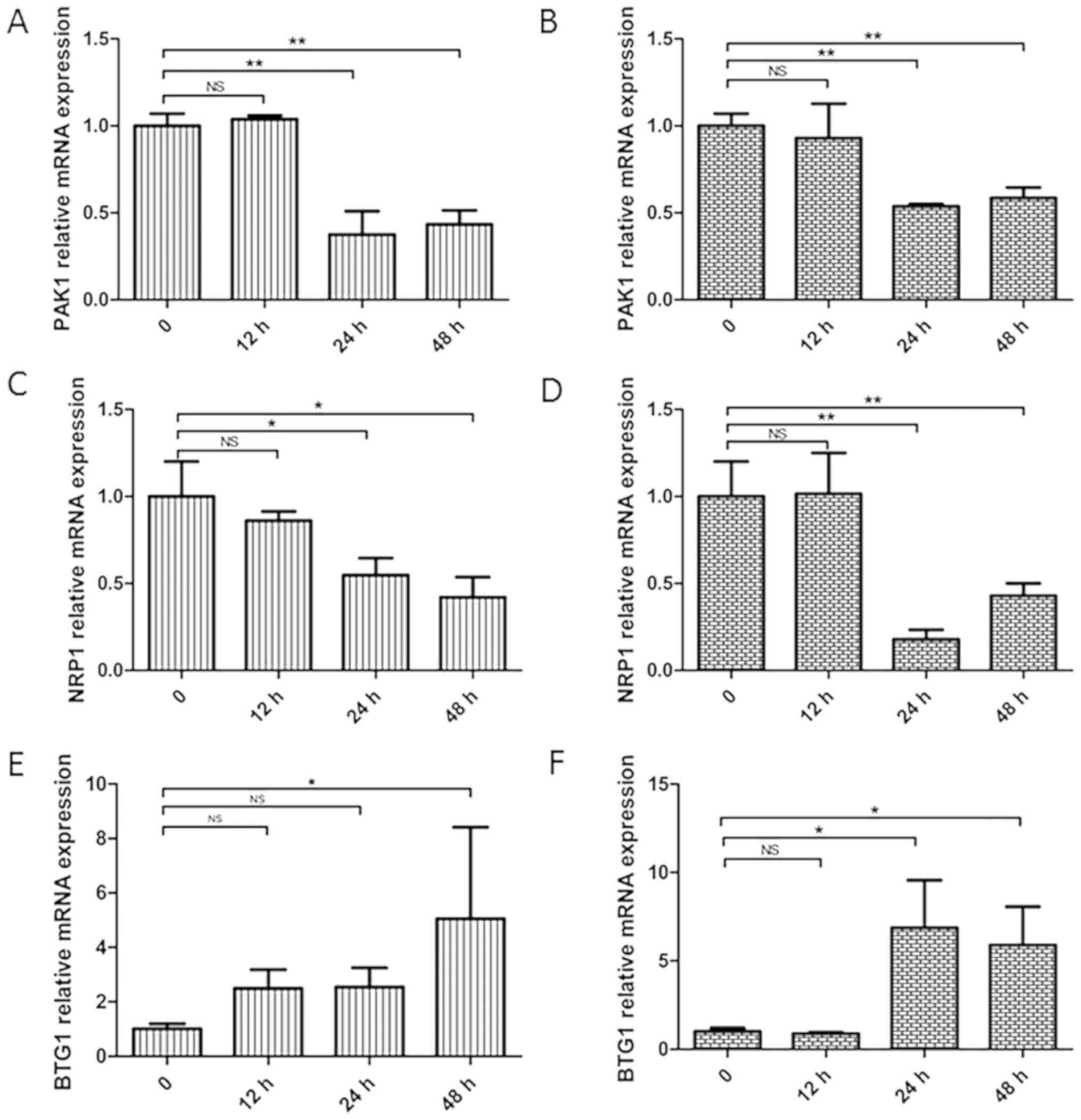 | Figure 6.Changes in the related genes, PAK1,
NRP1 and BTG1, after transfection of NB4 cells with siRNA-HLX1 or
siRNA-HLX2. Data at 0 h were the scramble siRNA and are used as the
control group. Changes in PAK1 gene expression in NB4 cells after
(A) siRNA-HLX1 or (B) siRNA-HLX2 transfection. Changes in NRP1 gene
expression in NB4 cells after (C) siRNA-HLX1 or (D) siRNA-HLX2
transfection. Changes in BTG1 gene expression in NB4 cells after
(E) siRNA-HLX1 or (F) siRNA-HLX2 transfection. *P<0.05,
**P<0.01. NS, not significant; HLX, H2.0-like homeobox gene;
siRNA, small interfering RNA; Neg, negative control; PAK1,
p21-activated kinase 1; BTG1, B-cell translocation gene 1; NRP1,
neuropilin 1. |
Discussion
HLX is located on chromosome 1q41-q421, and its
1467-bp open reading frame encodes a protein composed of 488 amino
acids (16). Previous studies have
reported that the HLX gene participates in a variety of processes,
including cell proliferation, differentiation and maturation
(17,18). Moreover, in combination with T-box
transcription factor 21 and other transcription factors, HLX
induces interferon-γ production and provides T helper Th-2 cells
with typical Th1-cell functions (19,20). At
present, previous studies have reported that the abnormal
expression of HLX is closely related to the development of
autoimmune diseases, such as Graves' disease, gastric cancer, colon
cancer and other solid tumors (21),
and that HLX plays an important role in the occurrence and
development of leukemia (10). HLX
is expressed in several types of leukemia but at different levels;
for example, the expression of HLX is high in myeloid leukemia but
low in lymphoid leukemia (11,17).
AML is a malignant tumor, and individuals <35
years of age have the highest morbidity and mortality (22). The pathological mechanism of AML
includes the abnormal clonal proliferation of leukemic cells, which
affects normal hematopoiesis and endangers some organs and systems
(10). Gene-targeting therapy has
been revealed to improve the cure rate of AML (22); thus, it is critical to identify new
gene targets for AML. By analyzing HLX gene expression data from
354 patients with AML in the USA, Kawahara et al (11) found that the HLX gene was
overexpressed in 87% of these patients and was associated with poor
prognosis. Thus, these findings demonstrated that HLX may be a
potential target in AML; however, to the best of our knowledge, few
studies have reported the specific mechanism of HLX in AML.
The aim of the present study was to investigate the
biological functions of HLX in AML cells. First, HLX expression was
analyzed in AML cell lines of different subtypes and HLX was found
to be differentially expressed in KG1a (AML/M0 subtype), NB4
(AML/M3 subtype) and THP-1 cells (AML/M5 subtypes), with the
highest expression in the NB4 cell line. Then, after knocking down
the HLX gene in NB4 cells using siRNA technology, the survival was
assessed at 12, 24, 48 and 72 h; cell proliferation was inhibited
by HLX knockdown. The cell cycle was analyzed by flow cytometry,
which identified an increased number of cells in
G0/G1 phase and a decreased number in S
phase, suggesting that the cell cycle was arrested at
G0/G1 phase. Collectively, the present
results indicated that the downregulation of HLX could block the
cell cycle in G0/G1 phase, thus inhibiting
the proliferation of AML cells.
The current study further investigated the signaling
pathway affected by HLX that was involved in AML cell cycle
regulation and proliferation. It was demonstrated that the
knockdown of HLX resulted in a decreased expression of STAT5 at the
protein level and of PAK1 and NRPl at the mRNA level, while BTG1
gene expression was increased. NRP1 is a receptor of
VEGF165 and can promote vascular proliferation via the
PI3K/Akt, JAK/STAT and Notch signaling pathways (23–25).
STAT5 is an important regulatory protein of the JAK/STAT signaling
pathway and is closely associated with hematological malignancies
(26). PAK1 is the downstream
effector of HLX, regulates the carcinogenic effects of STAT5 in
hematological disease (27,28), and is involved in the pathogenesis of
AML via the regulation of the MYC core network (12). Moreover, BTG1 is involved in a
translocation with c-Myc and functions as a tumor suppressor gene
in the BTG anti-proliferative protein family, leading to growth
arrest or apoptosis in tumor cells (13,14).
The JAK/STAT signaling pathway plays an important
role in the development and progression of AML. Epidermal growth
factor receptor (EGFR), a co-receptor of NRP1, is a receptor
tyrosine kinase located upstream of this signaling pathway
(24). Within this pathway, STAT5 is
an important regulatory protein, and PAK1 and c-Myc are notable
downstream target genes; furthermore, BTG1 is involved in the
translocation of c-Myc (27). The
findings of the present suggest that the HLX gene can regulate the
JAK/STAT signaling pathway in AML, and after silencing HLX, genes
associated with the JAK/STAT signaling pathway show altered
expression over time. NRP1 expression was decreased, STAT5 protein
expression was downregulated and the JAK/STAT signaling pathway was
blocked, resulting in a reduction in PAK1 expression. Furthermore,
under physiological conditions, STAT is known to regulate the
reticular system via positive or negative feedback mechanisms
involving the STAT5/c-Myc axis, and artificial mutagenesis of the
STAT-binding site on c-Myc inhibits the JAK/STAT signaling pathway
(29). Therefore, the increased
expression of BTG1 may also lead to the negative feedback
regulation of the JAK/STAT signaling pathway via the STAT5/c-Myc
network, thus leading to the synergistic inhibition of STAT5
phosphorylation, repression of cell proliferation and cell cycle
arrest.
Although the results are promising, the current
study has several limitations. Since the HLX gene is closely
related to genes such as BTG1, Forkhead Box O4, FYN proto-oncogene,
Src family tyrosine kinase, growth arrest and DNA damage inducible
α, ras homolog family member B, Tumor Protein P63, ZFP36 Ring
Finger Protein Like 1, Histone Deacetylase 7 and PAK1 (11), HLX may function similar to other
homeobox proteins in AML to regulate several linked signaling
pathways by affecting relevant upstream and downstream genes
(30). The present study only
examined the JAK/STAT pathway, and not investigate pathways that
could be regulated by HLX. Additionally, the effects of HLX on cell
differentiation and apoptosis were not fully elucidated. Thus,
future studies will assess these processes in relation to HLX. The
effects in the NB4 cell line was selected and the aim of the
current study was to examine the relationship between the HLX gene
and JAK signaling pathway. However, if the expression of HLX in
cell lines is not high, the changes in related genes may not be
obvious when HLX is downregulated. Thus, only the most
representative cell line (NB4) was selected to be presented. In a
subsequent study, HLX expression will be evaluated in AML primary
cells, especially in the M3 subtype. To confirm the JAK/STAT
signaling as the underlying mechanism of HLX, inhibitors of the
JAK/STAT pathway will be used in future studies after
downregulating HLX in AML cell lines, and the changes of JAK2
protein in NB4 cells will be measured after HLX knockdown. In
addition, further studies will measure the phosphorylated STAT5
protein; the protein levels of genes, such as PAK1, NRP1 and BTG1,
will also be evaluated.
In conclusion, the present results indicated that
the HLX gene may be an important therapeutic target in AML and that
it may play a critical role by regulating the JAK/STAT signaling
pathway to regulate cell proliferation and cell cycle progression.
Furthermore, the current study provides novel evidence of the
pathogenic mechanism of HLX in AML and thus may help improve the
treatment of AML (22).
Acknowledgements
The authors would like to thank Mr. Li-long Fan, Ms.
Li-Hua Chen and Ms. Hai-Yan Lv (all Department of Central
Laboratory, Taizhou Hospital of Zhejiang) for their instructive
advice and useful suggestions on the manuscript. The authors would
also like to thank Mr. Wei-Bo Zhao (Department of Orthopaedics,
Taizhou Hospital of Zhejiang), Ms. Shuai-Shuai Chen and Ms. Jia-Xi
Chen (both Department of Clinical Laboratory, Taizhou Hospital of
Zhejiang) for their advice concerning the English language.
Funding
This work was supported by the Zhejiang Scientific
Project of Health and Medicine, China (grant nos. 2014KYB309,
2017KY709 and 2017KY166), and the Zhejiang Province Foundation,
China (grant no. 2016C33233).
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
WDL designed most of the experiments. XYZ, QYG and
LZ carried out the cell experiments. BGC and MZ helped design the
experiments. LYW, DQZ and YPS analyzed the data. XYZ wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AML
|
acute myelogenous leukemia
|
|
HLX
|
H2.0-like homeobox gene
|
|
PAK1
|
p21-activated kinase 1
|
|
NRP1
|
neuropilin 1
|
|
BTG1
|
B-cell translocation gene 1
|
References
|
1
|
Seifert A, Werheid DF, Knapp SM and
Tobiasch E: Role of Hox genes in stem cell differentiation. World J
Stem Cells. 7:583–595. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Casaca VI, Illi S, Suttner K, Schleich I,
Ballenberger N, Klucker E, Turan E, Mutius EV, Kabesch M and Schaub
B: TBX21 and HLX1 polymorphisms influence cytokine secretion at
birth. PLoS One. 7:e310692012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rajaraman G, Murthi P, Pathirage N,
Brennecke SP and Kalionis B: Downstream targets of homeobox gene
HLX show altered expression in human idiopathic fetal growth
restriction. Am J Pathol. 176:278–287. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Prahst C, Kasaai B, Moraes F, Jahnsen ED,
Larrivee B, Villegas D, Pardanaud L, Pibouin-Fragner L, Zhang F,
Zaun HC, et al: The H2.0-like homeobox transcription factor
modulates yolk sac vascular remodeling in mouse embryos.
Arterioscler Thromb Vasc Biol. 34:1468–1476. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu ZY, Zhong NS, Xie Y and Hu PJ: Internal
Medicine. People's Health Press. (China). 563–674. 2011.(In
Chinese).
|
|
6
|
Leukemia & Lymphoma Group, Chinese
Society of Hematology, Chinese Medical Association, . The
Guidelines for Diagnosis and Treatment of Acute Myelogenous
Leukemia (Relapse/Refractory) in China (2017). Zhonghua Xue Ye Xue
Za Zhi. 38:183–184. 2017.(In Chinese). PubMed/NCBI
|
|
7
|
Luo B, Que ZJ, Zhou ZY, Wang Q, Dong CS,
Jiang Y, Hu B, Shi H, Jin Y, Liu JW, et al: Feiji recipe inhibits
the growth of lung cancer by modulating T-cell immunity through
indoleamine-2,3-dioxygenase pathway in an orthotopic implantation
model. J Integr Med. 19:283–289. 2018. View Article : Google Scholar
|
|
8
|
Ghasemi R, Struthers H, Wilson ER and
Spencer DH: Contribution of CTCF binding to transcriptional
activity at the HOXA locus in NPM1-mutant AML cells. Leukemia. May
12–2020.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Y, Zhang YM, Zhang YS, Tang GS,
Zhang WP, Yang JM, Wang JM and Hu XX: Prognostic significance of
minimal residual disease before post-remission therapy in younger
adult acute myeloid leukemia patients with intermediate risk and
negative of FLT3-ITD, NPM1 and biallelic CEBPA mutations. Zhonghua
Xue Ye Xue Za Zhi. 40:597–601. 2019.(In Chinese). PubMed/NCBI
|
|
10
|
Pandolfi A and Steidl U: HLX in AML: Novel
prognostic and therapeutic target. Oncotarget. 3:1059–1060. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawahara M, Pandolfi A, Bartholdy B,
Barreyro L, Will B, Roth M, Okoye-okafor UC, Todorova TI, Figueroa
ME, Melnick A, et al: H2.0-like homeobox (HLX) regulates early
hematopoiesis and promotes acute myeloid leukemia. Cancer Cell.
22:194–208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pandolfi A, Stanley RF, Yu Y, Bartholdy B,
Pendurti G, Gritsman K, Boultwood J, Chernoff J, Verma A and Steidl
U: PAK1 is a therapeutic target in acute myeloid leukemia and
myelodysplastic syndrome. Blood. 126:1118–1127. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Van Galen JC, Kuiper RP, Van EL, Levers M,
Tijchon E, Scheijen B, Waanders E, van Reijmersdal SV, Gilissen C,
van Kessel AG, et al: BTG1 regulates glucocorticoid receptor
autoinduction in acute lymphoblastic leukemia. Blood.
115:4810–4819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng HC, Li J, Shen DF, Yang XF, Zhao S,
Wu YZ, Takano Y, Sun HZ, Su RJ, Luo JS and Gou WF: BTG1 expression
correlates with pathogenesis, aggressive behaviors and prognosis of
gastric cancer: A potential target for gene therapy. Oncotarget.
6:19685–19705. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fröhling S: Widespread over-expression of
the non-clustered homeobox gene HLX in acute myeloid leukemia.
Haematologica. 97:14532012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Testori J, Schweighofer B, Helfrich I,
Sturtzel C, Lipnik K, Gesierich S, Nasarre P, Hofer-Warbinek R,
Bilban M, Augustin HG and Hofer E: The VEGF-regulated transcription
factor HLX controls the expression of guidance cues and negatively
regulates sprouting of endothelial cells. Blood. 117:2735–2744.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamakawa T, Sato Y, Matsumura Y, Kobayashi
Y, Kawamura Y, Goshima N, Yamanaka S and Okita K: Screening of
human cDNA library reveals two differentiation-related genes, HHEX
and HLX, as promoters of early phase reprogramming toward
pluripotency. Stem Cells. 34:2661–2669. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng WP, Zhao Q, Zhao X, Li B, Hubank M,
Schatz DG and Flavell RA: Up-regulation of Hlx in immature Th cells
induces IFN-gamma expression. J Immunol. 172:114–122. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu Y, Gao J, Su Z, Dai X, Li Y, Liu Y,
Chen J, Tong J, Zhang Y, Wu C, et al: Downregulation of Hlx closely
related to the decreased expressions of T-bet and Runx3 in patients
with gastric cancer may be associated with a pathological event
leading to the imbalance of Th1/Th2. Clin Dev Immunol.
2012:9498212012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morita M, Watanabe M, Inoue N, Inaoka C,
Akamizu T, Tatsumi KI, Hidaka Y and Iwatani Y: Functional
polymorphisms in TBX21 and HLX are associated with development and
prognosis of Graves' disease. Autoimmunity. 45:129–136. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Prada-Arismendy J, Arroyave JC and
Röthlisberger S: Molecular biomarkers in acute myeloid leukemia.
Blood Rev. 31:63–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li L, Jiang X, Zhang Q, Dong X, Gao Y, He
Y, Qiao H, Xie F, Xie X and Sun X: Neuropilin-1 is associated with
clinicopathology of gastric cancer and contributes to cell
proliferation and migration as multifunctional co-receptors. J Exp
Clin Cancer Res. 35:162016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hendricks C, Dubail J, Brohée L, Delforge
Y, Colige A and Deroanne C: A novel physiological
glycosaminoglycan-deficient splice variant of neuropilin-1 is
anti-tumorigenic in vitro and in vivo. PLoS One. 11:e01651532016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Piechnik A, Dmoszynska A, Omiotek M, Mlak
R, Kowal M, Stilgenbauer S, Bullinger L and Giannopoulos K: The
VEGF receptor, neuropilin-1, represents a promising novel target
for chronic lymphocytic leukemia patients. Int J Cancer.
133:1489–1496. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Z and Bunting KD: STAT5 activation in
B-cell acute lymphoblastic leukemia: Damned if you do, damned if
you don't. Cancer Cell Microenviron. 3:e11862016.PubMed/NCBI
|
|
27
|
Eswaran J, Li DQ, Shah A and Kumar R:
Molecular pathways: Targeting P21-activated kinase 1 signaling in
cancer-opportunities, challenges and limitations. Clin Cancer Res.
18:3743–3749. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chatterjee A, Ghosh J, Ramdas B, Mali RS,
Martin H, Kobayashi M, Vemula S, Canela VH, Waskow ER, Visconte V,
et al: Regulation of Stat5 by FAK and PAK1 in oncogenic FLT3- and
KIT-driven leukemogenesis. Cell Rep. 9:1333–1348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yamada O and Kawauchi K: The role of the
JAK-STAT pathway and related signal cascades in telomerase
activation during the development of hematologic malignancies.
JAKSTAT. 2:e252562013.PubMed/NCBI
|
|
30
|
Breitinger C, Maethner E, Garciacuellar MP
and Slany RK: The homeodomain region controls the phenotype of
HOX-induced murine leukemia. Blood. 120:4018–4027. 2012. View Article : Google Scholar : PubMed/NCBI
|